Abstract
A mycobacteriophage Ms6 strong promoter region (Plys) was isolated by using transcriptional fusions with the lacZ reporter gene. Two tandem σ70-like promoter sequences (P1 and P2) were found in this region. DNA sequencing of the promoter downstream region revealed a 214-bp leader sequence followed by five adjacent coding regions of 231 bp (ORF1), 1,152 bp (ORF2), 996 bp (ORF3), 231 bp (ORF4), and 372 (ORF5). ORF1 has the potential to encode a 77-amino-acid protein which revealed similarity to mycobacteriophage TM4 gp90, a predicted protein with unknown function. ORF2 encodes a 384-amino-acid protein which is related to several bacteriophage amidases. This protein induced cell lysis upon addition of chloroform, confirming its mureinolytic activity. ORF3 encodes a 332-amino-acid protein which is related to TM4 gp30, a protein with sequence similarity to amidases. ORF4 encodes a 77-amino-acid holin-like protein with significant similarity to the holin of Lactococcus lactis r1t bacteriophage. ORF5 encodes a 124-amino-acid protein which is related to mycobacteriophage L5 gp30, a protein with unknown function. These data indicate that the promoter region Plys drives the transcription of the Ms6 lysis genes. An intrinsic transcription termination signal was identified in the leader sequence. Experiments using lacZ fusions showed that β-galactosidase synthesis is inhibited when this transcription termination signal is present in the leader sequence. In conclusion, mycobacteriophage Ms6 cell lysis genes are expressed by their own promoter region, independently of virion structure and assembly protein genes. Moreover, an antitermination mechanism might be involved in their transcription regulation.
Double-stranded DNA (dsDNA) bacteriophages synthesize a mureinolytic enzyme, known as an endolysin, during late gene expression of the replication cycle, enabling the release of phage progeny. Phage-encoded lysins have several kinds of mureinolytic activities directed against the covalent linkages that maintain the cell wall integrity: (i) glycosylase and transglycosylase activity, targeting the glycosidic linkages; (ii) N-acetylmuramoyl-l-alanine amidase activity; and (iii) endopeptidase activity targeting the oligopeptide cross-links (17, 31).
To degrade the cell wall of the host, a second factor, designated holin, is required. The holin is a hydrophobic membrane protein that forms pores or lesions in the cell membrane through which the murein hydrolase is released to the periplasm and gains access to the peptidoglycan substrate. Such a dual-component lysis system has recently been discovered in many bacteriophages of both gram-negative and gram-positive bacteria (15, 17, 22, 25, 27, 32). The synthesis of the bacteriophage λ holin (protein S) is tightly controlled at the transcriptional, translational, and posttranslational levels because it determines the time of lysis. In phage λ, the lysis genes are the first genes of the late operon, which also encodes the structural proteins of the phage particle. Transcription of the cell lysis genes is initiated at the PR′ promoter, located downstream of gene Q, and is dependent on the antitermination protein Q. In the absence of protein Q, transcription from the PR′ promoter stops at the tR65 terminator signal located upstream of gene S, the first gene of the late operon (6a).
In bacteriophage L5, the best-studied mycobacteriophage, transcription terminator signals are rare and are found only in the middle of the genome, close to the ends of the left- and right-arm operons (12). The left-arm genes that encode host lysis and structural proteins of the virion are transcribed as a single operon from a late promoter located somewhere in the left end of the L5 DNA. In mycobacteriophages, the regulation of gene expression by an antitermination mechanism has not been reported yet.
In this study we describe the genetic organization of the lysis gene region of mycobacteriophage Ms6, a temperate phage that infects Mycobacterium smegmatis (21). We show that lysis genes are driven by two σ70-like promoters that are recognized by the host RNA polymerase. Moreover, a transcription termination signal was detected in the leader sequence upstream of the first open reading frame (ORF), suggesting that an antitermination mechanism is involved in Ms6 lysis gene transcription.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and plasmids.
The bacterial strains, bacteriophages, and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, bacteriophages, plasmids, and oligonucleotides used in this study
| Strain, bacteriophage, plasmid, or oligonucleotide | Descriptiona | Reference or source |
|---|---|---|
| Escherichia coli | ||
| TG1 | supE hsd Δ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | Gibson-Medical Research Council, Cambridge, England |
| K802 | F− e14− (McrA−) lacY1 or Δ(lac)6 supE44 galK2 galT22 rfbD1 metB1 mcrB1 hsdR2(rK− mK+) | 24 |
| Y1088 | e14 (mcrA) Δ(lac)U169 supE supF hsdR metB trpR tonA21 proC::Tn5 [pMC9] | Stratagene |
| Mycobacterium smegmatis mc2155 | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | 26 |
| Bacteriophages | ||
| Ms6 | Temperate bacteriophage from M. smegmatis | 21 |
| λgt11 | cI857 S100 | Stratagene |
| Plasmids | ||
| pQE30 | Expression vector | Qiagen |
| pJEM12 | Transcription-translation vector | 29 |
| pJEM15 | Transcription fusion vector | 29 |
| pMG1 | pJEM15 derivative carrying a 252-bp (128-379) SauIII fragment of the Ms6 promoter region Plys | This study |
| pMG19 | ORF1 N2 terminus-β-galactosidase fusion in pJEM12 | This study |
| pMG20 | ORF1 N2 terminus-β-galactosidase fusion in pJEM12 | This study |
| pMG21 | ORF1 N2 terminus-β-galactosidase fusion in pJEM12 | This study |
| pMG90 | pMG21 without the leader sequence | This study |
| pMG100 | pMG1 with the Ms6 lysis gene leader sequence cloned in the inverted direction, downstream of Plys | This study |
| pMG200 | pMG1 with the Ms6 lysis gene leader sequence cloned downstream of Plys | This study |
| pMG200a | pMG1 with the Ms6 lysis gene leader sequence (without the transcription terminator) cloned downstream of Plys | This study |
| pMG208 | pMG1 with the Ms6 lysis gene transcription terminator cloned downstream of Plys | This study |
| pMG220 | Plys fragment (9-359) in pJEM15 | This study |
| pMG221 | pMG220 with a P2−12C mutationb | This study |
| pMG222 | pMG220 with a P1−11C mutation | This study |
| pMG231 | ORF2 cloned into pQE30 | This study |
| pMG231A | ORF2 cloned into pQE30 | This study |
| pMP300 | ORF4 cloned into pMP231A | This study |
| pMP310 | ORF4 cloned into pQE30 | This study |
| pMPL1 | pJEM15 with the Ms6 leader sequence | This study |
| pMPLc1 | pJEM15 with the Ms6 leader sequence cloned in the inverted direction | This study |
| Oligonucleotides (5′-3′) | ||
| Pjem-1 | CCTTTAATAGATTATATTACTAATTAATTGGGG | This study |
| Pjem15-c1 | TTCCGATTCGTAGAGCCTCG | This study |
| P1-1 | CGCGGATCCGACAACGGTGTACGCCAAACTGC | This study |
| P1-c6 | GGGGTACCTGTCGATGATGTCCATGACTTGCGC | This study |
| PP1P2wt-c1 | CGATACCGAGGATAGTGTAGTTGTC | This study |
| PP2mut-12C-c1 | CGATACCGAGGATAGTGTGGTTGTC | This study |
| PP1mut-11C-c1 | CGATACCGAGGGTAGTGTAGTTGTC | This study |
| Porf2-1 | CGGGATCCGTGACCACGAAAGATCAAGTC | This study |
| Porf2-c1 | ATGCGAAGCTTCAGTGGCCCAACAGTTC | This study |
| Porf2-c2 | CCGTCGAGCTCTGCGCATCAGTGGC | This study |
| Porf4-1 | CTACGCGAGCTCACATAGGAGGCAC | This study |
| Porf4-c1 | GCACCGAAGCTTCCCAGATCATGCGG | This study |
| Ptertrans-1 | GATCACCCCGCCACCACCTGAGTGCGCGGGGTTTTTCTATG | This study |
| Ptertrans-c1 | GATCCATAGAAAAACCCCGCGCACTCAGGTGGTGGCGGGGT | This study |
| Pleader-1 | CCCCTGGATCCGGTCTCGGCC | This study |
| Pleader-2 | CCTGAGTGGATCCGCGGGGTTTTTCTATG | This study |
| PleaderBam-1 | CCTTTCCCCTGGATCCGGTCTCGG | This study |
| PleaderKpn-1 | CCTTTCCCCTGGTACCGGTCTCGG | This study |
| Pleader-c0 | CGGGATCCGTCGGGGTCCTTTCGGGCATAG | This study |
| Pleader-c1 | CATAGAAAAACCCCGCGGATCCACTCAGG | This study |
| P1del-c4 | GGCCGAGACCGGATCCAGGGG | This study |
| PleaderABC-c1 | CTCAGGTACCGGTGGCGGGGTCGATGG | This study |
| PleaderKpn-c0 | CCCGGTACCGCGCACTCAGGTGGTGG | This study |
| PleaderBam-c0 | CCCGGATCCGCGCACTCAGGTGGTGG | This study |
Underlined oligonucleotide bases were added to provide additional restriction sites.
P2−12C mutation, mutation (T→C) in the −12 base of promoter region P2.
Growth conditions.
M. smegmatis was grown in Myco broth (Middlebrook 7H9 [Difco], supplemented with 0.5% nutrient broth [Difco], 0.5% glucose, and 0.05% Tween 80) or in Myco agar plates (Myco broth with 1.2% Bacto agar [Difco], without Tween 80). Mycobacterium bovis BCG was grown in Myco broth or Myco agar plates supplemented with albumin, dextrose, and catalase enrichment (Difco). Escherichia coli strains were grown in Luria-Bertani (LB) medium. All strains were grown at 37°C, and kanamycin or ampicillin was added to the medium when appropriate.
DNA manipulation.
DNA manipulations, plasmid DNA extraction, electrophoresis, PCRs, Southern blotting, and hybridizations were carried out using standard techniques (24). Restriction endonuclease digestions, ligations with T4 DNA ligase, and reactions with T4 DNA polymerase were performed according to the manufacturers' specifications.
Electroporation of E. coli and mycobacteria.
One-liter cultures of E. coli or mycobacteria were grown to an optical density at 550 nm (OD550) of 0.8. The cultures were centrifuged at 6,000 × g for 10 min, and the pellets were suspended in 100 ml of ice-cold 10% glycerol and centrifuged for 10 min. The pellets were suspended again in 100 ml of 10% glycerol and centrifuged for 10 min. The pellets were then suspended in 2 ml of 10% glycerol and quickly frozen at −70°C. Electroporation conditions were 200 Ω, 25 μF, and 1,800 V. Time constants averaged 4 ms.
Isolation of Ms6 promoters.
Genomic DNA from mycobacteriophage Ms6 was partially digested with Sau3AI and size-fractionated by agarose gel electrophoresis. Fragments in the 0.2- to 1.5-kb size range were excised from the gel and ligated to pJEM15 cleaved with BamHI. The ligation mixtures were used to transform M. smegmatis mc2155. Transformants were selected on Myco agar with kanamycin (15 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (30 μg/ml). Blue colonies on this medium were selected.
Quantitative β-galactosidase assays.
β-Galactosidase activity was assayed in sonicated extracts of M. smegmatis and M. bovis BCG clones. Five milliliters of exponential-phase cultures (optical density at 600 nm, 0.30 to 0.40) was harvested, resuspended in 200 μl of phosphate-buffered saline (PBS), and then sonicated. The β-galactosidase activity of these extracts was measured by hydrolysis of ONPG (o-nitrophenyl-β-d-galactopyranoside; Sigma), at 37°C. Reaction mixtures, with 30 μl of extract, 201 μl of 0.1 M sodium phosphate (pH 7.5), 3 μl of 0.1 M MgCl2, 4.5 M β-mercaptoethanol, and 66 μl of ONPG (4 mg/ml in 0.1 M sodium phosphate, pH 7.5) were incubated at 37°C for 30 min and stopped by the addition of 500 μl of 1 M Na2CO3. Their optical densities were measured at 420 nm (24). Soluble protein in bacterial extracts was measured by the Bio-Rad protein assay. Units of β-galactosidase were calculated by the following formula: 200 × optical density at 420 nm per milligram of protein per minute (29). Experiments were carried out in triplicate. All standard deviations were ≤13% of the mean.
DNA cloning.
Unless indicated otherwise, the DNA fragments obtained by PCR amplification were amplified using Ms6 DNA as a template. In order to construct pMG220, pMG221, and pMG222, DNA fragments obtained by PCR amplification with primers P1-1 and PP1P2wt-c1, PP2mut-12C-c1, or PP1mut-11C-c1, respectively, restricted with the BamHI enzyme were cloned into pJEM15 previously restricted with KpnI, treated with enzyme T4 DNA polymerase, and then restricted with BamHI. In order to construct pMG231, the DNA fragment obtained by PCR amplification with primers Porf2-1 and Porf2-c1 was restricted with BamHI and HindIII and cloned into pQE30 (Qiagen) previously restricted with BamHI and HindIII. pMG231A was obtained by insertion of ORF2 amplified by PCR with primers Porf2-1 and Porf2-c2 and restricted with BamHI and SacI into the BamHI and SacI sites of pQE30.
To obtain plasmids pMP300 and pMP310, a DNA fragment carrying ORF4 was amplified by PCR using primers Porf4-1 and Porf4-c1, restricted with SacI and HindIII, and cloned into the corresponding sites of pMG231A and pQE30, respectively. In order to construct pMG208, primers Ptertrans-1 (0.1 pmol) and Ptertrans-c1 (0.1 pmol) were denatured at 95°C for 10 min, hybridized to each other at 25°C, forming BamHI and KpnI cohesive ends, and cloned into BamHI- and KpnI-cleaved pMG1. A DNA fragment with the promoter and the ORF1 translation signals, obtained by PCR amplification with primers P1-1 and P1-c6 and restricted with NruI, was cloned into transcription-translation probe vector pJEM12 previously restricted with ScaI, originating clone pMG21. This construction was restricted with BamHI or KpnI, treated with T4 DNA polymerase, and religated, creating pMG19 and pMG20, respectively. The leader sequence was amplified by PCR with primers Pleader-1 and Pleader-c0, restricted with BamHI, and cloned in opposite directions into the BamHI site of pMG1, generating plasmids pMG100 and pMG200.
To remove the leader sequence from pEA4 (2), a construction with a 2.5-kb Ms6 fragment carrying the promoter region Plys, two BamHI target sites were sequentially introduced, at both ends of the leader sequence, by site-directed mutagenesis, using the QuikChange site-directed mutagenesis kit (Stratagene). The primers used were Pleader-1/P1del-c4 and Pleader-2/Pleader-c1 (Table 1), and the reactions were performed according to the instruction manual. This construction was restricted with the BamHI enzyme and religated, creating pMG82. A DNA fragment obtained by PCR amplification from pMG82 with primers P1-1 and P1-c6, restricted with NruI, was cloned into the transcription-translation probe vector pJEM12 previously restricted with ScaI, creating clone pMG90. The leader sequence was amplified by PCR with primers PleaderBam-1 and PleaderABC-c1, restricted with BamHI and KpnI, and cloned into pMG1 previously restricted with BamHI and KpnI, generating plasmid pMG200a. In order to construct pMPL1 and pMPLc1, DNA fragments obtained by PCR amplification with primers PleaderBam-1/PleaderKpn-c0 or PleaderKpn-1/PleaderBam-c0 and restricted with BamHI and KpnI were cloned into pJEM15 previously restricted with BamHI and KpnI.
Complementation of phage lambda S100 lethal function.
E. coli K802 and K802 transformants containing plasmid pQE30 or pMP310 and the suppressing strain E. coli Y1088 were grown at 37°C in LB medium supplemented with 10 mM MgSO4 and 0.2% maltose. When the cultures reached an optical density at 600 nm of between 0.8 and 1, cells were pelleted at 500 × g for 10 min, resuspended in 10 mM MgSO4, and diluted to an OD600 of 0.5. Then 200 μl of host cells was infected at 37°C for 20 min with different dilutions of phage λgt11. Samples were than mixed with 4 ml of LB top agar and poured on LB plates. The plates were incubated at 37°C, and the number of plaques was determined.
DNA sequencing.
Sequencing of phage DNA and all the constructed plasmids was performed by thermal cycle sequencing with Taq DNA polymerase and fluorescent dye-labeled terminators in an ABI 377 DNA sequencer with primers Pjem-1 and/or Pjem15-c1.
Computer analysis of DNA sequences.
The ORFs were identified and aligned using the PCGene (IntelliGenetics) programs. Fasta3 (19) from EBI and Blast (1) from NCBI were used for database homology searches. Protein hydrophobicity profiles were determined with the ProtScale program from the Expasy server of the Swiss Institute of Bioinformatics using the Kyte and Doolittle amino acid hydropathicity scale (16). Analysis of the potential transmembrane regions was performed using the HMMTOP 2.0 prediction, TopPred2, and PredictProtein programs from the Expasy server of the Swiss Institute of Bioinformatics.
RNA extraction.
Total RNA was extracted from M. smegmatis using the RNeasy minikit (Qiagen). The eluate was treated with RNase-free DNase (Pharmacia) for 15 min at 37°C. After DNase digestion, the RNA was repurified with the RNeasy clean-up protocol (Qiagen).
Primer extension analysis.
Primer extension mapping was performed with the Promega avian myeloblastosis virus reverse transcriptase primer extension system. Total RNA was isolated from M. smegmatis carrying pJEM15 or pMG1. Primer extension reactions were carried out by the Promega protocol using the 32P-labeled primer Pjem15-c1, which hybridizes 108 or 113 bp downstream of the transcription start points. Reaction products were electrophoresed on a 6% polyacrylamide gel next to DNA sequencing reactions also generated with primer Pjem15-c1.
Nucleotide sequence accession number.
The nucleotide sequence of Plys and the downstream region, including ORF1, ORF2, ORF3, ORF4, ORF5, and ORF6, has been deposited in GenBank under accession number AF319619.
RESULTS
Isolation of a mycobacteriophage Ms6 strong promoter region.
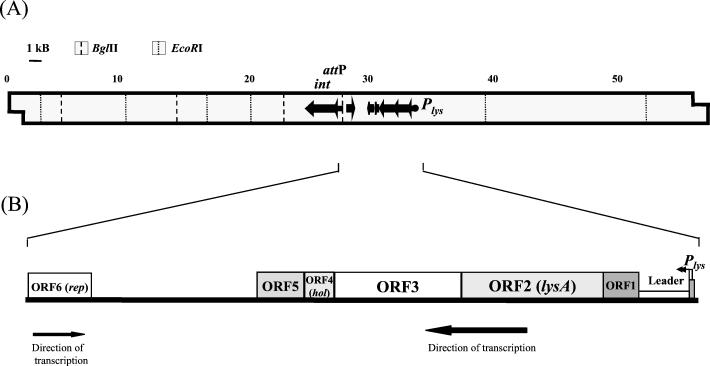
Identification of mycobacteriophage Ms6 promoter regions was carried out by constructing transcriptional lacZ fusions in the promoter probe vector pJEM15 (29). A library of Sau3AI Ms6 DNA fragments was inserted into a multiple cloning site adjacent to a promoterless lacZ reporter gene, and the plasmid constructs were electroporated into M. smegmatis. Transformants displaying dark blue color in plates with the chromogenic substrate X-Gal were selected for β-galactosidase activity measurement. The highest β-galactosidase activity was achieved with a plasmid construct, designated pMG1, containing a 252-bp Sau3AI insert. This sequence is located 6 kb upstream of the Ms6 integration locus (6) (Fig. 1A).
FIG. 1.
(A) Schematic representation of Plys on the 57-kb Ms6 DNA. Some pertinent restriction sites are shown. Plys was located on an 11.8-kb BglII-EcoRI Ms6 DNA fragment. The integrase gene and attP (6) are also represented. (B) Genetic organization of the Ms6 lysis locus.
Detection of the transcriptional start site.
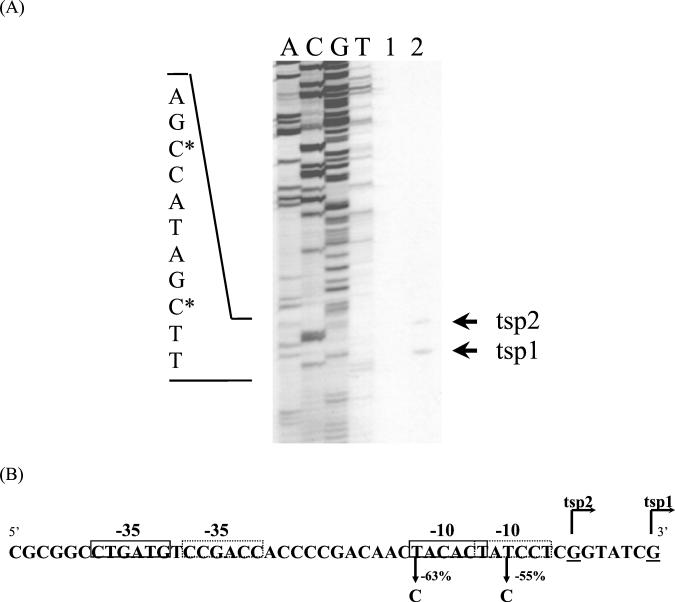
To identify the transcription start site of the isolated promoter region, a primer extension analysis was performed on RNA isolated from the M. smegmatis(pMG1) strain. As shown in Fig. 2A, two transcription start points (tsp1 and tsp2), separated by 5 bp, were detected. The tsp1 and tsp2 mapped to G residues on the nontranscribed strand, at bp 208 and bp 214, respectively, upstream from the ORF1 translation initiation codon.
FIG. 2.
(A) Primer extension analysis. Lanes: A, C, G, and T, DNA sequencing ladders; 1 and 2, primer extension products obtained with RNA from M. smegmatis bearing pJEM15 (negative control, without promoter) and pMG1, respectively. Primer extension products were synthesized as described in Materials and Methods and are marked by arrows (tsp1 and tsp2). DNA sequencing ladders were generated with the same primer as that used in the primer extension reactions. The nucleotide sequence of the complementary strand of the promoters is indicated to the left. The nucleotide designated +1 for each promoter is indicated with an asterisk. (B) Nucleotide sequence of the nontranscribed strand of P1 and P2. Transcription is initiated at two G residues indicated by tsp1 and tsp2. P1 hexameric −35 and −10 sequences are indicated by dashed boxes, and P2 hexameric −35 and −10 are indicated by solid-line boxes. Single nucleotide substitutions in the P1 and P2 −10 regions and the effect of each mutation on promoter function are indicated.
Two potential core promoter sequences, P1 and P2, with −10 and −35 hexamers similar to the canonical sequences recognized by E. coli σ70 (10, 13), Streptomyces σ70-like (28), and mycobacterial σ70-like (9) RNA polymerases, are present upstream of the transcription start points (Fig. 2B). In P1 the −10 hexamer is 8 bp upstream of tsp1 and is separated from the −35 hexamer by 16 bp. The P2-10 hexamer is 7 bp upstream of tsp2 and is separated from the −35 hexamer by 18 bp. To verify the importance of the −10 regions for the promoter strength, base substitutions were carried out in both hexamer sequences, and the β-galactosidase activity of the transcriptional fusions with lacZ reporter gene was measured. The replacement of T by C at position −11 of the P1-10 sequence resulted in a 55% decrease in β-galactosidase activity (4,148 to 1,867 U). On the other hand, the replacement of T by C at position −12 of P2 led to a 63% β-galactosidase activity decrease (4,148 to 1,532 U). These results indicate that the transcription start points identified by primer extension analysis are probably correct and that there might be two tandem promoters in this promoter region.
DNA sequence analysis.
To identify ORFs in the vicinity of this promoter, the nucleotide sequence from the promoter region to the integrative locus was determined. Within this region, with 63% G+C, six of the computer-predicted ORFs (PCGene; IntelliGenetics) exhibited significant similarities with other bacteriophage genes. ORF1, of 231 bp, is separated from the +1 position by a 214-bp leader sequence. ORF1 has the potential to encode a 77-amino-acid polypeptide which revealed similarity to the mycobacteriophage TM4 gp90 (5) (43% identity in a 53-amino-acid overlap) and the mycobacteriophage Bxb1 gp7 (18a) (39% identity in a 57-amino-acid overlap), two predicted polypeptides with unknown functions.
ORF2, of 1,152 bp, starts at a GTG codon that overlaps the ORF1 TGA stop codon, which is in a different reading frame. ORF2 has the potential to encode a 384-amino-acid polypeptide which revealed significant similarity to mureinolytic enzymes (lysins) of several bacteriophages: Lactobacillus casei bacteriophage A2 (7) (27% identity within 184 residues), lactococcal bacteriophages φUS3 (20) (29% identity within 119 residues), sk1 (3) (29% identity within 119 residues), and bIL170 (29% identity within 120 residues), and Listeria monocytogenes bacteriophage A511 (18) (27% identity within 142 residues). Lysins from bacteriophages φUS3, A511, sk1, and bIL170 were classified as amidases. Moreover, the bacteriophage A511 lysin has already been shown to be an N-acetylmuramoyl-l-alanine amidase (18). Therefore, ORF2 was designated lysA to indicate that it might represent an Ms6 lysin gene.
ORF3, with 996 bp, starts at an ATG codon that overlaps, in a different reading frame, the TGA stop codon of ORF2. ORF3 has the potential to encode a 332-amino-acid protein with similarity to the mycobacteriophage D29 gp12 (4) (43% identity within 252 residues), the mycobacteriophage L5 gp12 (11) (44% identity within 252 residues), the mycobacteriophage Bxb1 gp8 (18a) (46% identity within 322 residues), and the mycobacteriophage TM4 gp30 (5) (32% identity within 280 residues). The alignment of these five predicted proteins revealed some conserved domains which may be related to these proteins' functions. The N-terminal residues of the TM4 gp30 are similar to a family of proteins with N-acetylmuramoyl-l-alanine amidase activity (5), and the central regions of D29 gp10 and Bxb1 gp8 are similar to an endochitinase from Arabidopsis thaliana (12). These data led us to hypothesize that the potential protein encoded by ORF3 has a role in cell lysis. Further experiments must be carried out in order to clarify its function.
ORF4 begins with an ATG start codon located 10 nucleotides downstream of the ORF3 stop codon. ORF4 has the potential to encode a 77-amino-acid polypeptide which is highly related to the holin protein of Lactococcus lactis r1t bacteriophage (30) (49% identity within 68 residues) (Fig. 3). This polypeptide shares some structural characteristics with class II holins, which are usually hydrophobic in nature and small in size, with a highly hydrophilic carboxy-terminal domain and two potential transmembrane domains (31). Analysis of the predicted structure revealed two potential transmembrane regions according to the HMMTOP 2.0 prediction TopPred2 and PredictProtein programs from the Expasy server. Moreover, the ORF4 product is hydrophobic (average hydrophobicity = 0.5). On the basis of these structural features and taking into account that holin genes are usually closely associated to lysin genes, ORF4 was designated hol to indicate that it might represent the Ms6 holin gene.
FIG. 3.
Comparison of L. lactis bacteriophage r1t holin with the Ms6 ORF4-encoded 77-amino-acid protein. Numbers of the first and last residues of the aligned regions are given. Perfectly conserved residues are marked with asterisks; dots indicate positions with conservative substitutions.
ORF5, with 372 bp, starts at an ATG codon that overlaps the ORF4 TGA stop codon, which is in a different reading frame. ORF5 has the potential to encode a 124-amino-acid polypeptide which exhibits similarity (42.7% identity within 68 residues) to L5 gp30, a protein with unknown function. An ORF6, transcribed in the opposite direction, was also detected (Fig. 1B). ORF6 has the potential to encode a 169-amino-acid polypeptide which exhibits significant similarity to the Bxb1 repressor gp69 (18a) (97% identity within 170 amino acids) and L5 repressor gp71 (11) (45.5% identity within 156 residues). Therefore, ORF6 was designated rep to indicate that it might represent an Ms6 repressor gene. Between the terminus of ORF5 and ORF6, several potential ORFs were identified. However, none of them encodes a protein related to any of the database protein sequences. The identified promoter region was designated Plys, to indicate that it directs the expression of the Ms6 lysis genes.
Expression of Ms6 lysis genes in E. coli.
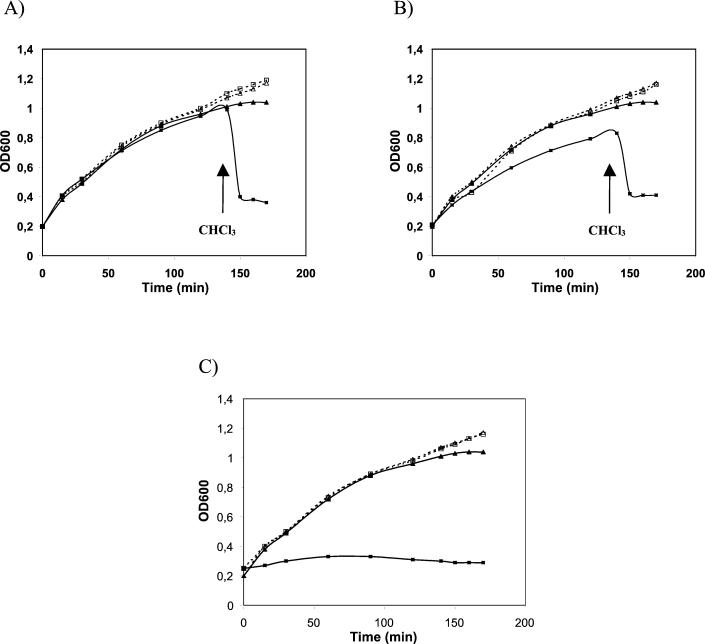
ORF2 and ORF4 were expressed in E. coli using the vector pQE30 under the control of a regulated T5 bacteriophage promoter. As shown in Fig. 4A, expression of the Ms6 lysA gene (pMG231) did not affect the E. coli growth rate. However, permeabilization of the plasma membrane by chloroform upon expression of the lysA gene resulted in immediate lysis. These observations are consistent with other studies, in which the overexpression of phage lysins in the absence of the respective holin functions does not inhibit E. coli growth unless chloroform has been added (3, 15). These results suggest that ORF2 encodes a lysin with mureinolytic activity that is functional in E. coli. This Ms6 lysin is the first one identified in mycobacteriophages.
FIG. 4.
Effects of lysA and hol genes on growth of E. coli. Strains were grown in LB broth at 37°C to an optical density at 600 nm (OD600) of 0.2. At time zero, transcription of cloned lysis genes was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside, 1 mM final concentration). At the time indicated by the arrow, CHCl3 (2% final concentration) was added to the induced cultures. (A) ▪, TG1(pMG231), induced; □, TG1(pMG231), uninduced; ▴, TG1(pQE30), induced; ▵, TG1(pQE30), uninduced. (B) ▪, TG1(pMP300), induced; □, TG1(pMP300), uninduced; ▴, TG1(pQE30), induced; ▵, TG1(pQE30), uninduced. (C) ▪, TG1(pMP310), induced; □, TG1(pMP310), uninduced; ▴, TG1(pQE30), induced; ▵, TG1(pQE30), uninduced.
When the Ms6 hol gene was expressed together with the lysA gene from a single plasmid (pMP300), the effect on E. coli growth was not significantly different from that obtained by the sole expression of lysA (Fig. 4B). A similar effect was also observed with Lactobacillus gasseri phage φ adh lysis genes (15). This result may be due to a reduced expression of the hol gene, seeing that it was cloned downstream of ORF2 and was transcribed using its own ribosome-binding site. The overexpression of the Ms6 hol gene (pMP310) had a severe effect on the E. coli growth rate (Fig. 4C). A lethal effect resulting from the overexpression of holins was also described for the S gene of phage λ (8) and phage φ29 gene 14 (27).
Ms6 hol gene complements a λ phage S mutant.
λgt11 carries an amber mutation in gene S (S100) and consequently cannot induce lysis of the infected host unless suppressing E. coli strains are used, since the holin is essential for the transit of lambda transglycosylase to murein (31). In order to verify the holin function of ORF4, the nonsuppressing E. coli strain K802 harboring plasmid pQE30 or the recombinant plasmid pMP310 (carrying the hol gene) was infected with λgt11. E. coli Y1088 produced 5 × 1010 PFU/ml, and K802 with no plasmid produced 2.4 × 105 PFU/ml. The number of PFU produced by E. coli K802(pMP310), 3 × 109 PFU/ml, was significantly higher than that by E. coli K802(pQE30) strain, 2.5 × 105 PFU/ml, and therefore the Ms6 hol gene can complement the defective S100 allele of phage λgt11. These data strongly suggest that ORF4 encodes the bacteriophage Ms6 holin.
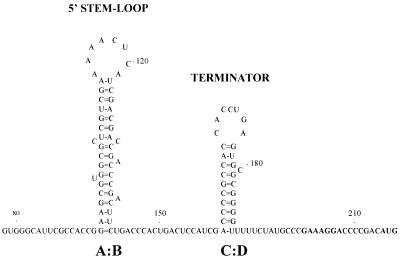
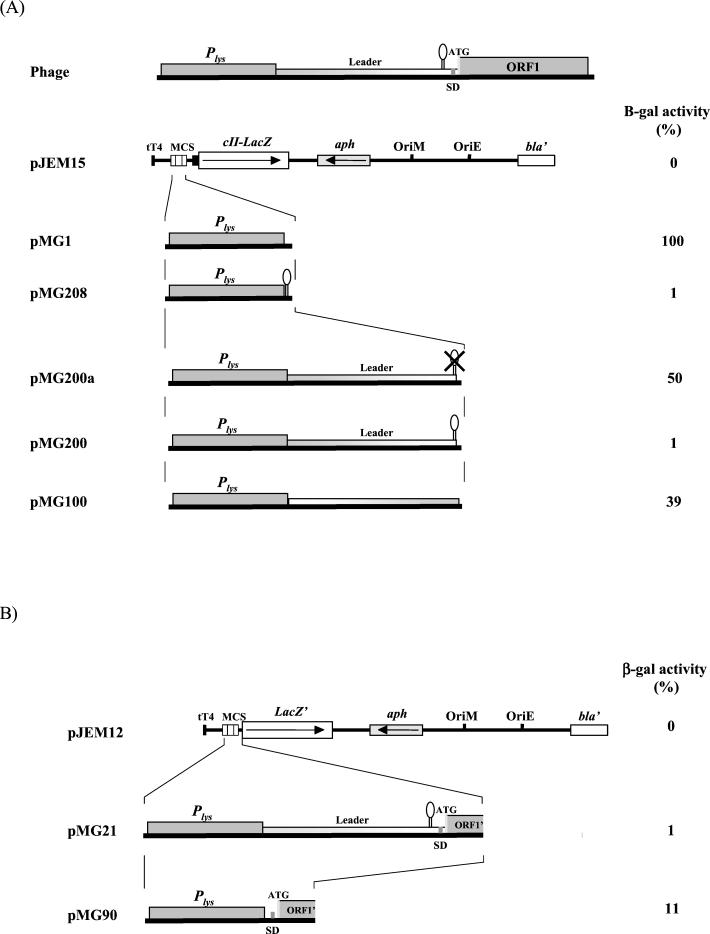
Intrinsic transcription termination signal is present in the leader sequence.
Secondary-structure analysis of the 5′ untranslated region predicted a stem-loop structure followed by a transcription termination signal located between bases 161 and 196 of the transcript (Fig. 5) (14). The inhibitory role of this transcription termination signal on gene expression was evaluated in M. smegmatis by using lacZ reporter gene fusions (Fig. 6A). All the transcriptional fusions were done using plasmid pJEM15 (29), which contains the promoterless lacZ gene and the pAL5000 origin of replication. M. smegmatis carrying pMG1, with the 252-bp promoter region cloned upstream of the start codon of lacZ, was considered to have 100% activity. Plasmid pMG208, derived from pMG1, contains the predicted transcription terminator sequence cloned between the promoter and the reporter gene.
FIG. 5.
Predicted secondary structure of the transcription terminator (C:D) and the 5′ stem-loop (A:B) in the leader transcript of the Ms6 lysis genes. Numbers indicate positions relative to the start of transcription. Translation start codon and potential Shine-Dalgarno sequence (SD) of ORF1 are in boldface.
FIG. 6.
(A) Schematic representation of the derivatives of the Ms6 lysis gene transcription regulation region fused to the lacZ reporter gene in the pJEM15 transcription fusion vector and their β-galactosidase activities. β-Galactosidase activity is presented as a percentage of the activity of pMG1 and was determined as described in Materials and Methods. The results are the averages for three independent experiments. (B) Schematic representation of the derivatives of the Ms6 lysis gene transcription regulation region and ORF1 translation signals fused to the lacZ reporter gene in the pJEM12 transcription-translation fusion vector and their β-galactosidase activities. β-Galactosidase activity is presented as a percentage of that of pMG1 and was determined as described in Materials and Methods. The results are the averages for three independent experiments.
Cell extracts of pMG208 clones exhibited only 1% (96 U) of the pMG1 β-galactosidase activity (8,967 U). A similar result was obtained with pMG200 (87 U), a plasmid fusion in which the entire leader sequence is flanked downstream by lacZ and upstream by the promoter region. However, when the predicted transcription terminator was deleted from the leader sequence (pMG200a), β-galactosidase activity increased to 4,478 U (50% of pMG1 β-galactosidase activity) (Fig. 6A). These results suggest that the predicted intrinsic transcription termination signal in the leader RNA transcript (structure C:D in Fig. 5) is functional. The β-galactosidase activity (3,512 U, 39% of pMG1 β-galactosidase activity) obtained using pMG100 indicates that the sequence direction of the leader region affects (as expected when an intrinsic transcription terminator is present) the expression of the downstream genes (Fig. 6A).
The presence of the 5′ stem-loop (structure A:B in Fig. 5) in the leader RNA does not inhibit the expression of the downstream genes (Fig. 6A). However, this structure may be involved in the stabilization of the leader RNA termination structure. On the other hand, gene fusions between the ORF1 5′ end and lacZ′ (a truncated lacZ gene which lacks the first seven codons) were constructed and expressed in M. smegmatis. As expected, plasmid pMG21, containing the in-frame fusion, exhibited low β-galactosidase activity (blue colonies; 1% activity, 91 U) (Fig. 6B). However, when the leader sequence, placed between the promoter region Plys and ORF1, was removed (plasmid pMG90), a significant β-galactosidase activity of 11% (1,030 U) emerged. Out-of-frame plasmid fusions (pMG19 and pMG20) did not present any β-galactosidase activity (white colonies; 0% activity) (data not shown). These experiments show that ORF1 translation signals are recognized by M. smegmatis, although with less efficiency then those of the cII-lacZ reporter gene (pJEM15).
These data led us to conclude that the Ms6 genetic region is transcriptionally active and that the predicted transcription termination signal of the RNA leader sequence is responsible for the inhibition of the downstream gene expression. In addition, it was demonstrated that the leader region does not have significant promoter activity. Two constructions in pJEM15, containing only the leader region in both orientations without the transcription terminator, did not present significant β-galactosidase activity (pMPL1 and pMPLc1 presented only 119 and 58 U, respectively).
DISCUSSION
In this study, we have described the genetic organization and some transcriptional control elements of the mycobacteriophage Ms6 lysis functions. We showed that transcription of the lysis genes is dependent on a strong promoter region, Plys, located about 6 kb away from the integration locus, which is located in the middle of the Ms6 DNA genome. The transcription initiated from this promoter goes towards the integration elements, and the mRNA transcript cannot be longer than 5.5 kb.
Plys fused with the reporter lacZ gene induces high level β-galactosidase activity in both M. smegmatis and M. bovis BCG (data not shown) cells multiplying exponentially. Genetic analysis of this promoter region showed the presence of two tandem σ70-like promoter sequences, P1 and P2. These two promoters can be grouped into group A of the mycobacterial promoter compilation, containing the σ70-like promoters (9). One of the requirements to belong to this group is the presence of −10 and −35 hexamers similar (with three or more identical bases in similar positions) to E. coli −10 and −35 consensus hexamers.
The DNA sequence of the Plys downstream region revealed the presence of five ORFs. Database searches indicated that these ORFs encode proteins involved in cell lysis. In agreement with what has been reported for other dsDNA phages, two of these ORFs encode essential proteins involved in cell lysis, a lysin (ORF2) and a holin (ORF4). ORF2 codes for a protein related to several bacteriophage endolysins with amidase activity. This protein, like the other reported phage lysins, induces cell lysis upon addition of chloroform, confirming its mureinolytic activity in E. coli (3, 15).
ORF4 encodes a holin-like protein with high similarity to the bacteriophage r1t holin. Our assumption that this ORF specifies the membrane-associated holin protein involved in host lysis by Ms6 derives support from the following observations: (i) the structural features of the polypeptide share characteristics with the class II holins (31); (ii) overexpression of the hol gene in E. coli led to a lethal phenotype, as observed with φ29 holin, explained by the introduction of nonspecific lesions in the cytoplasmic membrane; (iii) the product of the hol gene can complement a lambda S mutant, suggesting that this holin permits the nonspecific release of the murein hydrolases to the periplasm even in a heterologous system. Complementation in heterologous hosts has been also described for other holins, such as the bacteriophage φ adh holin (15).
One of the features common to holin proteins is the presence of a highly hydrophilic C terminus. The C-terminal domain of the Ms6 holin is not as charged as those from most of other holins. However, a similar observation has been reported for the Staphylococcus aureus bacteriophage 187 holin, which lacks a highly charged C terminus (17). Some holins have a dual-start motif that allows the synthesis of two products of different lengths, which are thought to be involved in the fine regulation of the pore-forming process (31). A dual-start motif was not found in ORF4. In contrast to what occurs in most bacteriophages, the hol gene does not map upstream but downstream of the lysin gene(s). However, different locations of the holin gene have been described in some bacteriophages. An inverted organization (lys upstream of hol) was reported for bacteriophage fOg44 (18b), while the Staphylococcus aureus phage 187 hol gene is fully embedded out of frame in the endolysin gene (17).
Although ORF3 encodes a protein with similarity to mycobacteriophage proteins related to lysins, its function remains to be determined. The majority of dsDNA bacteriophages possess only one lysin gene. However, two ORFs with the potential to encode proteins with sequence similarity to lysins were also described in the mycobacteriophages L5, Bxb1, and TM4 (12). Taking into account that we are studying a phage that infects a mycobacterial cell, which has a complex cell wall, we can speculate that additional lysis genes may be required to disrupt the cell wall. ORF1 and ORF5 code for proteins with unknown functions. We expect that the studies in course will lead to a better understanding of the role of each of the identified genes. In this sequence we did not find any ORF product related to phage structural and assembly proteins. The proteins encoded by ORF1, ORF3, and ORF5, which are related to mycobacterial proteins, could be mycobacterium-specific lysis factors.
In the mycobacteriophage L5, D29, and Bxb1 genomes, the left-arm genes code for the virion structure, assembly proteins, and potential amidases (12). These genes are expressed as a transcription unit from a late promoter located somewhere in the left end of the DNA genome. The bacteriophage λ late operon exhibits a similar organization of the late functions. Lysis and structural genes are clustered, and their transcription is initiated from the same strong promoter, PR′ (6a). It is noteworthy that the Ms6 lysis genes are expressed by their own promoter region (Plys) and are not expressed as a single operon with the virion structure and assembly protein genes. This separation of lysis genes from structural genes in two different operons indicates more complex transcriptional control of the late functions in bacteriophage Ms6.
Finally, we identified an intrinsic transcription terminator signal in the 214-bp leader sequence of the Ms6 lysis operon, immediately upstream of ORF1. Intrinsic terminators contain short GC-rich stem-loop structures, followed by a run of U residues. Stem-loop structures promote pausing of RNA polymerase, and the terminator has been proposed to destabilize the interaction of paused RNA polymerase with the template DNA (14). It was demonstrated that the identified transcription termination signal downregulates the expression of the downstream genes. In phage λ, a transcription termination signal is located upstream of gene S, the first gene of the late operon. The transcription of this operon is dependent on synthesis of the antitermination protein Q (23). Conversely, the few transcription termination signals identified in the L5 family of mycobacteriophages are located in the middle of the genome, close to the ends of the left- and right-arm operons, and no antitermination mechanism has yet been described (12).
These data led us to conclude that an antitermination mechanism might be involved in the regulation of Ms6 lysis gene transcription. During the early stages of the lytic cycle, an antiterminator factor should be synthesized in order to allow transcription to proceed beyond this termination signal. Therefore, lysis gene transcription might be dependent on an antiterminator factor. This is the first transcription antitermination mechanism reported in mycobacteriophages.
Acknowledgments
We thank Brigitte Gicquel for supplying plasmids pJEM12 and pJEM15.
M. Garcia is the recipient of a fellowship from Associação para o Desenvolvimento de Vacinas e Produtos Imunológicos-MICROFAR. This work was supported by PEDIP2-Projecto Imunopor-Cand 25/00254.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anes, E., I. Portugal, and J. Moniz-Pereira. 1992. Insertion into the Mycobacterium smegmatis genome of the aph gene through lysogenization with the temperate mycobacteriophage Ms6. FEMS Microbiol. Lett. 74:21-25. [DOI] [PubMed] [Google Scholar]
- 3.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 4.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143-164. [DOI] [PubMed] [Google Scholar]
- 5.Ford, M. E., R. W. Stenstrom, R. W. Hendrix, and G. F. Hatfull. 1998. Mycobacteriophage TM4: genome structure and gene expression. Tuber. Lung Dis. 79:63-73. [DOI] [PubMed] [Google Scholar]
- 6.Freitas-Vieira, A., E. Anes, and J. Moniz-Pereira. 1998. The site-specific recombination locus of mycobacteriophage Ms6 determines DNA integration at the tRNA(Ala) gene of Mycobacterium spp. Microbiology 144:3397-3406. [DOI] [PubMed] [Google Scholar]
- 6a.Friedman, D. I., and M. Gottesman. 1983. Lytic mode of lambda development, p. 21-51. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 7.Garcia, P., J. C. Alonso, and J. E. Suarez. 1997. Molecular analysis of the cos region of the Lactobacillus casei bacteriophage A2. Gene product 3, gp3, specifically binds to its downstream cos region. Mol. Microbiol. 23:505-514. [DOI] [PubMed] [Google Scholar]
- 8.Garret, J. M., and R. Young. 1982. Lethal action of bacteriophage lambda S gene. J. Virol. 44:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez, M., and I. Smith. 2000. Determinants of mycobacterial gene expression, p. 111-129. In G. F. Hatfull and W. R. Jacobs (ed.), Molecular genetics of mycobacteria. American Society for Microbiology, Washington, D.C.
- 10.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatfull, G. F., and G. J. Sarkis. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7:395-405. [DOI] [PubMed] [Google Scholar]
- 12.Hatfull, G. F. 2000. Molecular genetics of mycobacteriophages, p. 37-54. In G. F. Hatfull and W. R. Jacobs (ed.), Molecular genetics of mycobacteria. American Society for Microbiology, Washington, D.C.
- 13.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henkin, T. M. 1996. Control of transcription termination in prokaryotes. Annu. Rev. Genet. 30:35-57. [DOI] [PubMed] [Google Scholar]
- 15.Henrich, B., B. Binishofer, and U. Blasi. 1995. Primary structure and functional analysis of the lysis genes of Lactobacillus gasseri bacteriophage φ adh. J. Bacteriol. 177:723-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 17.Loessner, M. J., S. Gaeng, and S. Scherer. 1999. Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J. Bacteriol. 181:4452-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 18a.Mediavilla, J., S. Jain, J. Kriakov, M. E. Ford, R. L. Duda, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955-970. [DOI] [PubMed] [Google Scholar]
- 18b.Parreira, R., C. Sao-Jose, A. Isidro, S. Domingues, G. Vieira, and M. A. Santos. 1999. Gene organization in a central DNA fragment of Oenococcus oeni bacteriophage fOg44 encoding lytic, integrative and nonessential functions. Gene 226:83-93. [DOI] [PubMed] [Google Scholar]
- 19.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platteeuw, C., and W. M. de Vos. 1992. Location, characterization and expression of lytic enzyme-encoding gene, lytA, of Lactococcus lactis bacteriophage φ US3. Gene 118:115-120. [DOI] [PubMed] [Google Scholar]
- 21.Portugal, I., E. Anes, and J. Moniz-Pereira. 1989. Temperate mycobacteriophage from M. smegmatis. Acta Leprol. 7(Suppl. 1):243-244. [PubMed] [Google Scholar]
- 22.Rennell, D., and A. R. Poteete. 1985. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and lambda genes. Virology 143:280-289. [DOI] [PubMed] [Google Scholar]
- 23.Roberts, J. W. 1975. Transcription termination and late control in phage lambda. Proc. Natl. Acad. Sci. USA 72:3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schmidt, C., M. Velleman, and W. Arber. 1996. Three functions of bacteriophage P1 involved in cell lysis. J. Bacteriol. 178:1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 27.Steiner, M., W. Lubitz, and U. Bläsi. 1993. The missing link in phage lysis of gram-positive bacteria: gene 14 of Bacillus subtilis phage φ29 encodes the functional homolog of lambda S protein. J. Bacteriol. 175:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. Ruiters, and G. Venema. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 31.Young, I., I. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120-128. [DOI] [PubMed] [Google Scholar]
- 32.Ziermann, R., B. Bartlett, R. Calendar, and G. E. Christie. 1994. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J. Bacteriol. 176:4974-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]