Abstract
The bases of the mycobacterial SOS box important for LexA binding were determined by replacing each base with every other and examining the effect on the induction of a reporter gene following DNA damage. This analysis revealed that the SOS box was longer than originally thought by 2 bp in each half of the palindromic site. A search of the Mycobacterium tuberculosis genome sequence with the new consensus, TCGAAC(N)4GTTCGA, identified 4 sites which were perfect matches and 12 sites with a single mismatch which were predicted to bind LexA. Genes which could potentially be regulated by these SOS boxes were ascertained from their positions relative to the sites. Examination of expression data for these genes following DNA damage identified 12 new genes which are most likely regulated by LexA as well as the known M. tuberculosis DNA damage-inducible genes recA, lexA, and ruvC. Of these 12 genes, only 2 have a predicted function: dnaE2, a component of DNA polymerase III, and linB, which is similar to 1,3,4,6-tetrachloro-1,4-cylcohexadiene hydrolase. Curiously, of the remaining 10 genes predicted to be LexA regulated, 7 are members of the M. tuberculosis 13E12 repeat family, which has some of the characteristics of mobile elements.
The repair of damaged DNA is crucial to cell survival and replication. In bacteria the expression of a number of the genes responsible for DNA repair is induced following exposure to agents which cause such damage. This coordinated regulation of many genes at different loci on the genome was first established for Escherichia coli and was termed the SOS response (10, 14, 23). The SOS response has been studied in some detail in E. coli and the key regulatory components have been shown to be the proteins LexA and RecA (9, 14). LexA is a repressor protein which in the uninduced state binds to a specific sequence, termed the SOS box, upstream of the genes it regulates and so restricting expression (2, 15). When DNA becomes damaged, regions of single-stranded DNA arise, either from processing of the damaged region or from blockage of replication (25). RecA binds to these single-stranded regions, forming a nucleoprotein filament, and in this form it stimulates the autocatalytic cleavage of LexA (13). The cleaved fragments of LexA no longer bind to the SOS boxes (1), thus relieving repression and leading to increased expression of the genes of the SOS regulon.
The basic principles of this regulatory mechanism are found in many other species of bacteria, although the DNA sequence of the LexA binding site, or SOS box, varies. Thus, while the SOS box in E. coli and other enterobacteria has the consensus sequence taCTGTatatatatACAGta (where the bases in lowercase are less well conserved than those in uppercase) (12), it has been suggested that in rhizobia the SOS box is GAAC(N)7GTAC (29); in Bacillus subtilis the SOS box, originally thought to be GAAC(N)4GTTC (4, 5), has more recently been refined as CGAACRNRYGTTCG (30). A motif similar to the original short version of the B. subtilis SOS box has been found upstream of the recA and lexA genes and has been shown to bind LexA in mycobacteria (7, 18, 19). However, the specific bases required for LexA binding have not been determined, as demonstrated by the excessive number of hits found when this sequence was used to search the Mycobacterium tuberculosis genome sequence (3). A precise definition of the mycobacterial SOS box would allow the identification of LexA binding sites in the M. tuberculosis genome and thus aid in the discovery of other LexA-regulated genes. Therefore, we have undertaken an analysis of the effect of single base changes in the mycobacterial SOS box on LexA binding in vivo by comparing the induction ratios obtained, using a transcriptional fusion to a LexA-regulated promoter. Using the information obtained, we have been able to identify a number of novel LexA-regulated genes in M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains and media.
For general cloning E. coli strain DH5α was used, while for site-directed mutagenesis strain XL1Blue was used (24). The mycobacterial strains used were Mycobacterium smegmatis mc2155 (28) and M. tuberculosis H37Rv. E. coli was grown in L broth (24) and mycobacteria were grown in modified Dubos medium supplemented with albumin and 0.2% glycerol. E. coli and M. smegmatis were grown at 37°C with shaking. M. tuberculosis was grown at 37°C in a rolling incubator; under these conditions of growth the doubling time was 17 h. Antibiotics were added as appropriate: kanamycin was used at 50 μg ml−1 for E. coli and at 25 μg ml−1 for M. smegmatis.
Recombinant DNA techniques.
Plasmid DNA was prepared using SNAP Miniprep kits (Invitrogen). Site-directed mutagenesis was performed as described in the QuickChange Site-Directed Mutagenesis Kit (Stratagene). For other DNA manipulations, standard DNA protocols were followed (24). For each mutant made, the sequences of the promoter region and the junctions to the vector were determined on an ABI Prism 377 DNA sequencer using the ABI Prism dRhodamine dye terminator cycle sequencing kit (PE Applied Biosystems).
Introduction of clones into M. smegmatis and verification by PCR and sequencing.
Published protocols were followed for preparing electrocompetent cells of M. smegmatis mc2155 (22) and for electroporation (11). A preparation of total DNA suitable for PCR was isolated after streaking out the resulting transformants using an InstaGene matrix (Bio-Rad) according to the manufacturer's instructions except for using more bacteria. The insert and junctions of each clone were isolated as a PCR product using the primers PMINT2 (ACGAGGGGCATTCACACCAGATTG) and LACR (TTCCCAGTCACGACGTTGTAAAA) with 2.5 U of Pfu Turbo (Stratagene), and the cycle conditions were 94°C for 2 min and then 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, followed by 72°C for 7 min. Nucleic acid sequences of these PCR products were then determined on an ABI Prism 377 DNA sequencer using the ABI Prism dRhodamine dye terminator cycle sequencing kit (PE Applied Biosystems).
Induction conditions.
To induce DNA damage in M. smegmatis transformants, mitomycin C was added to one aliquot of a culture at an A600 of 0.4 to 0.5 to a final concentration 0.2 μg ml−1 and incubated for 5 h. An equal volume of the same culture was incubated for the same period of time without any addition, to provide an uninduced control. Following this, the bacteria were harvested, washed three times in Z buffer (17) without β-mercaptoethanol (Z*), and stored as a pellet at −20°C. For M. tuberculosis, cultures were induced at an A600 of ≈0.6 and induction was for 24 h prior to RNA isolation for microarray analysis as detailed below. In this case the uninduced control consisted of an equal volume of cells of the same culture harvested immediately before the addition of mitomycin C.
Preparation of cell extracts and β-galactosidase assays.
Untreated and mitomycin C-treated bacteria were resuspended in 1 ml of Z* buffer and lysed in the presence of glass beads (150 to 212 μm; Sigma) using a Ribolyser (Hybaid) at a speed setting of 6.5 for 30 s. The supernatant was collected by centrifugation for 5 min at full speed in a microcentrifuge, and the centrifugation was repeated to ensure no carryover of beads or cell debris occurred. An aliquot of the cell extract was used to determine its protein concentration using a bicinchoninic acid protein assay kit (Pierce). To the remaining extract, β-mercaptoethanol was added to a final concentration of 50 mM. These samples were then used to assay β-galactosidase activity as described previously (17) but with half-size reaction mixtures (500 μl) and reading of the absorbance of 300 μl of reaction mix in a flat-bottom microtiter plate at 405 nm. The specific activity in units per milligram of protein was calculated using the formula defined by Miller (17).
Computer searches.
Searches of the whole M. tuberculosis H37Rv genome were performed using the facilities provided at the TubercuList website (http://genolist.pasteur.fr/TubercuList/).
RNA extraction.
Commercially available kits were used for the isolation of total RNA (Hybaid Ribolyser Blue kit) from bacterial cultures (100 ml). Contaminating DNA in the RNA preparations was digested using RNase-free DNase (Roche), and the RNA was subsequently cleaned up using an RNeasy MiniKit (Qiagen). RNA concentrations were determined spectrophotometrically at 260 nm.
Microarray analysis.
A whole-genome DNA microarray for M. tuberculosis consisting of PCR products designed to minimize cross-hybridization and covering 90% of the predicted open reading frames (ORFs) was kindly supplied by J. Hinds, J. A. Mangan, and P. D. Butcher. Five micrograms of total RNA was used for first-strand cDNA synthesis using fluorescently labeled Cy3-dCTP and Cy5-dCTP (Amersham Pharmacia Biotech) in a standard reverse transcriptase reaction with Superscript II (Invitrogen). Briefly, total RNA with 6 μg of random primers (Invitrogen) in a reaction volume of 11 μl was denatured at 95°C for 5 min and snap-cooled on ice. Then, 5 μl of 5× first-strand buffer (Invitrogen), 2.5 μl of dithiothreitol (100 mM) (Invitrogen), 2.3 μl of deoxynucleoside triphosphate mix (5 mM concentrations of dATP, dGTP, and dTTP and 2 mM dCTP) (Amersham Pharmacia Biotech), 1.7 μl of Cy3- or Cy5-dCTP, and 2.5 μl of Superscript II were added. The labeling reaction mixtures were incubated at 25°C for 10 min and then at 42°C for 90 min.
Microarray slides were incubated in a prehybridization buffer (3.5× SSC buffer [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS], and 10 mg of bovine serum albumin/ml) at 60°C for 20 min. After incubation, slides were washed in distilled water for 1 min and then isopropanol for 1 min. Cy3- and Cy5-labeled RNA samples were combined and cleaned up using a MinElute column (Qiagen) and eluted in 13.5 μl of distilled water. After the addition of 4× SSC and 0.3% SDS, samples were denatured at 95°C for 2 min, cooled, and applied to the microarray slide, which was then covered with a glass coverslip. Each slide was placed in a waterproof hybridization chamber and submerged in a 60°C water bath overnight. After hybridization, slides were washed in 1× SSC plus 0.05% SDS for 2 min and then washed twice in 0.06× SSC for 2 min each.
Slides were scanned using a GenePix Axon 4000A scanner (Axon Instruments) at dual wavelengths and set to 600 V. The image data were quantified using GenePix Pro 3.0 software, and bad spots were removed. The data were further analyzed using Genespring 4.0.3 (Silicon Genetics). The data were normalized using each gene's measured intensity divided by its control channel value in each sample.
RESULTS
Effect of the immediate flanking bases on LexA binding.
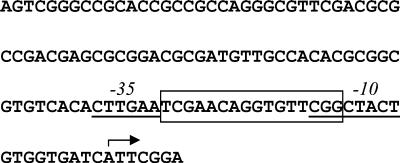
Previously, motifs matching the original B. subtilis SOS box had been found upstream of the recA and lexA genes of M. tuberculosis and purified M. tuberculosis LexA protein had been shown to bind to both these sites (18, 19). However, when we discovered a further motif identical to the original B. subtilis consensus sequence located downstream of the lexA gene and potentially regulating another gene, it became apparent that the M. tuberculosis SOS box was not fully defined as this site and did not bind M. tuberculosis LexA in a gel retardation assay (data not shown). Thus, bases outside that motif must be important for LexA binding. To address this issue we investigated the effects of changing bases in and around the M. tuberculosis recA SOS box. The M. tuberculosis recA gene is expressed from two promoters, only one of which is regulated by LexA (unpublished data), and so we used a construct containing only the LexA-regulated promoter contained in a 121-bp fragment (Fig. 1) transcriptionally fused to a lacZ reporter gene in the integrating plasmid pEJ414 (21). The SOS box is located between the predicted −35 and −10 elements of this promoter (Fig. 1).
FIG. 1.
The sequence of the DNA fragment upstream of the M. tuberculosis recA gene used in this study. The SOS box as defined in this study is boxed and the putative −10 and −35 elements are underlined. The transcriptional start site is indicated by the arrow.
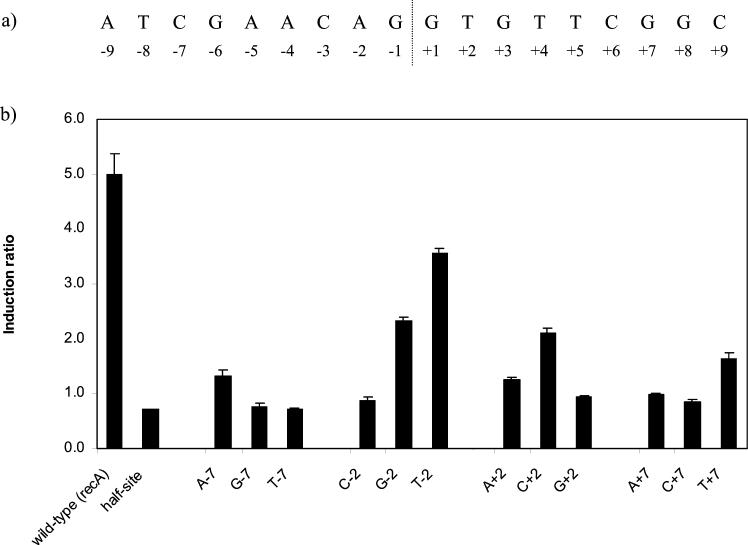
We initially focused on the bases at the immediate flanking positions of the mycobacterial SOS box. The wild-type DNA sequence of the recA SOS box along with the numbering system used to identify individual bases is shown in Fig. 2a. Thus, the positions investigated in this preliminary analysis were ±2 and ±7. We investigated expression in both the presence and absence of the DNA-damaging agent mitomycin C (0.2 μg/ml) for each construct. Single base changes were made at each of the ±2 and ±7 positions to each other nucleotide. In addition, double changes were made which maintained the ability to base pair at these positions by changing the internal A-T to T-A, C-G, and G-C and the external C-G to G-C, A-T, and T-A.
FIG. 2.
(a) The sequence of the recA SOS box along with the numbering system used to identify individual bases. (b) The effect of single base changes at ±2 and ±7 on the induction ratio. The base tested and its position is indicated below each bar. The wild-type sequence was the recA SOS box shown in panel a, and the half site consisted of four base changes from GAAC to TGGA. The graph shows the mean values obtained from duplicate assays of at least three independent inductions.
Many of these mutations affected the maximal level of expression seen. This effect on promoter strength is not surprising in view of the fact that the recA SOS box lies between the −10 and −35 sites of the recA promoter being used in this study. Owing to the variation in expression level, the easiest way to assess the effects of the mutations on LexA binding was to compare the induction ratios. If LexA is prevented from binding, repression will not occur under the uninduced conditions, so the expression level should be similar in the uninduced and induced samples, resulting in an induction ratio close to 1.
All of the double changes at both the ±2 and the ±7 positions eliminated DNA damage induction (data not shown), resulting in induction ratios of 0.7 to 0.9, similar to that seen with half of the original motif altered (Fig. 2b, half-site). A similar result was found for four of the six single changes at ±7, with only a very low residual degree of induction seen for the remaining two single changes at these positions (Fig. 2b). The induction ratios were 1.7 for T at +7 and 1.3 for A at −7, compared with an induction ratio of 5.0 for the wild-type sequence. Thus, the bases at the ±7 positions are indeed important in determining whether or not LexA will bind. Of the six single changes at ±2, two resulted in no DNA damage induction (G at +2 and C at −2), while four yielded levels of induction intermediate between that of the wild-type sequence and none (Fig. 2b). Thus, some bases at the ±2 positions are not compatible with LexA binding, while others permit LexA binding to a reduced extent.
Definition of the mycobacterial LexA binding site within the context of the recA SOS box.
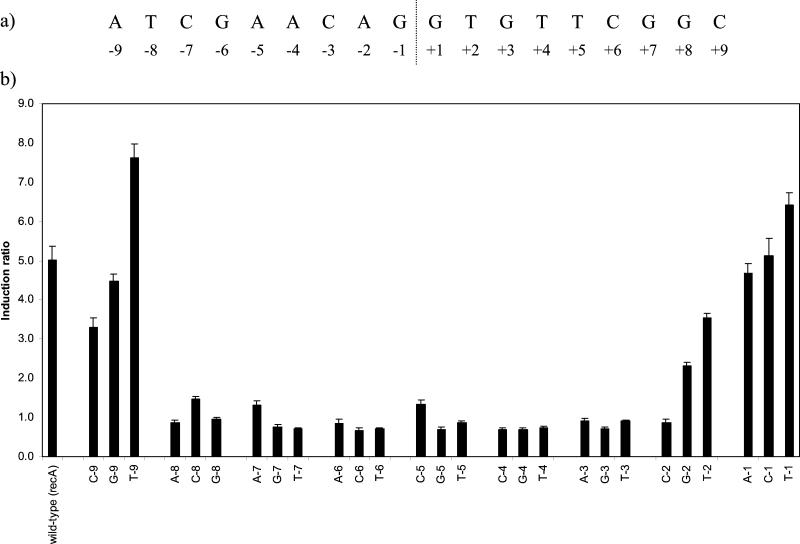
We then extended the study to examine the roles of individual bases throughout one-half of the symmetrical SOS box and further bases flanking it. M. tuberculosis LexA had previously been shown to bind only to this site within 390 bp upstream of recA by DNase I footprinting (19), where the protected region was clearly centered on the SOS box motif and extended to positions ±11 or 12 in the numbering system used here. With a single exception, the analysis of the ±2 and ±7 positions showed that comparable results were obtained when the reciprocal change was made in each half of the motif (Fig. 2b), so for this more extensive study we confined changes to one-half of the SOS box. Single changes to each of the other nucleotides were made at each position from −1 to −9 (Fig. 3a).
FIG. 3.
(a) Sequence of the recA SOS box along with the numbering system used to identify individual bases. (b) Effect on the induction ratio of single base changes from position −1 to −9 in the recA SOS box. The base tested and its position is indicated below each bar. The wild-type sequence was the recA SOS box shown in Fig. 1a. The graph shows the mean values obtained from duplicate assays of at least three independent inductions.
Each of the changes at positions −1 and −9 remained inducible to a similar degree as the wild-type sequence, suggesting that these locations define the boundary to the mycobacterial SOS box (Fig. 3b). With two exceptions, all of the changes at positions −3, −4, −5, −6, and −8 resulted in the abolition of DNA damage induction, while even in those two cases the induction ratios were severely reduced (to 1.3 for C at −5 and to 1.5 for C at −8). Whether such low levels of induction, as was also seen with A at −7, are significant in terms of the survival of the organism is doubtful. Thus, not only the bases of the originally defined motif (±3 to ±6) but also those at positions ±7 and ±8 are crucial for LexA binding.
Mismatches from the consensus which are functional.
The analysis of the recA SOS box had indicated that no substitutions of the G at position −6 were compatible with LexA binding. In view of the palindromic nature of the SOS box, this would imply that only a C at position +6 would be functional. However, the SOS box upstream of the lexA gene has a T at this position (+6) and has previously been shown to bind LexA in vitro (18) and more recently to confer DNA damage induction on LexA (unpublished results). These apparently contradictory observations can be resolved by examining the bases at ±8 which were previously not suspected of being important in determining LexA binding. When these bases are included in the analysis, we can see that the lexA SOS box has the optimum sequence at each position apart from that at +6 (T rather than C), yielding a functional site. However, the recA SOS box has already got a mismatch from the optimum LexA binding sequence at +8 (G rather than A), in the presence of which a further mismatch at ±6 is not tolerated.
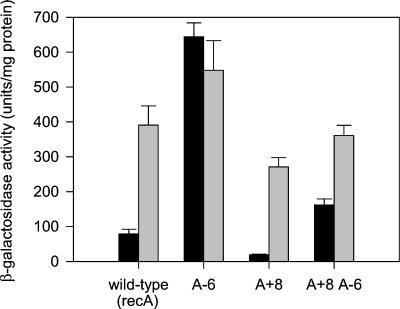
To confirm this hypothesis, we changed the base at position +8 in the recA SOS box to A, both in the wild-type sequence and in the sequence already containing an A at −6 (equivalent to the T at +6 in the lexA SOS box). We expected that in the presence of A at +8 LexA would be able to bind to the site containing A at −6, resulting in DNA damage-inducible expression. Furthermore, we would predict that the perfect palindrome created by the introduction of A at +8 in the wild-type sequence would yield a site with a higher affinity for LexA, which would be reflected in a lower expression level in the uninduced state and likely a higher induction ratio. Both of these predictions were fulfilled (Fig. 4). With the perfect palindrome (A + 8), the uninduced expression was reduced to 20 U of β-galactosidase/mg of protein, compared with ca. 80 for the wild-type recA SOS box, and the induction ratio increased from 5 to 14. In addition, the construct containing A at +8 and A at −6 was DNA damage inducible; although the induction seen was only 2.2-fold, this is to be compared with 0.9-fold for A at −6 in the native recA SOS box.
FIG. 4.
Comparison of the native recA SOS box (wild type) with a perfectly palindromic SOS box (A + 8) with and without the A −6 mutation. The β-galactosidase activity determined with (grey bars) or without (black bars) exposure to the DNA-damaging agent mitomycin C (0.2 μg/ml) is shown. The graph shows the mean values obtained from duplicate assays of at least three independent inductions, with the error bars indicating standard deviations.
Definition of the mycobacterial LexA binding site within the context of the perfect palindrome.
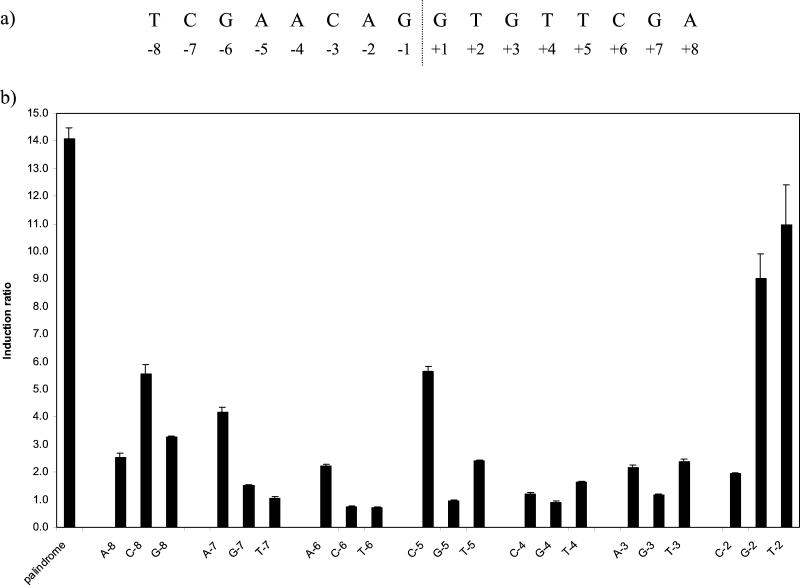
The experiments just described revealed that the recA SOS box itself contained a mismatch from the optimal LexA binding sequence and that, if this were corrected, a base change could be tolerated which originally had not been. Therefore, we proceeded to reexamine the roles of each of the bases throughout one-half of the perfectly palindromic SOS box created by changing the base at position +8 in the recA SOS box to A, in an otherwise identical series of constructs to those used initially. Single changes to each other nucleotide were made at each position from −2 to −8 in the presence of A at +8; positions −1 and −9 were not included, as base changes at these locations had been shown to be tolerated in the previous part of the study.
Many more changes from the optimal sequence were tolerated compared with the previous analysis insofar as expression remained DNA damage inducible, although to a reduced extent (Fig. 5b). The greatest degrees of induction were found with the changes to G or T at −2, although these changes had remained inducible even in the context of the recA SOS box. Although expression was also inducible with the remaining base change at this position (C −2), this mutation had a much stronger effect on the induction ratio, reducing it to only twofold. The change to C at −8, which is equivalent to the mismatch found in the native recA SOS box, resulted in ca. fivefold induction, which is also what was found above with the wild-type sequence. Two other changes also yielded ca. fivefold induction and might, therefore, be expected to occur in other native SOS boxes; these were A at −7 and C at −5. A number of changes permitted an induction of two- to threefold; in addition to A at −6 and C at −2, discussed above, these changes were A or G at −8, T at −5, and A or T at −3. As the lexA SOS box contains a mismatch from the optimal sequence equivalent to A at −6, it is likely that any of these differences might be found singly in SOS boxes for other genes. It is difficult to know whether the 1.6-fold induction seen with T at −4 and G at −7 is significant in terms of the response of the bacterium to DNA damage.
FIG. 5.
(a) Sequence of the perfectly palindromic SOS box along with the numbering system used to identify individual bases. (b) Effect on the induction ratio of single base changes from position −2 to −8 in a perfectly palindromic SOS box. The base tested and its position are indicated below each bar. The control palindromic sequence was the recA SOS box with the G at +8 changed to A. The graph shows the mean values obtained from duplicate assays of at least three independent inductions.
From this analysis it is possible to deduce an optimal sequence for the mycobacterial SOS box: TCGAACNNNNGTTCGA. Although when tested in the perfectly palindromic motif any base at position −8 appeared to result in a functional LexA binding site, the importance of the base at this position is demonstrated both by the significant reduction in the induction ratios for these sites and by the lack of tolerance of base changes at other positions in the presence of even the most favorable substitution at this position. Single deviations from the optimum sequence, as indicated in Table 1, might be expected to be functional sites. Increasing the spacing between the two halves of the palindrome by one or two bases also prevented LexA binding (data not shown).
TABLE 1.
Effect of single base changes in the perfect palindrome on induction
| Base at position | Fold inductiona | ||||||
|---|---|---|---|---|---|---|---|
| −8 | −7 | −6 | −5 | −4 | −3 | −2 | |
| T | C | G | A | A | C | A | 14 (wild type) |
| T | 11 | ||||||
| G | 9 | ||||||
| C | A | C | 5 | ||||
| G | 3 | ||||||
| A | A | T | A/T | C | 2 | ||
| G | T | 1.6 | |||||
The induction ratio obtained when each of the bases indicated on that line was tested individually as a single base change from the perfect palindrome, which is shown in the first line (wild type).
Use of the new consensus to identify potential SOS boxes in the genome sequence of M. tuberculosis.
While the detailed analysis of the SOS box described above was conducted in M. smegmatis and, therefore, defined the criteria important for M. smegmatis LexA binding, we believed that it was reasonable to extrapolate the main conclusions to M. tuberculosis for the following reasons. In a previous study (3) we had compared binding of purified M. tuberculosis LexA to a set of six mutant SOS boxes which exhibited various induction ratios in vivo. These six SOS box variants are those shown in Fig. 2b with changes at the ±2 position and have induction ratios that vary from 0.9 to 3.6. Gel-shift analysis (3) revealed that the degree of binding of M. tuberculosis LexA to these sequences paralleled the degree of induction, and hence the degree of LexA binding, seen in M. smegmatis. In addition, potential SOS boxes upstream of a number of homologs of genes which are members of the SOS regulon in E. coli were identified on the basis of the original short consensus sequence in that study, but their ability to bind M. tuberculosis LexA did not appear to correlate with the number of mismatches from that motif. However, when the sequences studied were compared with the newly defined consensus, with a single exception the sites which bound LexA had one or two mismatches while those which did not had three or more mismatches.
Previously, searching the genome sequence (6) of M. tuberculosis with the originally defined motif (GAACNNNNGTTC) identified 35 ORFs preceded by a motif which was a perfect match within 500 bp of the upstream sequence (3), which corresponded to 28 distinct sites. The study described here shows that the mycobacterial SOS box is actually more extensive than this, with the bases at positions ±7 and ±8 playing important roles in determining LexA binding. We therefore repeated this search of the M. tuberculosis genome with the optimum SOS box sequence defined above and with the constraint that the motif should be within 500 bp upstream of a start codon. The number of such ORFs was now reduced to 4 with no mismatches, corresponding to 3 sites, and 21 ORFs with a single mismatch, corresponding to 16 distinct sites, a much more plausible number.
We then used the newly defined consensus sequence for the mycobacterial SOS box to search the M. tuberculosis genome sequence with no constraints on the location of motif. This search identified 4 sites with no mismatches and 34 sites with a single mismatch. Of these 34 sites, 14 would not be expected to bind LexA at all, owing to the base which differs from the consensus. A further four sites may not be functional, as the induction ratio was only 1.6 in the test sequence containing those changes. In addition, six sites, including two of these four sites, have a C at −2 (or a G at +2), a substitution which was not tolerated in the presence of the native mismatch in the recA SOS box; thus, these sites may not be functional for LexA binding. Therefore, we would predict that there are a total of 16 functional LexA binding sites with zero or one mismatch from the newly defined consensus in the M. tuberculosis genome (Table 2).
TABLE 2.
SOS boxes identified in the M. tuberculosis genome and potentially LexA-regulated genesa
| No. of mismatches | Position of SOS box in genome sequenceb | Sequence of SOS boxg | Strand | Potential gene or ORF regulated | Location relative to ORFc | Induction ratiof |
|---|---|---|---|---|---|---|
| 0 | 1552510-1552525 | tcgaacacatgttcga | − | Rv1378c | +129 | 15.1 ± 7.0 |
| tcgaacatgtgttcga | + | Rv1379 (pyrR) | −144 | 1.7 ± 0.2 | ||
| 3436770-3436785 | tcgaacatgtgttcga | − | Rv3073c | −101 | 1.1 ± 0.1 | |
| tcgaacacatgttcga | + | Rv3074 | −8 | 21.3 ± 6.6 | ||
| 3784789-3784804 | tcgaacaattgttcga | − | Rv3370c (dnaE2) | −65 | 13.4 ± 7.6 | |
| tcgaacaattgttcga | + | Rv3371 | −142 | 1.3 ± 0.2 | ||
| 4221081-4221096 | tcgaacgtatgttcga | + | Rv3776 | −8 | 13.1 ± 4.7 | |
| 1, predicted to bind LexA | 79480-79495 | tcgaaTatgagttcga | + | Rv0071 | −6 | 2.1 ± 0.8 |
| 400160-400175 | tcgaacatacTttcga | − | Rv0335c | −125 | 1.6 ± 0.4 | |
| tcgaaAgtatgttcga | + | Rv0336 | −32 | 16.1 ± 3.1 | ||
| 606519-606534 | tcgaaAgtatgttcga | + | Rv0515 | −32 | 13.6 ± 3.0 | |
| 1117141-1117156 | tcgaacgaatgtGcga | − | Rv1000c | −8 | 6.7 ± 1.4 | |
| tcgCacattcgttcga | + | Rv1001 (arcA) | −44 | 0.9 ± 0.1 | ||
| 1928566-1928581 | tcgaacatgtAttcga | − | Rv1702c | −6 | 3.1 ± 0.6 | |
| 2358495-2358510 | tcgCacacatgttcga | + | Rv2100 | +106 | 6.8 ± 1.3 | |
| 2903540-2903555 | tAgaacggttgttcga | − | Rv2578c | −24 | 2.9 ± 0.7 | |
| tcgaacaaccgttcTa | + | Rv2579 (linB) | −99 | 7.9 ± 2.5 | ||
| 2925405-2925420 | tcgaacgattgttcgG | − | Rv2594c (ruvC) | −37 | 6.4 ± 1.4 | |
| Ccgaacaatcgttcga | + | Rv2595 | −87 | 1.3 ± 0.2 | ||
| 3031739-3031754 | tcAaacatgtgttcga | − | Rv2719c | −218d | 18.1 ± 8.7 | |
| tcgaacacatgttTga | + | Rv2720 (lexA) | −105 | 7.2 ± 1.6 | ||
| 3051531-3051546 | tcgaacaggtgttcgG | − | Rv2737c (recA) | −123 | 17.3 ± 3.1 | |
| 3453546-3453561 | tcgaacgggagttcgC | + | ||||
| 3811650-3811665 | tcgaacatatTttcga | − | Rv3395c | +240 | 16.8 ± 3.8 | |
| 1, thought unlikely to bind LexA | 6327-6342 | Gcgaaccgcagttcga | + | |||
| 2265299-2265314 | tGgaactgcggttcga | + | Rv2018 | +19 | 1.8 ± 0.5 | |
| 2358405-2358420 | Gcgaacgacggttcga | − | Rv2099c | −214 | 1.7 ± 0.2 | |
| tcgaaccgtcgttcgC | + | Rv2100 | +16e | 6.8 ± 1.3 | ||
| 2950938-2950953 | tcgaacccaagAtcga | + | ||||
| 3534096-3534111 | tcgCacgacggttcga | − | Rv3164c (moxR3) | −207 | 0.9 ± 0.1 | |
| 3640311-3640326 | tcgaTcacttgttcga | − | Rv3260c (whiB2) | −186 | 0.5 ± 0.1 | |
| tcgaacaagtgAtcga | + | Rv3261 | −231 | 0.8 ± 0.1 | ||
| 3866007-3866022 | tcgaacggccgAtcga | + | ||||
| 3973763-3973778 | Acgaaccgcagttcga | − | Rv3534c | −187 | 1.2 ± 0.2 |
Those potentially LexA-regulated genes which were DNA damage inducible and their induction ratios are highlighted in bold.
The coordinates in the M. tuberculosis genome sequence of the motif matching the SOS box.
The location of the first base of the SOS box relative to the predicted translational start site of the ORF is given; a negative number indicates that the SOS box is upstream and a positive number indicates that it is within the coding region.
Although Rv2719c is DNA damage inducible, it is not regulated by this site (unpublished data).
Althought Rv2100 is DNA damage inducible, it may not be regulated by this site, as the site at 2358495 is also within the coding region of Rv2100.
Mean ± standard deviation.
The consensus sequence used to search the genome is in bold, with mismatches from it in uppercase.
We next looked for genes potentially regulated by these sites, initially assuming that the SOS boxes would lie just upstream of such genes. Surprisingly, of the three such genes identified adjacent to sites with no mismatches, only one was DNA damage inducible (see next section). However, two of the sites overlapped predicted translational start sites of other genes and one was within a predicted coding sequence, and these genes were inducible. We therefore included in our analysis ORFs for which the SOS box was located between 300 bp upstream (−300) and 300 bp downstream (+300) of the predicted translation initiation codon; these ORFs are listed in Table 2.
Are the genes with predicted SOS boxes DNA damage inducible in M. tuberculosis?
As part of a separate study we are seeking to identify DNA damage-inducible genes of M. tuberculosis empirically by using microarray analysis. Therefore, we examined the microarray data specifically for the genes listed in Table 2 as potentially being regulated by LexA binding to the identified SOS boxes to see which ones were DNA damage inducible. For many of the SOS boxes there were two potentially regulated genes, one on each strand, although in most cases we would only expect one of each pair to be regulated by a particular site.
As alluded to above, only one of the four genes preceded by an SOS box with no mismatches (Rv3370c or dnaE2) was induced following mitomycin C treatment of M. tuberculosis. However, all three of the genes where the perfect SOS box overlapped or was contained within the coding sequence were induced (Table 2). Thus, it would appear that all four LexA binding sites with no mismatches from the consensus are involved in regulating gene expression following DNA damage.
A gene which was induced following DNA damage was found for 10 of the 12 SOS boxes with a mismatch from the consensus which were expected to bind LexA. In addition, an ORF, Rv0071, which might be inducible but for which the data were not clear, was appropriately positioned by one of the remaining two sites. In contrast, only one gene, Rv2100, located near the sites thought unlikely to bind LexA, was DNA damage inducible; this gene may not be regulated by that site, as another site is also situated within its coding sequence. Those sites for which no potentially regulated ORF is given in Table 2 were positioned further into an ORF, which was in any case not DNA damage inducible (data not shown). The majority of the motifs found in the search for which the mismatch was predicted to be incompatible with LexA binding were located far from ORFs, with only one being within 300 bp of a predicted gene and three within 400 bp. None of these genes exhibited DNA damage-inducible expression (data not shown). Thus, it would appear that the newly defined mycobacterial SOS box has good predictive power in terms of identifying LexA binding sites in the M. tuberculosis genome.
In one case, two divergently transcribed genes (Rv2578c and Rv2579 or linB) are apparently both regulated by a single SOS box located between them. Although this also appears to be the case for Rv2719c and Rv2720 or lexA, we have shown in a separate study (unpublished data) that Rv2719c is not regulated by this SOS box. Thus, the locations of the SOS boxes which look to be functional range between 123 bp upstream of the translational start site and 240 bp into the coding sequence, although it should be emphasized that these coding regions are predicted ones and some of these ORFs may actually have alternative start sites.
The induction ratio for recA observed in the microarray analysis is somewhat higher than that found with the lacZ reporter fusion to the wild-type sequence upstream of recA which was used in the analysis of the SOS box. This is likely to be due to the combined effects of a number of factors. Firstly, the reporter construct used contains only one of the two recA promoters, whereas obviously both are involved in expressing recA from the genome in the global analysis. Secondly, the reporter construct was assayed in M. smegmatis, whereas the microarray measured expression in M. tuberculosis and there may be a difference in the degree of induction between the two species. Thirdly, the uninduced control for the β-galactosidase assays was incubated in parallel with the induced sample, while for the microarray analysis it consisted of a zero time point. Finally, and linked to the above, in the microarray analysis we were measuring RNA rather than the more stable β-galactosidase protein, accumulation of which from low-level expression during the uninduced incubation would reduce the apparent induction ratio.
DISCUSSION
By analyzing the effects of single base changes in the M. tuberculosis SOS box on the expression of a LexA-regulated promoter linked to a lacZ reporter gene, we have been able to define a consensus sequence for the SOS box in M. smegmatis. This has also allowed us to determine what changes from the consensus are tolerated while retaining function in gene regulation. The mycobacterial SOS box defined here is longer than originally thought by two bases in each half of the palindromic site. It was clear that the end of the site had been reached in the mutational analysis as with any base at the next adjacent position similar induction ratios were obtained. Interestingly, the new mycobacterial site is also longer than the redefined B. subtilis SOS box (30) to which it is similar, although in that case analysis was not pursued until there was no effect on induction when a base was substituted by any other. Although the sequences recognized in B. subtilis and M. smegmatis are related, there are significant species-specific determinants of LexA binding. For example, the motif with a T at −7 was fully functional in B. subtilis, whereas the same change completely blocked LexA binding in M. smegmatis, while the change to C at −5 prevented LexA binding in B. subtilis but it remained functional in M. smegmatis.
For the reasons stated above in the Results section, we considered it likely that the same motif would be recognized in M. tuberculosis as in M. smegmatis. Therefore, we went ahead with searching the M. tuberculosis genome sequence with the new consensus and identified 16 sites which we expected to be functional for LexA binding. The subsequent discovery in M. tuberculosis of DNA damage-inducible genes associated with all but 2 of these 16 sites supports this hypothesis. Furthermore, of eight sites identified with single mismatches predicted to be incompatible with LexA binding, only one had a gene nearby which was induced by DNA damage, and that gene was also associated with another site which was expected to bind LexA. Taken together, these results suggest that the SOS box defined in M. smegmatis also applies to M. tuberculosis.
In some cases, there were ORFs in both orientations on the chromosome within a reasonable distance of the SOS box, either (or rarely, both) of which might be regulated, while in a few cases there was no identified ORF in a suitable location. In these latter cases it might be either that the identified SOS box is not performing a regulatory function or that there could be an as-yet-unidentified ORF nearby. Small ORFs in particular are difficult to predict in genome annotation projects, and some small M. tuberculosis proteins have been identified recently which do not have annotated ORFs in the genome sequence (16). By examining the expression before and after DNA damage of the alternative ORFs near SOS boxes, 15 genes were identified whose expression is most likely regulated by LexA. For eight of these genes, the SOS box was located upstream of the ORF, as expected by analogy with LexA regulation in E. coli (26). Surprisingly, however, four of the SOS boxes overlapped the predicted translational start site and a further three (or possibly four, depending on whether both SOS boxes within Rv2100 are functional) SOS boxes were located within the coding sequence. This positioning would imply that in M. tuberculosis LexA can repress expression by interfering with transcription after it has begun as well as by affecting transcription initiation, as is the case in E. coli (26). However, it is possible that translation may actually begin further downstream than these predicted start sites and, thus, all the SOS boxes could in fact be upstream of the genes they regulate.
Interestingly, only 5 of the 15 genes predicted to be LexA regulated have known functions, with the other 10 genes representing novel DNA damage-inducible LexA-regulated genes. Of the five named genes, three (recA, lexA, and ruvC) were already known to be DNA damage inducible in M. tuberculosis (3, 7, 19). The remaining two are dnaE2, which encodes the α chain of DNA polymerase III, and linB, the product of which is similar to 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase. Curiously, of the 10 novel genes predicted to be LexA regulated, 7 (Rv0336, Rv0515, Rv1378c, Rv1702c, Rv2100, Rv3074, and Rv3776) appear to be members of the M. tuberculosis 13E12 repeat family, which has some of the characteristics of mobile elements (6). Thus, it may be that these elements have tapped into the LexA regulatory system to allow them to become active when DNA damage occurs, conditions when their own existence might be threatened. Interestingly, of the remaining three genes, Rv1000c is similar to a Xylella fastidiosa protein (BLASTP score, 6 × 10−21) described as a DNA repair system specific for alkylated DNA (27), although this is based on sequence similarity to alkB of Brucella melitensis. Although Rv3395c is annotated as having some similarity with the RecA protein of Thiobacillus ferrooxidans (http://genolist.pasteur.fr/TubercuList), this is somewhat limited (BLASTP score, 0.27) and is more likely to reflect a similar domain than a related function for the gene product. Finally, Rv2578c bears some resemblance to a member of the moaA/nifB/pqqE family (CC1330, BLASTP score of 10−32) from Caulobacter crescentus (20); although this particular example is of unknown function, other members of this family appear to be involved in the biosynthesis of cofactors containing metal (particularly molybdenum).
At first sight, it may seem surprising that other known DNA repair genes which are part of the SOS regulon in E. coli were not identified in this analysis. However, in a previous study where we specifically examined a number of such genes in M. tuberculosis for DNA damage induction and LexA binding (3), we found that some of the M. tuberculosis homologs were not induced following exposure to mitomycin C (recN, dinP, dinG), while some others did exhibit DNA damage-inducible expression but no binding of LexA was detectable to DNA from 500 bp upstream of the coding region to 100 bp within it (uvrA, ssb). Thus, it appears likely that there is an alternative mechanism of DNA damage induction in M. tuberculosis as well as that controlled by LexA.
This study has identified a set of 15 DNA damage-inducible genes which are most likely regulated by LexA in M. tuberculosis. A similar approach used for E. coli brought the total number of LexA-regulated genes in that species to 31 (8). We do not expect the list of genes identified here to be exhaustive, as we have only considered sites with up to one mismatch from the optimal sequence here. There could be some functional SOS boxes with more than one mismatch. Indeed, Rv2719c appears to be regulated by multiple SOS boxes each containing two or more mismatches (unpublished data). Our search of the M. tuberculosis genome revealed the presence of 351 sites containing two mismatches. Elimination of those sites where one of the mismatches alone would prevent LexA binding leaves 96 potential sites, of which we would expect only a small number to be functional. However, identifying which of these sites might be functional is not straightforward with the information currently available, as we cannot tell which combinations of mismatches which individually permit LexA binding would retain that ability and which would not. Our initial analysis using the native recA SOS box suggests that very few pairs of mismatches may result in a functional site.
Acknowledgments
We thank J. Hinds, J. A. Mangan, and P. D. Butcher for supplying us with microarrays, and we are indebted to Roger Buxton and Lisa Rickman for setting up the microarray system in the division. We also thank Patricia Brooks and Bosco Chan for technical support and M. J. Colston for critical reading of the manuscript.
REFERENCES
- 1.Bertrand-Burggraf, E., S. Hurstel, M. Daune, and M. Schnarr. 1987. Promoter properties and negative regulation of the uvrA gene by the LexA repressor and its amino-terminal DNA binding domain. J. Mol. Biol. 193:293-302. [DOI] [PubMed] [Google Scholar]
- 2.Brent, R., and M. Ptashne. 1981. Mechanism of action of the lexA gene product. Proc. Natl. Acad. Sci. USA 78:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks, P. C., F. Movahedzadeh, and E. O. Davis. 2001. Identification of some DNA damage-inducible genes of Mycobacterium tuberculosis: apparent lack of correlation with LexA binding. J. Bacteriol. 183:4459-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheo, D. L., K. W. Bayles, and R. E. Yasbin. 1991. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J. Bacteriol. 173:1696-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheo, D. L., K. W. Bayles, and R. E. Yasbin. 1993. Elucidation of regulatory elements that control damage induction and competence induction of the Bacillus subtilis SOS system. J. Bacteriol. 175:5907-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Durbach, S. I., S. J. Andersen, and V. Mizrahi. 1997. SOS induction in mycobacteria: analysis of the DNA-binding activity of a LexA-like repressor and its role in DNA damage induction of the recA gene from Mycobacterium smegmatis. Mol. Microbiol. 26:643-653. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez de Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg, E., G. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 10.Gudas, L. J., and A. B. Pardee. 1975. Model for regulation of Escherichia coli DNA repair functions. Proc. Natl. Acad. Sci. USA 72:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, L. K., G. R. Harlow, L. A. Gregg-Jolly, and D. W. Mount. 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507-523. [DOI] [PubMed] [Google Scholar]
- 13.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 14.Little, J. W., and D. W. Mount. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11-22. [DOI] [PubMed] [Google Scholar]
- 15.Little, J. W., D. W. Mount, and C. R. Yanisch-Perron. 1981. Purified lexA protein is a repressor of the recA and lexA genes. Proc. Natl. Acad. Sci. USA 78:4199-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattow, J., P. R. Jungblut, E. C. Muller, and S. H. Kaufmann. 2001. Identification of acidic, low molecular mass proteins of Mycobacterium tuberculosis strain H37Rv by matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. Proteomics 1:494-507. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Movahedzadeh, F., M. J. Colston, and E. O. Davis. 1997. Characterization of Mycobacterium tuberculosis LexA: recognition of a Cheo (Bacillus-type SOS) box. Microbiology 143:929-936. [DOI] [PubMed] [Google Scholar]
- 19.Movahedzadeh, F., M. J. Colston, and E. O. Davis. 1997. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J. Bacteriol. 179:3509-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papavinasasundaram, K. G., C. Anderson, P. C. Brooks, N. A. Thomas, F. Movahedzadeh, P. J. Jenner, M. J. Colston, and E. O. Davis. 2001. Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology 147:3271-3279. [DOI] [PubMed] [Google Scholar]
- 22.Papavinasasundaram, K. G., M. J. Colston, and E. O. Davis. 1998. Construction and complementation of a recA deletion mutant of Mycobacterium smegmatis reveals that the intein in Mycobacterium tuberculosis recA does not affect RecA function. Mol. Microbiol. 30:525-534. [DOI] [PubMed] [Google Scholar]
- 23.Radman, M. 1975. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 5:355-367. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 26.Schnarr, M., P. Oertel-Buchheit, M. Kazmaier, and M. Granger-Schnarr. 1991. DNA binding properties of the LexA repressor. Biochimie 73:423-431. [DOI] [PubMed] [Google Scholar]
- 27.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 28.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 29.Tapias, A., and J. Barbe. 1998. Mutational analysis of the Rhizobium etli recA operator. J. Bacteriol. 180:6325-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winterling, K. W., D. Chafin, J. J. Hayes, J. Sun, A. S. Levine, R. E. Yasbin, and R. Woodgate. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]