SUMMARY
Plant flavonoids are polyphenolic compounds commonly found in vegetables, fruits and many food sources that form a significant portion of our diet. These compounds have been shown to interact with several ATP-Binding Cassette transporters that are linked with anticancer and antiviral drug resistance and as such, may be beneficial in modulating drug resistance. The present study investigates the interactions of six common polyphenols; quercetin, silymarin, resveratrol, naringenin, daidzein and hesperetin with the multidrug resistance associated proteins, MRP1, MRP4 and MRP5. At non-toxic concentrations, several of the polyphenols were able to modulate MRP1-, MRP4- and MRP5- mediated drug resistance though to varying extents. The polyphenols also reversed resistance to NSC251820, a compound that appears to be a good substrate for MRP4 as had been predicted by data mining studies. Furthermore, most of the polyphenols showed direct inhibition of MRP1-mediated [3H]-dinitrophenyl S-glutathione and MRP4-mediated [3H]-cGMP transport in inside-out vesicles prepared from human erythrocytes. Additionally, both quercetin and silymarin were found to inhibit MRP1-, MRP4-, and MRP5-mediated transport from intact cells with high affinity. They also had significant effects on ATPase activity of MRP1 and MRP4 without having any effect on [α-32P]8-azidoATP binding to these proteins. This suggests that these flavonoids most likely interact at the transporter’s substrate-binding sites. Collectively, these results suggest that dietary flavonoids such as quercetin and silymarin can modulate transport activities of MRP1, 4 and 5. Such interactions could influence bioavailability of anticancer and antiviral drugs in vivo and thus, should be considered for increasing efficacy in drug therapies.
Keywords: ABC transporters; drug resistance; Plant polyphenols; MRP 1, 4 and 5; red blood cells
Abbreviations: ABC, ATP binding cassette; BeFx, Beryllium fluoride; calcein-AM, calcein acetoxymethylester; BCECF, 2′, 7′-bis(2-carboxyethyl)-5-(6)-carboxyfluorescein; DNP-SG, dinitrophenyl S-glutathione conjugate; FACS, Fluorescence Activated Cell Sorter; GSH, reduced glutathione; GSSG, oxidised glutathione; HEK, human embryonic kidney; MRP, multidrug resistance protein; MK-571, (3-(3-(2-(7-chloro-2-quinolinyl)ethenyl)phenyl) ((3-(dimethyl amino-3-oxo propyl)thio)methyl)thio)propanoic acid; PAGE, polyacrylamide gel electrophoresis; PGE1,Prostaglandin E1
INTRODUCTION
Multidrug resistance (MDR) is associated with the over-expression of ATP-binding cassette (ABC) transporters such as P-glycoprotein (Pgp), multidrug resistance proteins (MRPs), or ABCG2 (also called BCRP or MXR) [1, 2]. These transporters efflux a wide range of compounds and anticancer agents out of the cells; thus, inhibition of these pumps is crucial to overcome drug resistance. MRP1, MRP4, and MRP5 belong to the MRP family (ABCC subfamily); some members of which are ubiquitously expressed and known to transport a vast variety of substrates across cell membranes [3-5]. Over-expression of these transporters is known to cause resistance to doxorubicin, etoposide, 9-(2-phosphonyl-methoxyethyl) guanine (PMEG), and thioguanine [6–8].
Plant polyphenols such as flavonoids and stilbenes are abundant in vegetables, fruits, and many of the plant products consumed daily. The average U.S. diet supplies about 200 milligrams of polyphenols daily; however, it is possible for an adult to ingest more than 1 gram of polyphenols per day depending on the types of food consumed [9]. Many of these compounds are also found in herbal medicines. A number of polyphenols cause carcinogen inactivation, antiproliferation, cell cycle arrest, and inhibition of angiogenesis [10, 11]. Polyphenols are predominantly in sugar-conjugated forms but undergo enzymatic cleavage into free aglycone forms after ingestion. These free aglycones are then absorbed through the gut wall. After Phase I and II metabolism, the polyphenols can either remain as free aglycones or as glucoronidated, methylated or sulphated metabolites [12]. Bioavailability of polyphenols depends highly on the chemical structure of the polyphenol and physical variations within individuals [9]. Although plasma concentrations of polyphenols are usually less than 1 nM, local concentrations within the intestine should be substantially greater and can reach 3mM following a meal containing 500 mg of polyphenols [9]. Since MRP1, 4 and 5 are located in the intestine [2], it is likely that they can be exposed to such high polyphenol concentrations. Furthermore, recent studies show correlation between the in vitro effects of flavonoids at the low micromolar range and in vivo work using oral solutions of flavonoids [13, 14].
Many of these plant polyphenols may modulate the activities of the multidrug transporters. It has previously been reported that silymarin and several other flavonoids can increase daunomycin accumulation in Pgp expressing cells in a manner that depends on both the concentration of the flavonoids and the level of Pgp expression. It was proposed that the flavonoids interacted directly with Pgp substrate binding because they potentiated doxorubicin cytotoxicity, inhibited Pgp ATPase activity, and inhibited [3H]-azidopine photoaffinity labelling of Pgp [15]. Interactions of polyphenols with MRP1 have also been reported. Versantvoort et al (1993) demonstrated that genistein could increase daunorubicin accumulation in non-Pgp expressing MDR cell lines that were later revealed to overexpress MRP1, and subsequently, other flavonoids were found to modulate the activities of MRP1 [16, 17]. Leslie et al. used membrane vesicle preparations to demonstrate that flavonoids could directly inhibit MRP1-mediated LTC4 transport and to a lesser extent 17β-estradiol 17β-(D-glucoronide) transport. Since these inhibitory effects were enhanced by GSH, it was proposed that GSH might be co-transported with the polyphenolic compounds. Because there are variations in activity profiles for these flavonoids, it has been proposed that they may interact with different sites on the MRP1 molecule. Similar results were reported in another study in which several different flavonoids were used [18]. More recently, several flavonoids were shown to reverse breast cancer resistance protein (BCRP; ABCG2)-mediated transport and multidrug resistance [19, 20] as well as to activate the cystic fibrosis transmembrane conductance regulator (CFTR; ABCC7) chloride channel [21].
Despite the numerous studies investigating the interactions between polyphenols with Pgp, BCRP, and MRP1, the possible interaction of these compounds with MRP4 and MRP5 has not been studied until now. Unlike MRP1, MRP4 and MRP5 are able to transport cyclic nucleotides such as cGMP and cAMP [22, 23], antiviral drugs and prostaglandins [5, 24]. In the present study, we have investigated the six most common plant polyphenols for their ability to modulate the function of MRP1, 4 and 5 at the low micromolar range. Our results demonstrate that these plant polyphenols interact with MRP4 and MRP5 and affect their transport function to a greater extent than the transport function of MRP1. Some polyphenols are high affinity inhibitors, whereas others may be substrates themselves. Since polyphenols are relatively non-toxic, they may be valuable in reversing resistance to various drug therapies because of their abundance in commonly consumed nutritional products. In addition, we also demonstrate that sensitivity to NSC251820, a compound which has been predicted by data mining to be a substrate for MRP4 [25], is significantly lower in cells expressing this transporter. This suggests that NSC251820 may be a good substrate for this transporter, and polyphenols also reverse the resistance to this compound in MRP4-expressing cells.
Results
Characterization of the mRNA expression of selected ABC transporters in transfected HEK293 cells
To determine the relative mRNA expression of the various ABC transporters of interest in the cell lines utilized in this work, we isolated total RNA from each of the cell lines and performed quantitative real time RT-PCR (sequence of specific primer sets given in Table 1). The expression levels for each of the ABC transporters in the transfected HEK293 cells were normalized to the levels within the parental HEK293 cells. These studies confirmed that each of the MRP transfectants show overexpression of only that particular MRP (Fig. 1); for example MRP4-expressing HEK293/4.63 cells have nearly 100-fold more MRP4 than the parental HEK293 cells. It is also clear from the analyses that the selection with G418 (transfected HEK293 cells) does not result in overexpression of other ABC drug transporters. These results correlate well with Western blotting results which were previously reported for these three transfected cell lines [26, 27].
Table 1.
List of oligonucleotide primer sequences for the ABC transporters for quantitative real time RT-PCR reactions
| ABC transporter | Position of primer | Forward oligo sequence | Reverse oligo sequence |
|---|---|---|---|
| ABCB1 | 834–1086 | GCCTGGCAGCTGGAAGACAAATAC | ATGGCCAAAATCACAAGGGTTAGC |
| ABCC1 | 1119–1670 | AGTGGAACCCCTCTCTGTTTAAG | CCTGATACGTCTTGGTCTTCATC |
| ABCC4 | 3880–4124 | TGATGAGCCGTATGTTTTGC | CTTCGGAACGGACTTGACAT |
| ABCC5 | 3692–3864 | AGAGGTGACCTTTGAGAACGCA | CTCCAGATAACTCCACCAGACGG |
| ABCC11 | 3025–3560 | CCACGGCCCTGCACAACAAG | GGAATTGCCAAAAGCCACGAACA |
Figure. 1. Characterization of expression of selected ABC transporters in HEK293 transfectants.
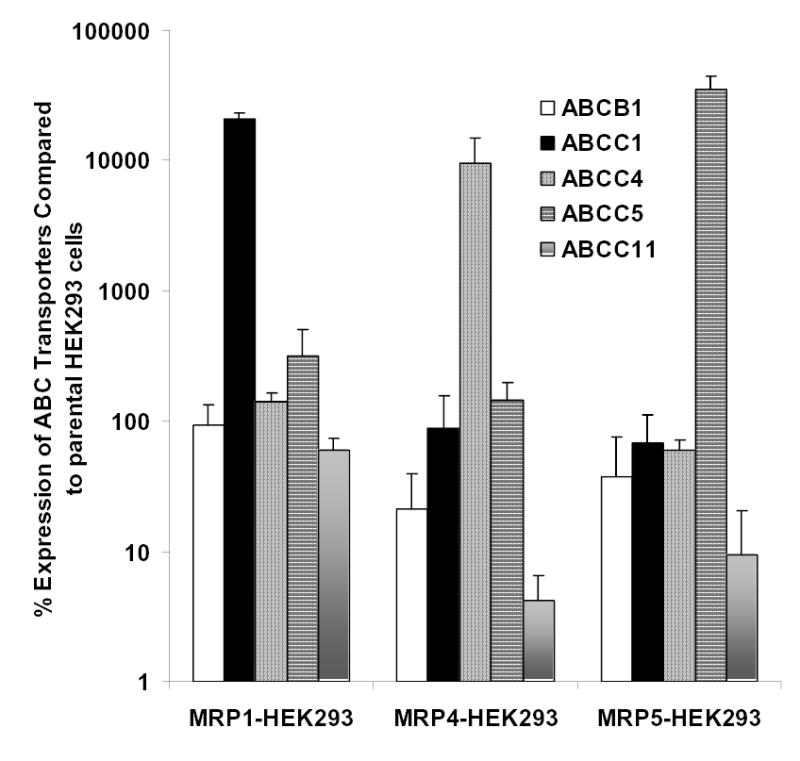
Real time RT-PCR using SYBR green was performed on all of the cell lines utilized in this work. The mRNA expression values for MDR1 (ABCB1), MRP1 (ABCC1), MRP4 (ABCC4), MRP5 (ABCC5), and MRP8 (ABCC11) were determined for each cell line. Following normalization to GAPDH, the expression values for each transfectant was compared to the expression of each transporter within the parental HEK293 cells. The values represent the mean, and the error bars are standard deviation (n=4).
Sensitivities of parental and MRP1-, 4-, and 5-expressing HEK293 cells to plant polyphenols
The relative sensitivities of the parental and the various MRP-expressing HEK293 cell lines to the six plant polyphenols under investigation were determined following exposure for 72 h. IC50 values were calculated from the cell survival curves; these are summarized in Table 2. For each polyphenol tested, the IC50 values for parental and for vector alone transfected-HEK293 cells were similar, with naringenin being the least toxic and resveratrol the most toxic. IC50 values for naringenin, hesperetin, silymarin and daidzein obtained in the MRP1-, MRP4- and MRP5-expressing cells did not differ significantly from those obtained in the parental HEK293 cells. By contrast, in MRP1-expressing cells the IC50 values for quercetin were lower and those for resveratrol were higher; i.e., these cells were more sensitive to quercetin but more resistant to resveratrol than the parental HEK293 cells. In MRP4- and 5-expressing cells, the IC50 values for both quercetin and resveratrol were higher, suggesting both cell types to be more resistant to these polyphenols. Such observations hint at the possibility of these particular polyphenols being expelled from the cells, i.e., being substrates for MRP4 and 5.
Table 2.
Sensitivity of parental and MRP1, 4 and 5 expressing HEK293 cells to selected plant polyphenols.
|
IC50 [μM]a |
|||||
|---|---|---|---|---|---|
| Polyphenols | pcDNA-HEK293 | MRP1-HEK293 (MRP1) | HEK293 | HEK293/4.63 (MRP4) | HEK293/5I (MRP5) |
| Quercetin | 40.9 ± 5.6 | 24.1 ± 5.7 * | 38.6 ± 5.4 | 108.5 ± 20.3 # | 90.9 ± 9.6 # |
| Silymarin | 103.9 ± 35.9 | 152.6 ± 57.5 | 130.6 ± 41.6 | 180.6 ± 70.8 | 143.9 ± 36.7 |
| Daidzein | 84.4 ± 21.3 | 141.6 ± 30.6 | 157.6 ± 48.8 | 161.7 ± 50.4 | 151.5 ± 40.8 |
| Naringenin | 314.4 ± 70.8 | 252.3 ± 55.0 | 266.9 ± 78.4 | 309 ± 86.5 | 338.2 ± 86.8 |
| Hesperetin | 207.9 ± 51.5 | 131.7 ± 19.8 | 200.8 ± 49.3 | 162.4 ± 34.4 | 180.4 ± 34.1 |
| Resveratrol | 16.7 ± 4.8 | 37.7 ± 10.2 * | 17.4 ± 4.6 | 37.1 ± 11.7* | 39.5 ± 10.7 * |
IC50 values are mean ± S.D. in the presence and absence of flavonoids. The IC50 values were calculated from dose response curves obtained from three independent experiments,
p value < 0.05
p value < 0.01
Effect of plant polyphenols on etoposide and vinblastine cytotoxicity in MRP1-HEK293 cells
To investigate whether the polyphenols were able to modify MRP1-mediated resistance, the sensitivity of MRP1-expressing cells to etoposide and to vinblastine, two known MRP1 substrates [7], was evaluated. MRP1-HEK293 cells were found to be approximately 138- and 4-fold more resistant to etoposide (Table 3) and vinblastine (data not shown) respectively than control pcDNA-HEK293 cells. Non-toxic concentrations of each polyphenol were used in combination with increasing concentrations of etoposide to determine the effects of the polyphenols on IC50 values and relative resistance (Table 3). Silymarin, hesperetin, resveratrol, MK-571 and naringenin significantly enhanced the sensitivity of MRP1-HEK293 cells to etoposide in a concentration-dependent manner, though silymarin and MK-571 also enhanced etoposide sensitivity in HEK293 cells (data not shown).
Table 3.
Reversal effect of plant polyphenols on etoposide toxicity in parental pcDNA-HEK293 and MRP1-expressing MRP1-HEK293 cells
|
IC50 [μM]a |
||||
|---|---|---|---|---|
| Drug tested | [Conc.] | pcDNA-HEK293 | MRP1-HEK293 | R.Rb |
| Etoposide alone | - | 0.28 ± 0.07 | 38.8 ± 5.6 | 138.6 |
| + Quercetin | 10 μM | 0.27 ± 0.03 | 55.5 ± 6.8 * | 205.6 |
| + Silymarin | 10 μM | 0.15 ± 0.02 | 35.8 ± 4.3 | 238.7 |
| 20 μM | 0.12 ± 0.03 | 21.7 ± 3.3 * | 180.8 | |
| 50 μM | 0.12 ± 0.03 | 15.4 ± 1.9 # | 128.3 | |
| + Daidzein | 10 μM | 0.26 ± 0.04 | 39.5 ± 4.6 | 151.9 |
| 20 μM | 0.21 ± 0.04 | 45.2 ± 8.1 | 215.2 | |
| + Naringenin | 10 μM | 0.27 ± 0.05 | 36.2 ± 4.0 | 134.1 |
| 20 μM | 0.18 ± 0.03 | 30.2 ± 5.6 | 167.8 | |
| 50 μM | 0.21 ± 0.04 | 22.7 ± 3.8 * | 108.1 | |
| + Hesperetin | 10 μM | 0.25 ± 0.02 | 45.2 ± 6.7 | 180.8 |
| 20 μM | 0.19 ± 0.03 | 24.7 ± 3.8 * | 130.0 | |
| + Resveratrol | 10 μM | 0.32 ± 0.05 | 50.5 ± 3.9 * | 157.8 |
| + MK571 | 50 μM | 0.15 ± 0.03 | 7.9 ± 1.1 # | 52.7 |
IC50 values are mean ± S.D in the presence and absence of flavonoids, which were calculated from dose response curves obtained from three independent experiments
p value < 0.05
p value<0.01.
Relative resistance values were obtained by dividing the IC50 value of the MRP1-HEK293 cells by the IC50 value of the empty vector (pcDNA3.1) transfected cell line.
Effect of polyphenols on MRP4- and MRP5-mediated resistance to thioguanine and NSC251820
To examine the potential of the polyphenols at concentrations below their IC50 values to reverse MRP4- and MRP5- mediated resistance, the sensitivity to thioguanine, a known substrate of MRP4 and MRP5 [5, 22, 24], was first evaluated in MRP4- and MRP5-expressing HEK cells. These cells were revealed to be approximately 4-fold and 3-fold more resistant than parental HEK293 cells, respectively (Fig. 2). These data are comparable to values reported previously [5]. Quercetin, hesperetin, and MK-571 enhanced the sensitivity of MRP4-expressing cells while quercetin, daidzein, naringenin and hesperetin enhanced sensitivity of MRP5 expressing cells toward thioguanine. Silymarin (and/or its metabolites) produced the opposite effect, actually increasing resistance, rather as if it were enhancing thioguanine efflux, perhaps by stimulating transporter activity or by a co-transport mechanism (Table 4).
Figure. 2. Sensitivity of control HEK293 and MRP4- and 5-expressing cells to thioguanine and NSC251820.
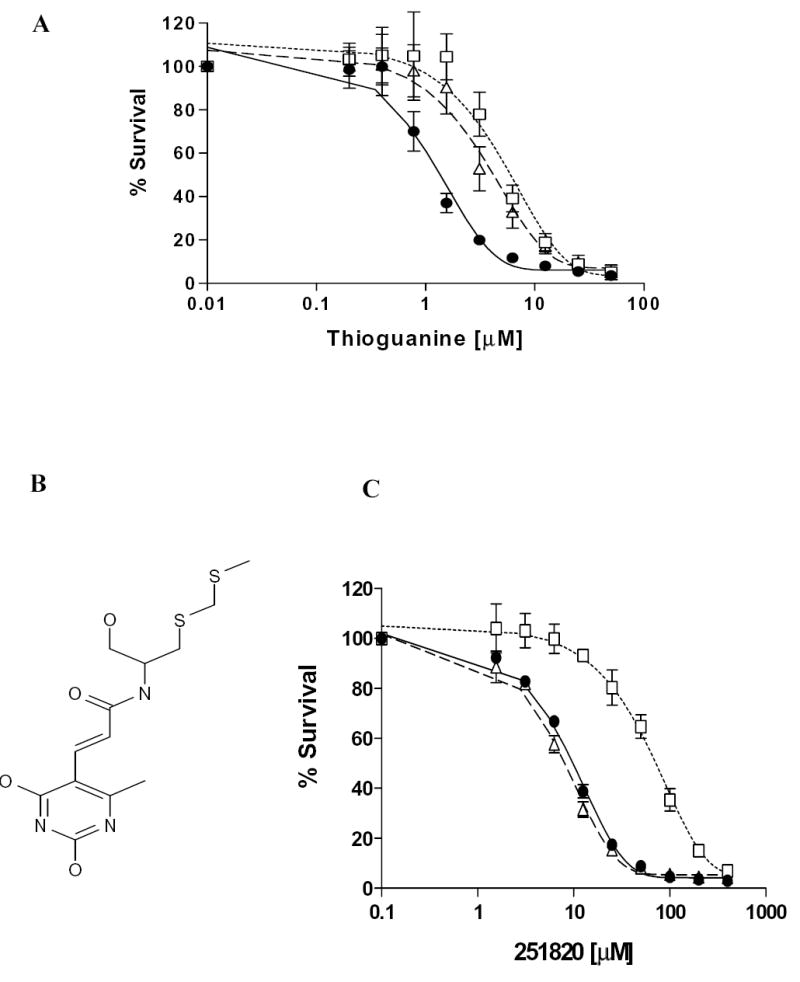
Cytotoxicity assays were used to determine the sensitivity of control HEK293 (filled circles), MRP4-expressing HEK293/4.63 (open squares) and MRP5-expressing HEK293/5I (open triangles) to (A) thioguanine and predicted substrate of MRP4 based on data-mining NSC 251820 (C) as described previously [25]. The structure of NSC251820 is shown in (B). Cells (5.0 x 103 cells) were plated into 96 well plates, cultured overnight and then exposed to thioguanine for 72 h. Viable cells were determined by the Cell Counting Kit (CCK) technique as detailed in the Material and Methods section. The mean values from three independent experiments are shown with error bars as S.D.
Table 4.
Effect of polyphenols on the sensitivities of parental HEK293, MRP4-expressing (HEK293/4.63) and MRP5-expressing (HEK293/5I) HEK293 cells to thioguanine.
|
IC50 ± S.D [μM]a |
||||
|---|---|---|---|---|
| Drug tested | [Conc.] | HEK293 | HEK293/4.63 (MRP4) | HEK293/5I (MRP5) |
| Thioguanine alone | - | 1.1 ± 0.3 | 4.8 ± 1.3 | 3.4 ± 0.7 |
| + Quercetin | 5 μM | 1.0 ± 0.2 | 3.5 ± 1.0 | 2.0 ± 0.2 * |
| 10 μM | 0.8 ± 0.1 | 1.9 ± 0.6 * | 0.9 ± 0.3 # | |
| + Silymarin | 5 μM | 1.1 ± 0.3 | 8.9 ± 3.4 | 3.4 ± 0.7 |
| 10 μM | 0.9 ± 0.2 | 8.9 ± 3.1 | 4.0 ± 1.0 | |
| 20 μM | 1.3 ± 0.3 | 9.4 ± 3.5 | 6.3 ± 1.1 * | |
| 30 μM | 2.0 ± 0.4 | 13.0 ± 3.2 * | 10.7 ± 1.9 # | |
| + Daidzein | 20 μM | 0.9 ± 0.2 | 3.3 ± 0.6 | 1.9 ± 0.2 * |
| + Naringenin | 20 μM | 0.8 ± 0.1 | 2.8 ± 0.4 | 1.8 ± 0.3 * |
| + Hesperetin | 10 μM | 0.9 ± 0.2 | 4.2 ± 0.9 | 3.1 ± 0.5 |
| 20 μM | 0.7 ± 0.1 | 2.5 ± 0.5 * | 1.6 ± 0.2 * | |
| + Resveratrol | 5 μM | 1.2 ± 0.2 | 4.4 ± 0.8 | 2.8 ± 0.4 |
| 10 μM | 1.9 ± 0.4 | 3.3 ± 0.7 | 3.1 ± 0.4 | |
| + MK-571 | 50 μM | 1.2 ± 0.2 | 1.6 ± 0.2 * | 2.5 ± 0.2 |
Values are mean IC50 values ± SD in the presence and absence of flavonoids. The IC50 values were calculated from dose response curves obtained from six independent experiments
p value < 0.05
p value < 0.01.
To study further the effect of polyphenols on MRP4, the sensitivity of MRP4-expressing cells to NSC251820 in the presence of polyphenols was also examined. NSC251820 (Fig. 2B) is a compound that by data mining [25] has been predicted to be a potential MRP4 substrate. MRP4-expressing cells were shown to be highly resistant to this compound (approximately 7.5-fold) compared to their lower resistance to thioguanine (approximately 3-fold). Interestingly, the MRP5-expressing cells did not show resistance to NSC251820 (Fig. 2C), suggesting that NSC251820 and/or its metabolites are not transported by MRP5. All polyphenols tested, apart from daidzein, reduced the Relative resistance values to NSC251820 in MRP4-expressing cells (Table 5), and among these, quercetin, hesperetin, and resveratrol were the most effective.
Table 5.
Effect of polyphenols on the sensitivities of parental HEK293 and MRP4-expressing (HEK293/4.63) HEK293 cells to NSC251820
|
IC50 [μM]a |
||||
|---|---|---|---|---|
| Drug tested | [Conc.] | HEK293 | HEK293/4.63 (MRP4) | R.Rb |
| NSC251820 alone | - | 7.9 ± 1.2 | 58.6 ± 8.8 | 7.4 |
| + Quercetin | 10 μM | 8.3 ± 0.9 | 23.4 ± 4.0 # | 2.8 |
| + Silymarin | 20 μM | 6.7 ± 1.1 | 27.6 ± 3.5 # | 4.1 |
| 50 μM | 3.8 ± 0.6 | 11.5 ± 1.3 + | 3.0 | |
| + Daidzein | 20 μM | 6.7 ± 0.9 | 46.9 ± 6.0 | 7.0 |
| + Naringenin | 20 μM | 5.4 ± 0.7 | 24.8 ± 3.4 # | 4.6 |
| 50 μM | 3.8 ± 0.4 | 15.5 ± 2.2 # | 4.1 | |
| + Hesperetin | 20 μM | 4.3 ± 0.9 | 13.3. ± 1.9 + | 3.1 |
| + Resveratrol | 10 μM | 6.9 ± 1.0 | 20.7 ± 4.1 # | 3.0 |
| + MK-571 | 25 μM | 3.3 ± 0.4 | 9.2 ± 0.7 + | 2.8 |
Values are mean IC50 values ± SD in the presence and absence of flavonoids. The IC50 values were calculated from dose response curves obtained from six independent experiments
p value < 0.05
p value < 0.01
p value < 0.001.
Relative resistance values were obtained by dividing the IC50 value of the MRP1-HEK293 cells by the IC50 value of the empty vector (pcDNA3.1) transfected cell line.
Inhibition of [3H]-DNP-SG and [3H]-cGMP transport in human erythrocytes by polyphenols
Human erythrocytes are known to express not only MRP1, but also MRP4 and MRP5. Inside-out vesicles were prepared from red blood cells and used in uptake experiments to assess the direct inhibitory effects of polyphenols on transport mediated by these MRPs, so avoiding possible interference by potential polyphenol metabolites. It has been shown previously that ATP-dependent transport of high affinity [3H]-DNP-SG in human erythrocyte vesicles is MRP1-mediated and linear for at least 60 minutes; however, ATP-dependent transport of 3.3 μM [3H]-cGMP is most likely to be MRP4-mediated and linear for at least 30 minutes [28]. Concentrations of polyphenols were tested in the range of 0–200 μM. The rate of 3 μM [3H]-DNP-SG uptake was inhibited by all polyphenols tested except daidzein (Fig. 3A), and the rate of 3.3 μM [3H]-cGMP uptake was inhibited by all 6 polyphenols tested (Fig. 3B). In Fig. 3A, the results suggest that a fraction of DNP-SG may be transported by unknown transporters other than MRP 4 or 5 which the polyphenols do not affect. The IC50 values are summarised in Table 6. Apart from silymarin, all polyphenols tested produced inhibitory effects on transport, in general by inhibiting cGMP transport at lower concentrations than those required to block DNP-SG transport. Silymarin, by contrast, inhibited DNP-SG transport with very high affinity compared to cGMP transport (IC50 values 0.26 and 0.91 μM, respectively).
Figure. 3. Plant polyphenols inhibited uptake of [3H]-DNP-SG and [3H]cGMP into membrane vesicles prepared from human erythrocytes.
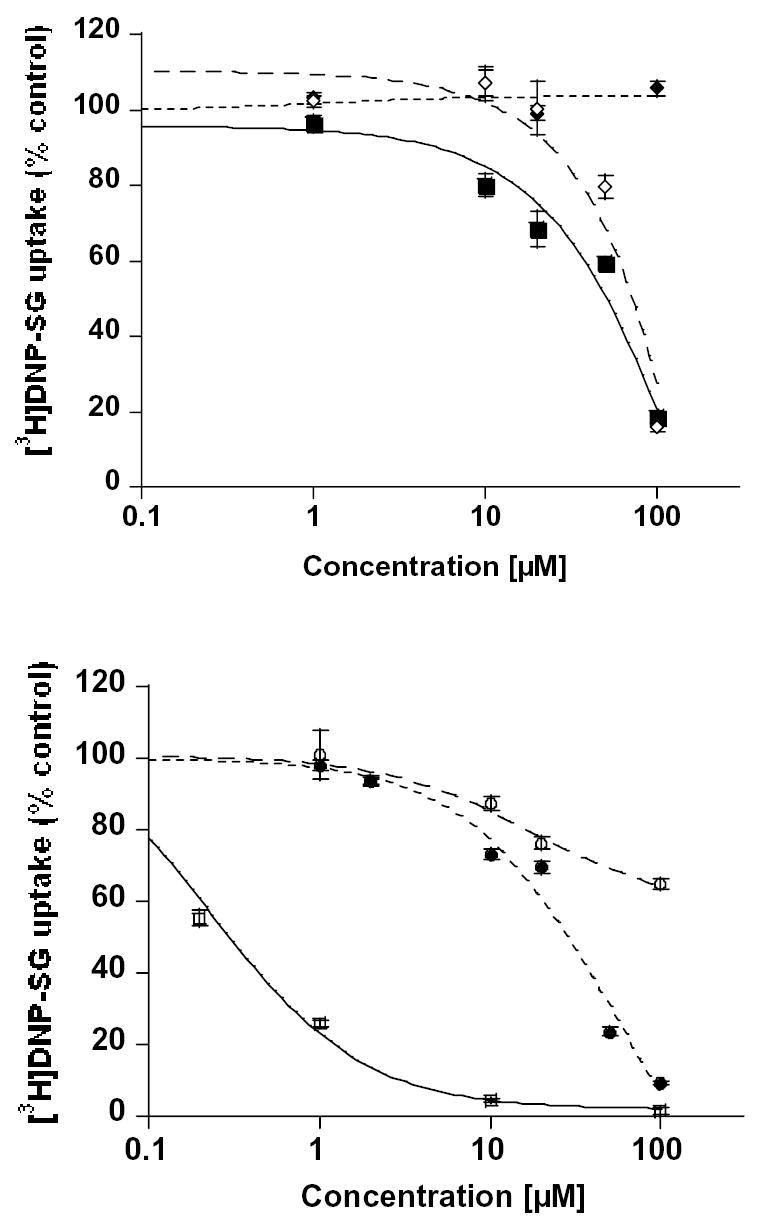
ATP-dependent uptake at 37°C for 30 minutes in erythrocytes membrane vesicles using 3 μM [3H]-DNP-SG or 3.3 μM [3H]-cGMP was carried out as described in Materials and Methods. (A) [3H]-DNP-SG and (B) [3H]-cGMP uptake, quercetin (filled squares), hesperetin (open diamonds), daidzein (filled diamonds), silymarin (open squares), resveratrol (open circle) and narigenin (filled circle). The mean values from six independent experiments are shown with error bars as S.E.M.
Table 6.
Effect of plant polyphenols on MRP-mediated transport in membrane vesicles prepared from human erythrocytes
| Polyphenol | MRP1-mediated DNP-SG transportaIC50[μM]b | MRP4-mediated cGMP transportaIC50[μM]b |
|---|---|---|
| Quercetin | 45.12 ± 11.93 | 1.16 ± 0.17 |
| Silymarin | 0.26 ± 0.06 | 0.91 ± 0.11 |
| Naringenin | 23.70 ± 5.99 | 3.37 ± 0.28 |
| Hesperetin | 70.18 ± 40.24 | 2.46 ± 0.14 |
| Resveratrol | 169.9 ± 53.0
17.12 ± 6.15 (58.3 ± 4.6 % UI) |
1.66 ± 0.14 |
| Daidzein | No effect | 9.67 ± 1.55 |
The transport of [3H]DNP-SG and [3H]cGMP in inside-out membrane vesicles of human erythrocytes was determined in the presence and absence of indicated polyphenols as described in the experimental procedures.
IC50 values are mean ± S.D in the presence and absence of flavonoids. The IC50 values were calculated from dose response curves obtained from three independent experiments.
Effect of polyphenols on fluorescent substrate accumulation and MRP-mediated efflux
The effects of polyphenols on efflux of fluorescent substrates from MRP-expressing cells were analysed using flow cytometry, where levels of accumulation in control and MRP-expressing HEK293 cells were assessed in the absence or presence of polyphenols. Cells (5x105) were incubated with non-fluorescent precursors, and the intensity of the fluorescence of accumulated substrates was then analysed by FACS. Calcein-AM which becomes hydrolysed to the fluorescent MRP1 substrate calcein, was used to measure MRP1-mediated transport whilst BCECF-AM, which is hydrolysed to the fluorescent MRP5 substrate BCECF [29] was used to study MRP5-mediated transport. The results of the 50 μM polyphenol treatments are shown in Fig. 4 and 5, respectively. Quercetin and silymarin dramatically increased the accumulation of the fluorescent substrates in both MRP1- and MRP5-expressing cells in a concentration-dependent manner (data not shown), with concentrations needed to achieve 50% of the maximum inhibitable portions of between 50–75 μM for MRP1, and 25–50 μM for MRP5, respectively. By contrast, hesperetin, resveratrol, daidzein, and naringenin at concentrations up to 50 μM had no significant effects on MRP1 substrate accumulation (Fig. 4), but a small effect on MRP5 substrate accumulation (Fig. 5). The LTD4 antagonist MK-571 (25 μM) completely blocked MRP1-mediated calcein efflux (Fig. 4B), while only having a moderate effect on MRP5-mediated BCECF efflux (Fig. 5B).
Figure. 4. Effect of selected polyphenols on calcein accumulation in MRP1-HEK293 cells.
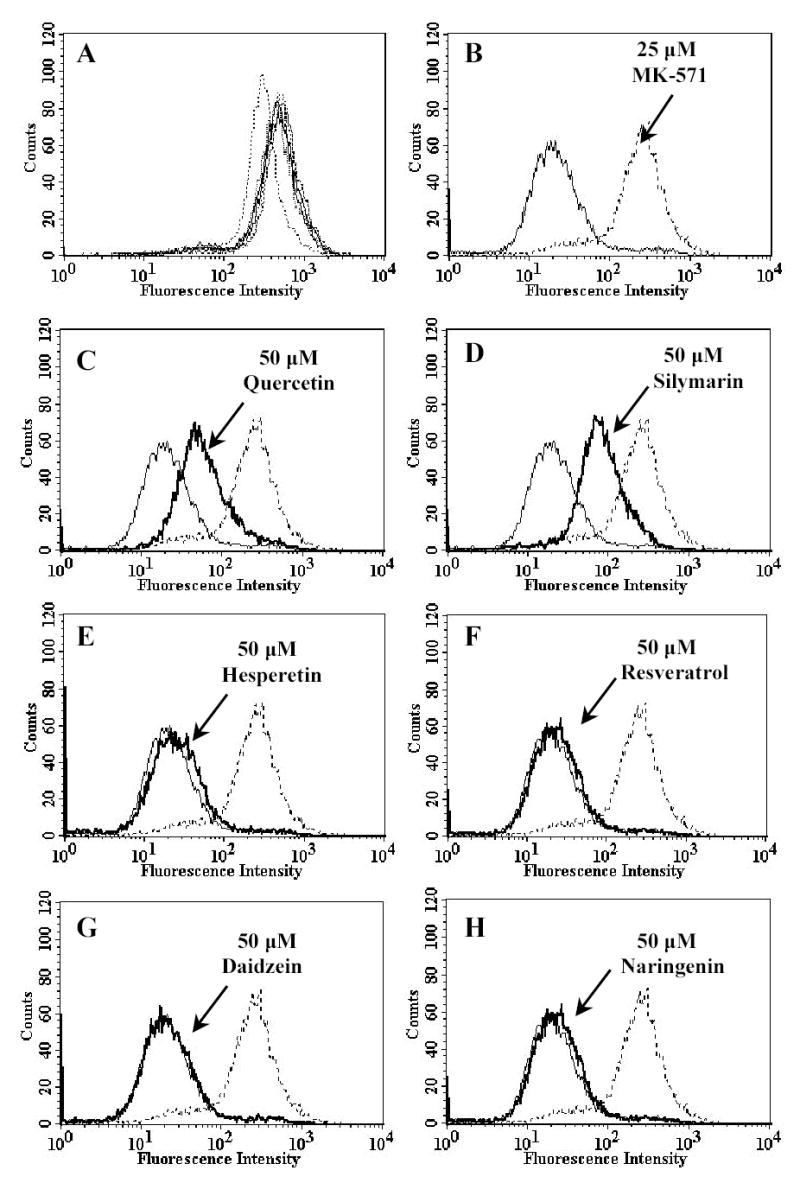
Cells (control pcDNA-HEK293 and MRP1-transfected MRP1-HEK293) were resuspended in IMDM supplemented with 5% FBS. 0.25 μM calcein-AM was added to 3x105 cells in 4 ml of IMDM in the presence or absence of MK-571 and polyphenols. The cells were incubated at 37°C in the dark for 10 minutes. The cells were pelleted by centrifugation at 500 x g and resuspended in 300 μl of PBS containing 0.1% BSA. Samples were analysed immediately by using flow cytometry. Panel A shows that except 50 μM of silymarin (dotted line), MK571 and other polyphenols had no effect on control HEK293 cells. Panel B-H, thin solid line represent MRP1-overexpressing MRP1-HEK293 cells, dotted line represents MRP1-HEK293 cells in the presence of 25 μM of MK-571 and bold solid line represents MRP1-HEK293 cells in the presence of various polyphenols. Panel B: 25 μM of MK-571, Panel C: 50 μM quercetin, Panel D: 50 μM silymarin, Panel E: 50 μM hesperetin, Panel F: 50 μM resveratrol, Panel G: 50 μM daidzein and Panel H: 50 μM naringenin. Representative histograms of three independent experiments are shown.
Figure. 5. Effect of various polyphenols on BCECF accumulation and MRP5-HEK293 cells.
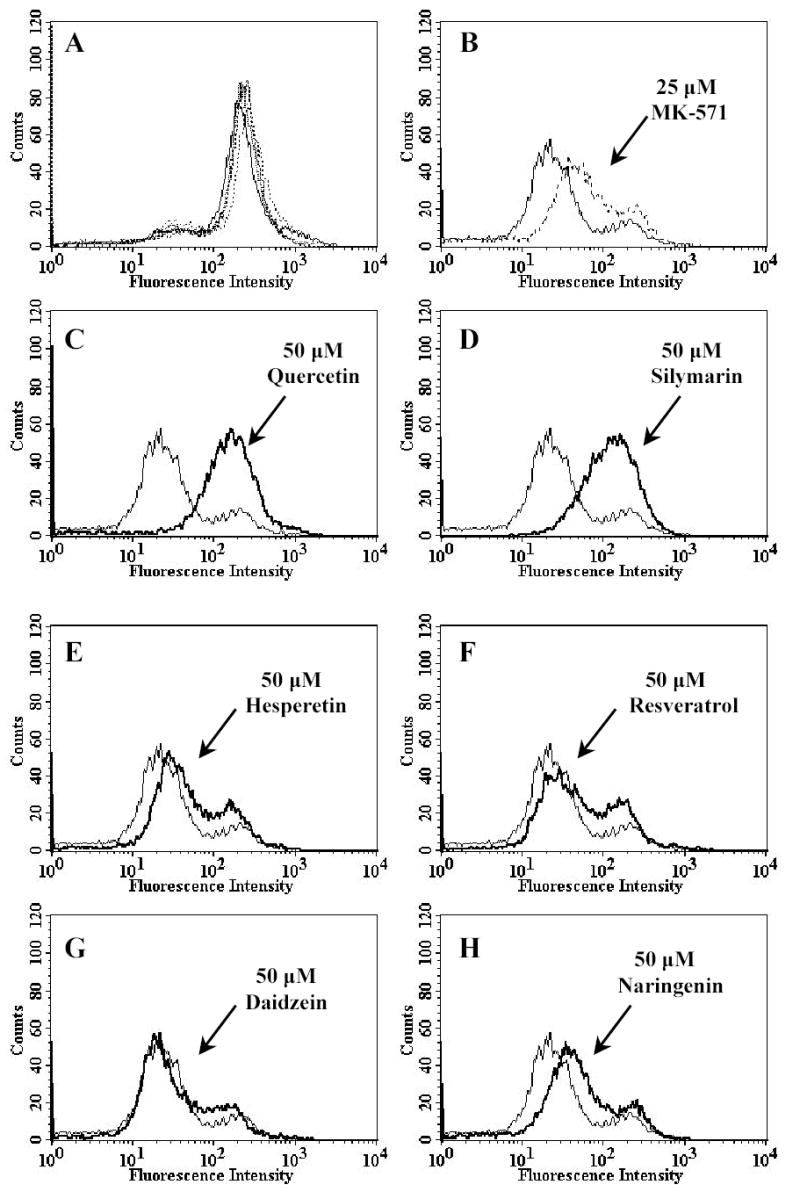
Cells (control HEK293 and MRP5-transfected HEK293/5I) were resuspended in IMDM supplemented with 5% FBS. 0.25 μM BCECF-AM was added to 3x105 cells in 4 ml of IMDM in the presence or absence of MK-571 and polyphenols. The cells were incubated at 37°C in the dark for 10 min and pelleted by centrifugation at 500 x g and resuspended in 300 μl of PBS containing 0.1% BSA. Samples were analysed immediately by flow cytometry. Panel A shows that all polyphenols and MK-571 had no effect on control HEK293 cells. Panel B-H, thin solid line and bold solid line represent MRP5-overexpressing HEK293/5I cells in the absence and presence of drugs tested, respectively. Panel B: 25 μM of MK-571 (dotted line), Panel C: 50 μM quercetin, Panel D: 50 μM silymarin, Panel E: 50 μM hesperetin, Panel F: 50 μM resveratrol, Panel G: 50 μM daidzein and Panel H: 50 μM naringenin. Representative histograms of three independent experiments are shown.
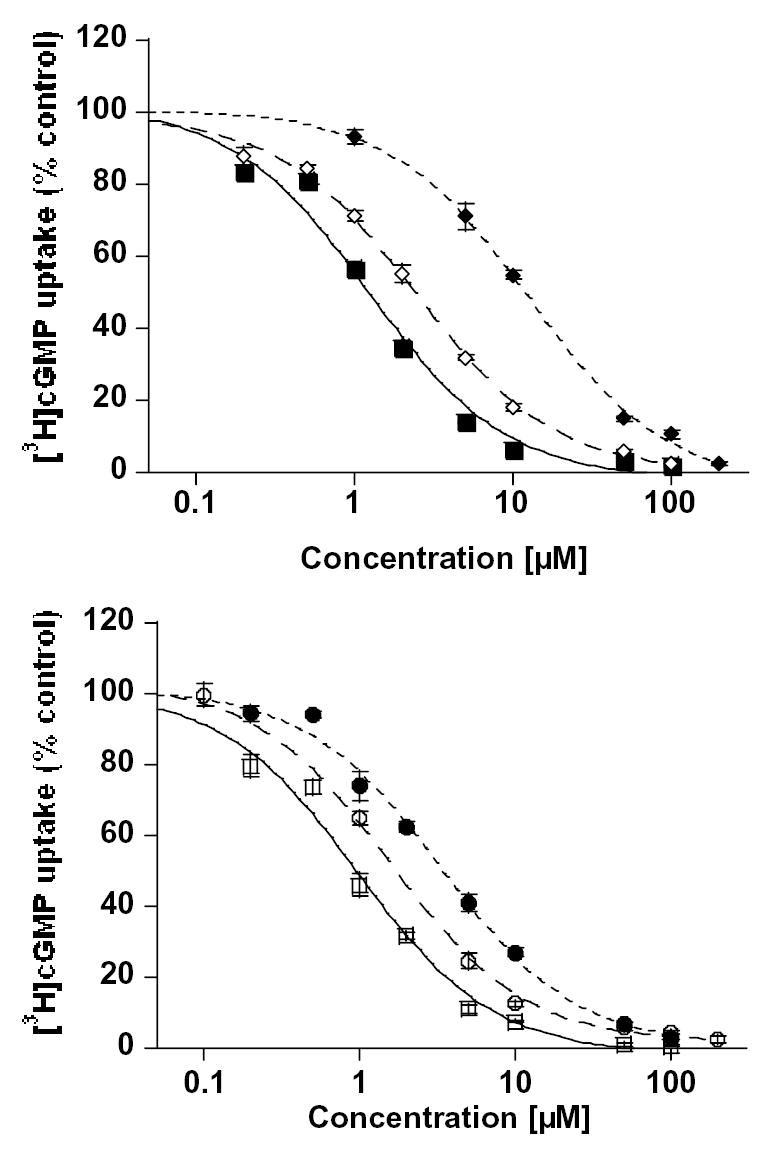
Effect of polyphenols on MRP1- and MRP4-mediated ATP hydrolysis
The effects of the polyphenols on the MRP1- and MRP4-mediated ATP hydrolysis were also examined (results summarized in Table 7). Hesperetin, naringenin, daidzein, and resveratrol had moderate effects on the ATPase activities of both MRPs. Plant polyphenols exerted maximum stimulation on MRP1-mediated ATPase activity at 100 μM for hesperetin (15%), 50 μM for naringenin (7%), 5 μM for daidzein (35%), and 30 μM for resveratrol (49%) (Fig. 6). On the other hand, flavonoids had maximum stimulation on MRP4-mediated ATPase activity at various concentrations; 20 μM for hesperetin (33%), 10 μM for naringenin (9%), 200 μM for daidzein (34%), and 23 μM for resveratrol (10%) (Fig. 7A). Quercetin had a biphasic effect on both MRP1- and MRP4-mediated ATP hydrolysis, which indicates that it stimulated ATPase activity at low concentrations, whereas it inhibited the activity at higher concentrations. The stimulatory effect suggests that quercetin is likely to be a substrate of both MRP1 and MRP4. Quercetin had maximum stimulation at 10 μM for MRP1 of 101% and 61% for MRP4, and it had maximum inhibitory effects of 25% for MRP1 at 100 μM and 55% for MRP4 at 200 μM. Conversely, silymarin inhibited both MRP1 (60% at 100 μM) and MRP4 (72% at 200 μM) ATPase activity. To assess whether polyphenols affect ATPase activity by interacting at the substrate site, we tested the effect of quercetin and silymarin on substrate-stimulated ATP hydrolysis by MRP4. Both quercetin and silymarin were able to inhibit PGE1 stimulated MRP4-mediated ATP hydrolysis (Fig. 7B). PGE1 has been shown to be a MRP4 substrate that stimulates its ATPase activity [24, 30]. These results suggested that quercetin and silymarin do interact at the same MRP4 substrate-binding sites as PGE1. Quercetin inhibited 95% of the stimulated MRP4 ATPase activity, and silymarin inhibited 62% of this activity.
Table 7.
Effect of polyphenols on the beryllium-fluoride-sensitive ATPase activity measured in crude membranes prepared from High Five insect cells expressing human MRP1 or MRP4.
| Drug | Concentration tested [μM] | Effect on basal ATPase activity | Maximum inhibition or stimulation (%) | na |
|---|---|---|---|---|
| MRP1 | ||||
| Quercetin | 5–100 | Stimulation | 86 | 4 |
| Inhibition | 22 | |||
| Quercetin + GSH* | 2–100 | Stimulation | 19 | 4 |
| Inhibition | 72 | |||
| Silymarin ± GSH | 5–100 | Inhibition | 60 | 3 |
| Hesperetin ± GSH | 5–100 | No effect | - | 3 |
| Daidzein ± GSH | 5–100 | No effect | - | 3 |
| Naringenin ± GSH | 5–100 | No effect | - | 6 |
| Resveratrol ± GSH | 5–100 | Stimulation | 49 | 3 |
| Misc. | ||||
| Reduced GSH | 3000 | Stimulation | 68 | 3 |
| Methotrexate | 5–100 | No effect | - | 3 |
| Folinic acid | 5–100 | No effect | - | 3 |
| Verapamil | 5–100 | Stimulation | 35 | 4 |
| Sodium arsenite (± GSH) | 1–500 | No effect | - | 3 |
| MRP4 | ||||
| Quercetin | 1–200 | Stimulation | 61 | 4 |
| Inhibition | 55 | |||
| Silymarin | 1–200 | Inhibition | 72 | 6 |
| Hesperetin | 1–200 | No effect | - | 6 |
| Daidzein | 1–200 | Stimulation | 34 | 6 |
| Naringenin | 1–200 | No effect | - | 5 |
| Resveratrol | 1–200 | Stimulation | 23 | 4 |
| Quercetin + PGE1 | 1–200 | Inhibition | 93 | 3 |
| Silymarin + PGE1 | 1–200 | Inhibition | 62 | 3 |
| Daidzein + PGE1 | 1–100 | No effect | - | 4 |
| Hesperetin + PGE1 | 1–200 | No effect | - | 3 |
| Naringenin + PGE1 | 1–100 | No effect | - | 3 |
| Resveratrol + PGE1 | 1–100 | Inhibition | 25 | 3 |
| Misc. | ||||
| PGE1 | 1–200 | Stimulation | 66 | 6 |
| DHEAS | 1–200 | Stimulation | 42 | 7 |
| GSH | 1–5 | No effect | - | 3 |
| Ibuprofen | 1–100 | No effect | - | 3 |
| Topotecan | 1–100 | No effect | - | 3 |
| Dipyridamole | 1–100 | Inhibition | 19 | 4 |
3 mM of reduced glutathione (GSH) was used where indicated
The mean values were calculated from at least three independent experiments
Figure. 6. Effect of various polyphenols on MRP1-mediated ATP hydrolysis.
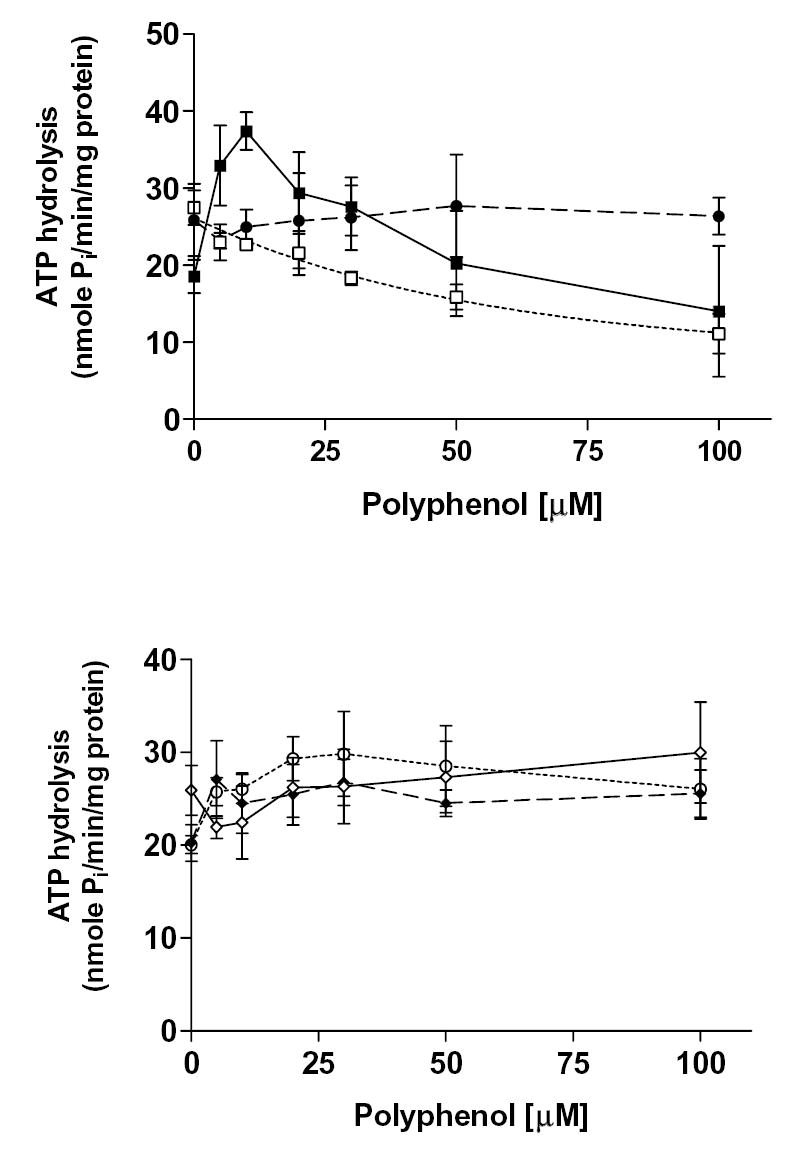
Crude membranes of MRP1 baculovirus infected High Five insect cells (100 μg/ml protein) were incubated at 37°C for 5 minutes with polyphenols in the presence and absence of BeFx. The reaction was initiated by addition of 5 mM ATP and terminated with SDS (2.5% final concentration) after 20 min incubation at 37°C. The amount of Pi released was quantitated using a colorimetric method [30, 34]. MRP1-specific activity was recorded as the BeFx-sensitive ATPase activity. Top panel: quercetin (filled squares), silymarin (open squares) and naringenin (filled circle); Bottom panel: hesperetin (open diamonds), daidzein (filled diamonds) and resveratrol (open circle). Values represent mean ± S.D from at least three independent experiments.
Figure. 7. Effect of various bioflavonoids on basal and PGE1-stimulated MRP4 ATPase activity.
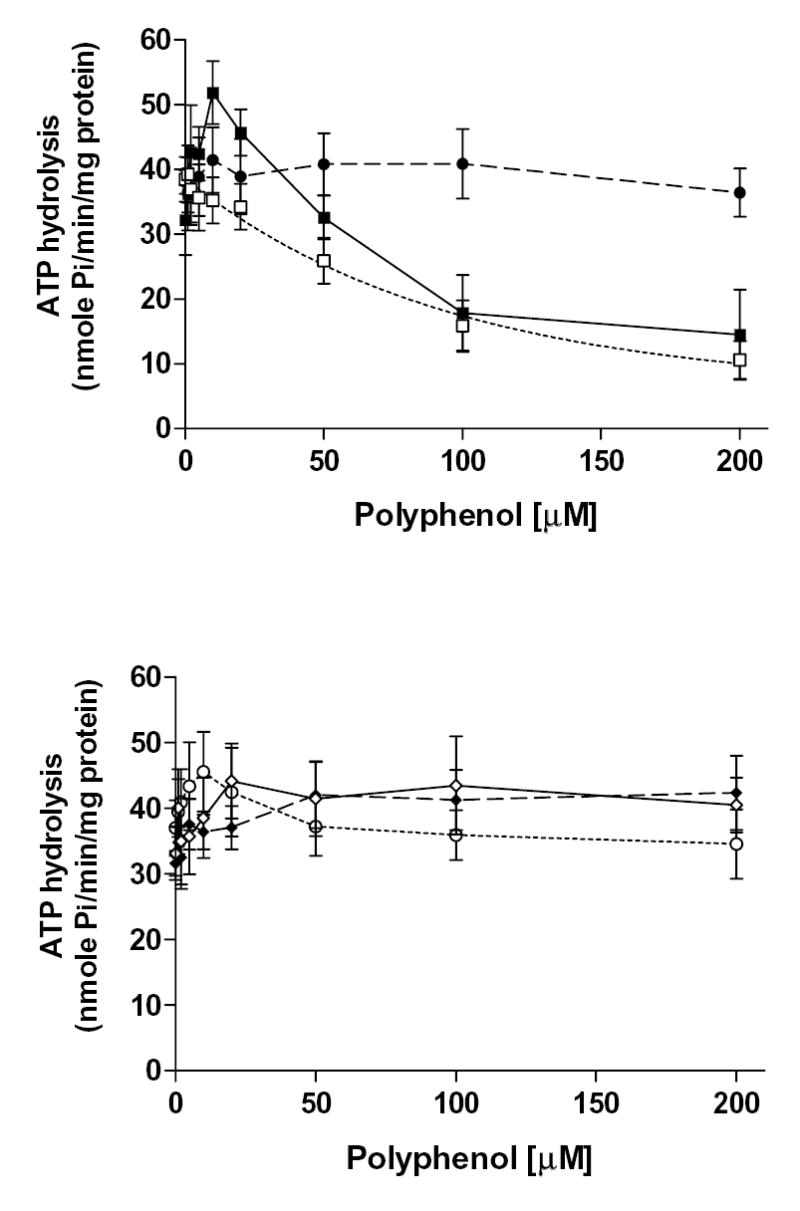
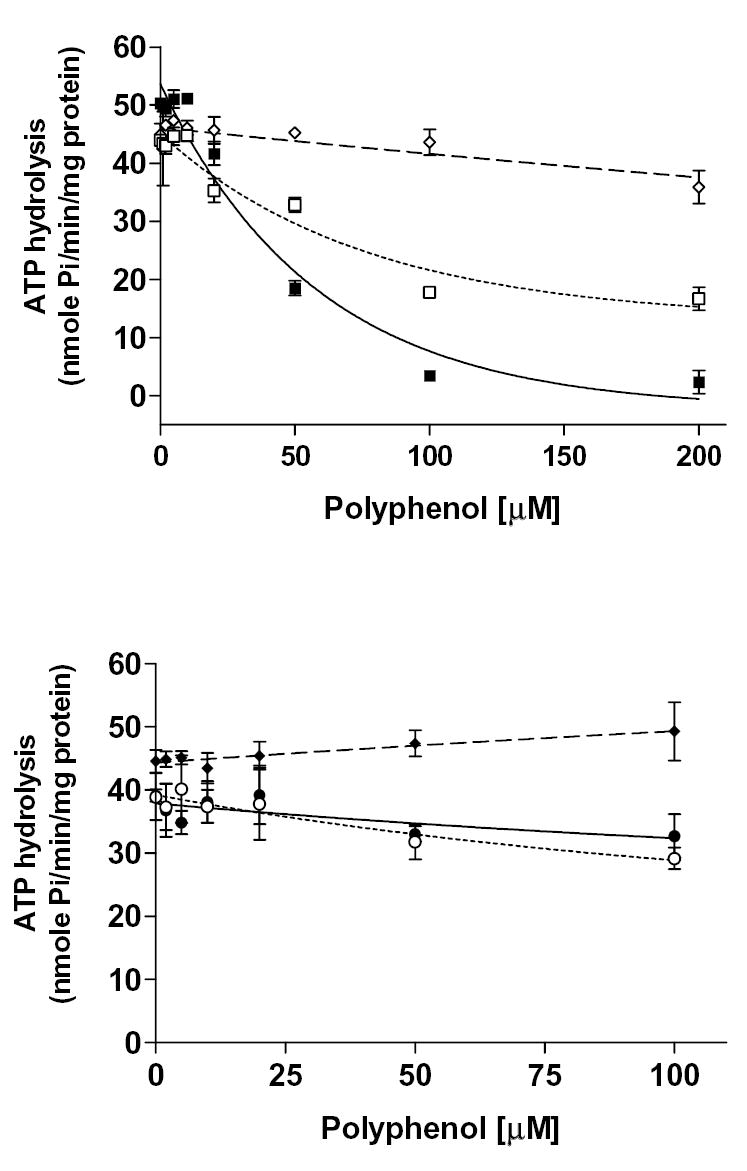
(A) Crude membranes of MRP4 baculovirus infected High Five insect cells (100 μg protein/ml) were incubated at 37°C for 5 min with polyphenols in the presence and absence of BeFx. The reaction was initiated by addition of 5 mM ATP and terminated with SDS (2.5% final concentration) after 20 min incubation at 37°C. The MRP4-specific activity was determined as described in the legend to Fig. 7. (B) MRP4 substrate PGE1 stimulates MRP4 ATPase activity [30]. Briefly, crude membranes were incubated with 20 μM of PGE1 in the absence or presence of polyphenols at indicated concentrations, and the ATPase assay was carried out as described above. In A and B panels: quercetin (filled squares), silymarin (open squares), naringenin (filled circle), hesperetin (open diamonds), daidzein (filled diamonds), and, resveratrol (open circle). Values represent mean ± S.D from at least three independent experiments.
Effects of quercetin and silymarin on photoaffinity labelling of MRP1 and MRP4 with [α-32P] 8-azidoATP
To determine whether silymarin and quercetin bind to nucleotide (ATP) binding sites on MRP1 and MRP4 (thus inhibiting ATPase activity), the effects of these two flavonoids on the photoaffinity labelling of MRP1 and MRP4 with [α-32P]8-azidoATP were examined [18]. The 8-azidoATP, an analog of ATP, has been shown previously to bind specifically to the nucleotide binding domain of Pgp and MRPs [30, 31]. At tested concentrations (10, 50 and 100 μM), neither quercetin nor silymarin had any effect on [α-32P]8-azidoATP labelling (Fig. 8). This suggests that these flavonoids more likely bind to the transport-substrate binding site(s) rather than the nucleotide binding sites to cause inhibition of the ATP hydrolysis. Note that lane 9 in panel A and B of Fig. 8 represents the displacement of the [α-32P]8-azidoATP labelling by the presence of excess ATP (10 mM), as expected.
Figure. 8. Quercetin and silymarin do not inhibit photoaffinity labelling of MRP1 or MRP4 with [α-32P]8-azidoATP.
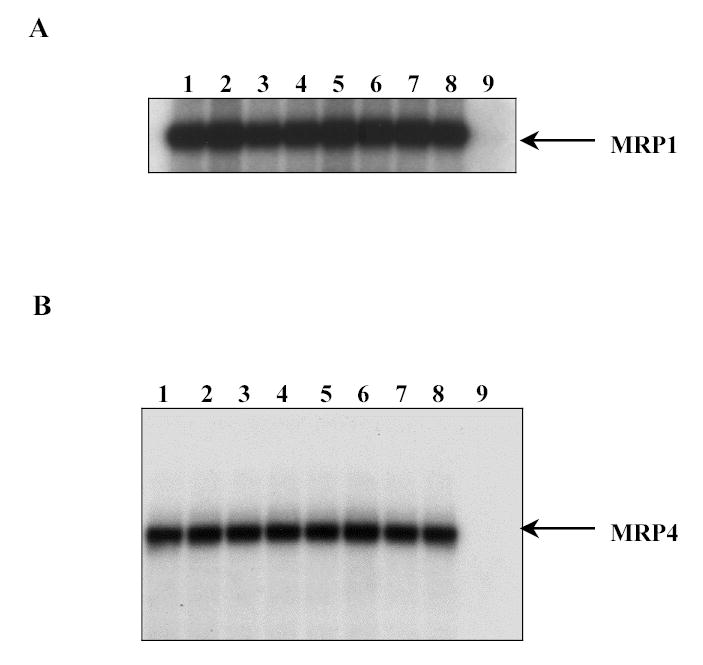
Crude membranes (50–75 μg protein) of MRP1 or MRP4 baculovirus infected High Five insect cells were incubated at 4°C for 5 minutes with 10 μM [α-32P]8-azidoATP (10 μCi/nmole) in the presence and absence of quercetin or silymarin. The photocrosslinking with 365 nm UV light was carried out on ice for 10 minutes as described previously [30]. Incorporation of [α-32P]8-azidoATP detected by phosphorimaging and by exposure to X-ray film at −70C for 2-8 h after gel electrophoresis. A, Photolabeling of MRP1 and B, MRP4, respectively. In both panels, lane 1 and 5, membranes exposed to [α-32P]8-azidoATP alone, lanes 2, 3 and 4 membranes treated with 10 nM, 50 μM and 100 nM quercetin, respectively and lanes 6, 7 and 8, with 10 nM, 50 μM and 100 nM silymarin, respectively. Lane 9, crude membranes were incubated with 10 μM [α-32P]8-azidoATP in the presence of 10 mM ATP-Mg2+.
Discussion
The present study was undertaken to determine whether six of the most abundant plant polyphenols found in commonly consumed foods have modulatory effects on MRP1-, 4- and 5-mediated transport. Some of these compounds have already been shown to interact with other ABC transporters, e.g. Pgp, MRP1 and ABCG2 [15, 17–20].
The transfected cell lines used in the study were first characterized by real time RT-PCR to confirm that only the MRPs of interest and no other ABC drug transporters with similar substrate specificities were expressed at a significant level. This has allowed us to study the effect of flavonoids on a given transporter without any detectable contribution by other ABC transporters. Sensitivities to the polyphenols were assessed using cell survival assays. These revealed variation between cell types, the MRP1- overexpressing cells being more resistant than the untransfected HEK293 cells to silymarin and resveratrol whereas MRP4- and MRP5-overexpressing cells being more resistant than untransfected HEK293 cells to quercetin, silymarin, naringenin, and resveratrol (Table 2). Such data suggest that these particular polyphenols might be substrates for the MRPs.
Non-toxic concentrations of the polyphenols were chosen to investigate their potential in reversing MRP-mediated drug resistance. MRP1-expressing HEK293 cells are known to be highly resistant to etoposide [26]. In the present study, it was seen that silymarin, naringenin, and hesperetin could reduce this resistance in these cells by enhancing sensitivity to etoposide in a concentration-dependent manner, with silymarin being the most potent (Table 3). Similar results were also obtained when vinblastine was used as the cytotoxic agent (data not shown). MRP4- and MRP5-expressing cells are known to show resistance to the chemotherapeutic agent, thioguanine [22], and in the present study resistance factors of 4.4 and of 3 for MRP4-expressing HEK293/4.6 and MRP5-expressing HEK293/5I, respectively, were obtained (Fig. 2A and Table 4). These values are comparable to values reported previously [22]. The polyphenols quercetin and hesperetin significantly enhanced the sensitivity towards thioguanine in MRP4-expressing cells while quercetin, daidzein, naringenin and hesperetin did so in MRP5-expressing cells, though resveratrol had only a moderate effect (Table 4). Interestingly, silymarin had the opposite effect, reducing the toxicity of thioguanine in MRP4- and MRP5-expressing cells. This may indicate that silymarin in some way is able to enhance efflux of thioguanine. It is however possible that other action(s), unconnected with efflux, could account for such an effect. This requires further investigation in the future to clarify.
The effect of polyphenols on resistance of MRP4- and MRP5-expressing cells to another putative substrate, NSC251820, was also examined. This compound though predicted to be a substrate for MRP4 has never been shown experimentally to be so [25]. Results in the present study suggest very strongly that NSC251820 may indeed be a good MRP4 substrate since the MRP4-expressing cells but not the MRP5-expressing cells were more resistant to this compound than the untransfected HEK293 cells (see Fig. 2 and Table 5). Sensitivity of MRP4-expressing cells to NSC251820 was significantly restored by relatively low concentration of polyphenols (see Table 5).
To obtain more direct evidence of flavonoid interactions with MRP-mediated transport, studies were conducted to examine their effects on uptake of the MRP1 substrate, DNP-SG and the MRP4 substrate, cGMP into inside-out vesicles prepared from human erythrocyte membranes. All 6 polyphenols showed high potencies and comparable IC50 values for inhibition of MRP4-mediated cGMP uptake whereas they were of limited potency against MRP1 (Table 6).
Data from flow cytometry, which assessed the effects of polyphenols on the accumulation of fluorescent substrates into intact cells provided further support for interactions between the polyphenols and MRPs. Silymarin and quercetin were the best inhibitors for both MRP1- and MRP5-mediated efflux. Naringenin, hesperetin, resveratrol, and daidzein at 50 μM had moderate to no effect on MRP1- and MRP5-mediated efflux (Fig. 4 and 5). No flow cytometry studies were performed on MRP4-expressing cells since no suitable fluorescent MRP4 substrate could be identified. Several fluorescent compounds, including calcein-AM, BCECF-AM, Fluo4-AM and Alexa Fluor 647-cAMP, were tested but none showed any differences in accumulation between untransfected cells and MRP4-overexpressing cells suggesting that none were being effluxed preferentially by MRP4 (data not shown).
ATPase assays showed that the polyphenols other than quercetin and silymarin moderately stimulated MRP1- and MRP4-mediated ATP hydrolysis (for summary see Table 7). This confirmed the interactions between the polyphenols and MRP1 or MRP4 since ATP hydrolysis and transport are closely linked [32]. Exposure to drug substrates can lead to stimulation or inhibition of ATPase activity of the ABC transporters [33, 34]. Quercetin had a biphasic effect on the ATPase activity of MRP1 and MRP4. ATPase activities were stimulated at lower concentrations but inhibited at higher concentrations. Silymarin, on the other hand, significantly inhibited MRP1 and MRP4 ATPase activity. It was previously shown that a known MRP4 substrate PGE1 strongly stimulated MRP4 ATP hydrolysis [30]. Quercetin and silymarin were able to completely inhibit PGE1-stimulated MRP4 ATPase activity, which suggested that most likely PGE1, quercetin and silymarin shared the same MRP4 substrate binding pocket(s). Hesperetin, naringenin, and resveratrol only partially inhibited the PGE1-stimulated MRP4 ATP hydrolysis, whereas daidzein had no effect.
Previous studies suggested that a silybin analog [18] can bind to ATP binding sites of MRP1 thus affecting hydrolysis of ATP. Since silybin is a major component of silymarin, we examined whether silymarin or quercetin binds to ATP sites of MRP1 and MRP4 by using photoaffinity analog of ATP, [α-32P]8-azidoATP. At the concentrations that cause stimulation and inhibition of ATPase activity, both quercetin and silymarin were unable to affect crosslinking of [α-32P]8-azidoATP to either MRP1 or MRP4 (Fig. 8). This provided further evidence that changes in MRP1 and MRP4 ATP hydrolysis were caused by polyphenols possibly binding to the substrate binding sites and not the nucleotide binding sites.
The fact that quercetin inhibited MRP1-, MRP4-, and MRP5-mediated transport in inside-out vesicle uptake and flow cytometry studies and stimulated MRP1 and MRP4 ATPase activity, suggests that quercetin is an MRP substrate. However, in a long-term (72 h) cell toxicity assay, resistance to quercetin was not observed in MRP1-expressing cells. This suggests that quercetin or some of its metabolites might be better substrates for MRP4 and MRP5 than MRP1. Silymarin, conversely, behaves as a typical inhibitor in short term uptake assays, flow cytometry studies, and ATPase assays, and it significantly inhibited the MRP1 and MRP4 ATPase activity. In 72 h cell toxicity assays, silymarin metabolites were formed and are likely to be substrates of MRP1, MRP4 and MRP5. It is clear that additional work on the effect of metabolites of flavonoid needs to be carried out.
In conclusion, our results indicate that polyphenols interact directly with MRP1, MRP4, and MRP5, and that some of them i.e., quercetin and silymarin, may well prove to be substrates for MRPs. They modulate both transport function and ATPase activities of MRP1 and MRP4. Despite the fact that these polyphenols belong to the same class of compounds and are structurally similar, they all have unique properties and should be studied individually. Given the amounts of polyphenols ingested daily, it is likely that the transporters in vivo would be exposed to relatively high concentrations and become susceptible to modulation of both function and expression. This in turn could influence bioavailability, distribution and transport of various dietary toxins and chemotherapeutics handled by these transporters. Understanding the interactions of these polyphenols with MRPs may be useful for improving the efficacy of anticancer as well as antiviral drug therapies.
Materials and Methods
Chemicals
[Glycine-2-3H]-glutathione (1.9 TBq/mmol) and [8-3H]-guanosine-3′, 5′-cyclic phosphate, NH4 salt (cGMP) (0.559 TBq/mmol) were purchased from Perkin-Elmer LAS Ltd (Boston, MA) and Amersham Biosciences (Piscataway, NJ), respectively. ATP, ATP-γ-S, 1-chloro-2, 4-dinitrobenzene (cDNB), creatine phosphokinase, creatine kinase, 3′, 5′-cyclic guanosine monophosphate (cGMP), doxorubicin, quercetin, silymarin, hesperetin, daidzein, resveratrol, naringenin, reduced glutathione (GSH) and glutathione S-transferase were all obtained from Sigma Chemicals (Poole, Dorset, UK). Acetoxy-methyl esters of calcein (calcein-AM) and of BCECF (BCECF-AM) were purchased from Molecular Probes (Eugene, OR). GSH stock solutions were freshly prepared on the day of each experiment. [α-32P] 8-azidoATP (15-20 Ci/mmole) and 8-azido ATP were obtained from Affinity Labelling Technologies, Inc. (Lexington, KY). Dulbecco’s modified Eagle’s medium (DMEM), Iscove’s modified Dulbecco’s medium (IMDM), L-glutamine, and penicillin/streptomycin were obtained from Invitrogen (Carlsbad, CA). Cell Counting Kit-8 was purchased from Dojindo Molecular Technologies, Inc. (Gaithersburg, MD). [3H]-DNP-SG was synthesised enzymatically as previously described [35, 36]. NSC251820 compound was obtained from the drug synthesis and chemistry branch, DCTD, NCI.
Cell Lines
Parental HEK293 cells (293 Human Embryonic Kidney cells), HEK293/5I cells transduced with MRP5, and the MRP4-overexpressing HEK293/4.63 cells [27, 37] were generous gifts of P. Borst (Division of Molecular Biology and Centre for Biomedical Genetics, The Netherlands Cancer Institute, Amsterdam, The Netherlands). HEK293/4.63 and HEK293/5I cells were reported to express significantly more MRP4 and MRP5, respectively [5]. Parental HEK293 cells and all transfectants were grown in DMEM, supplemented with 10% fetal calf serum and 100 units of penicillin/streptomycin per ml (Invitrogen, Carlsbad, CA), at 37°C in 5% CO2 humidified air. 80 μg/ml of G418 was added to the MRP1-HEK293 cell culture medium [26].
RNA Isolation
RNA was isolated from cells grown in 6-well plates to characterize ABC transporter expression in parental HEK293 cells and all transfected HEK293 cell lines. The medium and any detached cells were first removed from the wells, and RNA isolation was performed on the cells that remained attached using the Qiagen Rneasy kit (Valencia, CA), as per the manufacturer’s protocol. RNA samples were isolated in duplicate. The pure RNA was quantified using a spectrophotometer. The integrity of the RNA was verified using the Agilent 2100 Bioanalyzer (Palo Alto, CA) with the Eukaryote Total RNA assay. The RNA samples were stored at −80°C until needed.
Quantitative RT-PCR
Real-time quantitative RT-PCR was used to measure the mRNA expression levels of the selected ABC transporters. The LightCycler RNA Master SYBR Green Kit and LightCycler machine (Roche Biochemicals, Indianapolis, IN) were utilized in these studies. Specific PCR primer sequences for all ABC transporters except for ABCC5 were generously provided by G. Szakacs and M. Gottesman (Laboratory of Cell Biology, National Cancer Institute, NIH, Bethesda, MD) [25]. Primers for ABCC5 were designed using the LightCycler Probe Design Software 2.0 (Roche Biochemicals). All primer sets were tested prior to use in this work to ensure that only a single product of the correct size was amplified for all ABC transporter primer sets (Table 1). The RT-PCR reaction was performed on 300 ng total RNA with 250 nM specific primers under the following conditions: reverse transcription (20 min at 61°C), one cycle of denaturation at 95°C for 30 seconds, and PCR reaction of 45 cycles with denaturation (15s at 95°C), annealing (30s at 58°C), and elongation (30s at 72°C with a single fluorescence measurement). The PCR reaction was followed by a melting curve program (65–95°C with a heating rate of 0.1°C per second and a continuous fluorescence measurement) and then a cooling program at 40°C. Negative controls consisting of no-template (water) reaction mixtures were run with all reactions. PCR products were also run on agarose gels to confirm the formation of a single product at the desired size. Crossing points for each transcript were determined using the 2nd derivative maximum analysis with the arithmetic baseline adjustment. Data were normalized to expression of only a single convenient reference gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are presented as a comparison of gene expression for the transfectants relative to that for the parental HEK293 cells.
Preparation of red cell plasma membrane vesicles and vesicle transport assays
Membrane vesicles from human erythrocytes were isolated as previously described and ATP-dependent transport of [3H]-DNP-SG or [3H]-cGMP into membrane vesicles was measured by a rapid filtration technique [28]. Thawed membrane vesicles were diluted in transport buffer, and all standard transport assays were carried out at 37°C. 50 ng of membrane vesicles from human erythrocytes was added to a buffer system (55 μl final volume) containing 1 mM ATP, 10 mM MgCl2, 10 mM creatine phosphate, 100 μg/ml creatine kinase, 10 mM Tris-HCl (pH 7.4). At indicated time points, samples were taken from the mixture, diluted in 1 ml of ice cold stop solution (10 mM Tris-HCl, pH 7.4) and subsequently filtered through nitrocellulose filters (Whatman 0.2 μm pore size, pre-soaked overnight in 3% (w/v) bovine serum albumin). The filters were rinsed with 3 ml of ice-cold stop solution, and the tracer retained on the filter was determined by liquid scintillation counting. All transport data are presented as the difference of the values measured in the presence and absence of ATP, and ATP regenerating system was expressed relative to the protein concentration of the membrane vesicles. All data were corrected for the amount of radiolabel that remained bound to the filter in the absence of vesicle protein. Inhibitors were added to the uptake buffer solution immediately prior to the addition of the vesicles.
Cytotoxicity assay
Sensitivities of cell lines to various chemicals were examined using the Cell-Counting Kit (CCK) technique as detailed previously [38]. This technique detects the activities of dehydrogenases in viable cells, converting a colourless tetrazolium salt to a yellow formazan product soluble in the culture medium. Briefly, cells were plated at a density of 2000–5000 cells/well in 96 well plates containing 100 μl of culture medium. After 24 h incubation at 37°C in the humidified tissue-culture chamber, drugs were added into wells to a final volume of 200 μl per well and incubated for an additional 72 hours. CCK reagent was then added into each well and incubated for 4 hours before reading at a wavelength of 450 nm. IC50 values were calculated from dose-response curves obtained from at least three independent experiments. Comparisons between IC50 values were made using a 2-tailed unpaired t-test with combined estimate of the variance [39].
Effect of polyphenols on fluorescent substrate accumulation detected by FACS (Fluorescence Activated Cell Sorter)
A FACSort flow cytometer equipped with Cell Quest software (Becton-Dickinson, Franklin Lakes, NJ) was used for FACS analysis as described previously [31]. Two fluorescent substrates were used for the efflux assays. Calcein was used to study MRP1-mediated efflux (MRP1-HEK293), whereas BCECF was utilized for MRP5-mediated efflux studies (HEK293/5I). Briefly, cells were harvested after trypsinization by centrifugation at 500x g and then resuspended in IMDM supplemented with 5% fetal bovine serum. 0.25 μM calcein-AM or BCECF-AM was added to 3 x 105 cells in 4 ml of IMDM in the presence or absence of MK-571 and various polyphenols. The cells were incubated in a water bath at 37°C in the dark for 10 minutes for calcein and BCECF efflux assays prior to being pelleted by centrifugation at 500x g. The cell pellet was then resuspended in 300 μl of PBS containing 0.1 % BSA and then analyzed immediately using the flow cytometer.
Preparation of crude membranes from High Five insect cells infected with recombinant baculovirus carrying the human MRP1 or MRP4 cDNA
High Five insect cells were infected with the recombinant baculovirus carrying the human MRP1 cDNA with a 10 histidine tag at the C-terminal end (BV-MRP1 (H10)) or human MRP4 cDNA (pVL1393-MRP4 plasmid was provided by Dr. Gary Kruh, Fox Chase Cancer Centre, PA). Crude membranes were prepared from these cells as described previously [30, 34]. The protein content was determined by the Amido-black B protein estimation method [34]. The crude membranes were quickly frozen in dry ice and stored at −70°C.
ATPase assays
ATPase activities of MRP1 and MRP4 in crude membranes of High Five cells were measured by endpoint, Pi assay as previously described [30, 40]. MRP-specific activity was recorded as beryllium fluoride (BeFx)-sensitive ATPase activity. The assay measured the amount of inorganic phosphate released over 20 minutes at 37°C in the ATPase assay buffer (50 mM MES-Tris, pH 6.8, 50 mM KCl, 5 mM NaN3, 1 mM EGTA, 1 mM ouabain, 2 mM DTT and 10 mM MgCl2) in the absence and presence of 2.5 mM NaF and 0.2 mM beryllium sulfate. The assay was initiated by the addition of 5 mM ATP in the presence and absence of test compounds or transport substrates and quenched with SDS (2.5% final concentration). The amount of Pi released was quantified using a colorimetric method [40].
Photoaffinity labelling MRP1 and MRP4 with [α-32P]8-azidoATP
Crude membranes (1mg/ml protein) were incubated in the ATPase assay buffer containing 10 nM [α-32P]8-azidoATP (10 nCi/nmol) on ice in the dark on ice for 5 min and then 4°C for 5 min in the presence or absence of indicated concentrations of tested compounds. The samples were then illuminated with a UV lamp assembly (365 nm) for 10 min on ice (4°C). Ice-cold ATP (12.5 mM) was added to displace excess non-covalently bound [α-32P]8-azidoATP. After SDS-PAGE on an 7% Tris-glycine gel at constant voltage, gels were dried and exposed to Bio-Max MR film at −70°C for the required period of time (2–12 hours) as described previously [41]. The gels were also exposed to a phosphorimager screen for quantitation of the incorporation of [α-32P]8-azidoATP in the presence and absence of test compounds as described previously [41].
Acknowledgments
We thank Dr. Piet Borst (The Netherlands Cancer Institute) for the HEK294/4.63 and HEK293/5I cells and Dr. Gary Kruh (Fox Chase Cancer Centre) for MRP4 plasmid. We also thank Developmental Therapeutics Program, DCT&D, NCI for providing NSC251820 and Mr. George Leiman (LCB, NCI) for editorial help. CPW was supported by Cambridge Commonwealth Trust Fund and by a visiting preCRTA award from the NCI, NIH. This research was supported in part by the Intramural Research Program of the National Cancer Institute, NIH.
References
- 1.Allen JD, Brinkhuis RF, van Deemter L, Wijnholds J, Schinkel AH. Extensive contribution of the multidrug transporters P-glycoprotein and Mrp1 to basal drug resistance. Cancer Res. 2000;60:5761–5766. [PubMed] [Google Scholar]
- 2.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 3.Borst P, Oude-Elferink R. Mammalian ABC Transporters in health and disease. Annual Review of Biochemistry. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 4.Cole SP, Sparks KE, Fraser K, Loe DW, Grant CE, Wilson GM, Deeley RG. Pharmacological characterization of multidrug resistant MRP-transfected human tumor cells. Cancer Res. 1994;54:5902–10. [PubMed] [Google Scholar]
- 5.Reid G, Wielinga P, Zelcer N, De Haas M, Van Deemter L, Wijnholds J, Balzarini J, Borst P. Characterization of the transport of nucleoside analog drugs by the human multidrug resistance proteins MRP4 and MRP5. Mol Pharmacol. 2003;63:1094–103. doi: 10.1124/mol.63.5.1094. [DOI] [PubMed] [Google Scholar]
- 6.Breuninger LM, Paul S, Gaughan K, Miki T, Chan A, Aaronson SA, Kruh GD. Expression of multidrug resistance-associated protein in NIH/3T3 cells confers multidrug resistance associated with increased drug efflux and altered intracellular drug distribution. Cancer Res. 1995;55:5342–7. [PubMed] [Google Scholar]
- 7.Grant CE, Valdimarsson G, Hipfner DR, Almquist KC, Cole SP, Deeley RG. Overexpression of multidrug resistance-associated protein (MRP) increases resistance to natural product drugs. Cancer Res. 1994;54:357–61. [PubMed] [Google Scholar]
- 8.Schuetz JD, Connelly MC, Sun D, Paibir SG, Flynn PM, Srinivas RV, Kumar A, Fridland A. MRP4: A previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat Med. 1999;5:1048–51. doi: 10.1038/12487. [DOI] [PubMed] [Google Scholar]
- 9.Scalbert A, Williamson G. Dietary Intake and Bioavailability of Polyphenols. J Nutr. 2000;130:2073S–2085. doi: 10.1093/jn/130.8.2073S. [DOI] [PubMed] [Google Scholar]
- 10.Kim HP, Son KH, Chang HW, Kang SS. Anti-inflammatory Plant Flavonoids and Cellular Action Mechanisms. J Pharmacol Sci. 2004;96:229–45. doi: 10.1254/jphs.crj04003x. [DOI] [PubMed] [Google Scholar]
- 11.Neuhouser ML. Dietary flavonoids and cancer risk: evidence from human population studies. Nutr Cancer. 2004;50:1–7. doi: 10.1207/s15327914nc5001_1. [DOI] [PubMed] [Google Scholar]
- 12.Murota K, Terao J. Antioxidative flavonoids quercetin: implication of its intestinal absorption and metabolism. Arch Biochem Biophys. 2003;417:12–17. doi: 10.1016/s0003-9861(03)00284-4. [DOI] [PubMed] [Google Scholar]
- 13.Verbeek R, Plomp A, van Tol E, van Noort J. The flavones luteolin and apigenin inhibit in vitro antigen-specific proliferation and interferon-gamma production by murine and human autoimmune T cells. Biochem Pharmacol. 2004;68:621–9. doi: 10.1016/j.bcp.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Verbeek, R., van Tol, E. A. F. & van Noort, J. M. Oral flavonoids delay recovery from experimental autoimmune encephalomyelitis in SJL mice, Biochemical Pharmacology. In Press, Corrected Proof. [DOI] [PubMed]
- 15.Zhang S, Morris ME. Effects of the flavonoids biochanin A, morin, phloretin and silymarin on P-glycoprotein-mediated transport. TPET. 2003;304:1258–1267. doi: 10.1124/jpet.102.044412. [DOI] [PubMed] [Google Scholar]
- 16.Bobrowska-Hagerstrand M, Wrobel A, Mrowcynska L, Soderstrom T, Shirataki Y, Motohashi N, Molnar J, Michalak K, Hagerstrand H. Flavonoids as inhibitors of MRP1-like efflux activity in human erythrocytes. A structure-activity relationship study. Oncol Res. 2003;13:463–469. doi: 10.3727/000000003108747983. [DOI] [PubMed] [Google Scholar]
- 17.Leslie EM, Mao Q, Oleschuk CJ, Deeley RG, Cole SP. Modulation of multidrug resistance protein 1 (MRP1/ABCC1) transport and ATPase activities by interaction with dietary flavonoids. Mol Pharmacol. 2001;59:1171–1181. doi: 10.1124/mol.59.5.1171. [DOI] [PubMed] [Google Scholar]
- 18.Trompier D, Baubichon-Cortay H, Chang XB, Maitrejean M, Barron D, Riordon JR, Di Pietro A. Multiple flavonoid-binding sites within multidrug resistance protein MRP1. Cell Mol Life Sci. 2003;60:2164–2177. doi: 10.1007/s00018-003-3177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooray H, Janvilisri T, van Veen H, Hladky S, Barrand M. Interaction of the breast cancer resistance protein with plant polyphenols. Biochem Biophys Res Commun. 2004;317:269–75. doi: 10.1016/j.bbrc.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Yang X, Morris M. Flavonoids are inhibitors of breast cancer resistance protein (ABCG2)-mediated transport. Mol Pharm. 2004;65:1208–1216. doi: 10.1124/mol.65.5.1208. [DOI] [PubMed] [Google Scholar]
- 21.Andersson C, Servetnyk Z, Roomans GM. Activation of CFTR by genistein in human airway epithelial cell lines. Biochem Biophys Res Commun. 2003;308:518–22. doi: 10.1016/s0006-291x(03)01436-0. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZS, Lee K, Kruh GD. Transport of cyclic nucleotides and estradiol 17-beta-D-glucuronide by multidrug resistance protein 4. Resistance to 6-mercaptopurine and 6-thioguanine. J Biol Chem. 2001;276:33747–54. doi: 10.1074/jbc.M104833200. [DOI] [PubMed] [Google Scholar]
- 23.Jedlitschky G, Burchell B, Keppler D. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J Biol Chem. 2000;275:30069–74. doi: 10.1074/jbc.M005463200. [DOI] [PubMed] [Google Scholar]
- 24.Reid G, Wielinga P, Zelcer N, van der Heijden I, Kuil A, de Haas M, Wijnholds J, Borst P. The human multidrug resistance protein MRP4 functions as a prostaglandin efflux transporter and is inhibited by nonsteroidal antiinflammatory drugs. PNAS. 2003;100:9244–9249. doi: 10.1073/pnas.1033060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szakacs G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, Reinhold W, Guo Y, Kruh GD, Reimers M, Weinstein JN, Gottesman MM. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–37. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Müller M, Yong M, Peng XH, Petre B, Arora S, Ambudkar SV. Evidence for the role of glycosylation in accessibility of the extracellular domains of human MRP1 (ABCC1) Biochemistry. 2002;41:10123–32. doi: 10.1021/bi026075s. [DOI] [PubMed] [Google Scholar]
- 27.Wielinga PR, Reid G, Challa EE, van der Heijden I, van Deemter L, de Haas M, Mol C, Kuil AJ, Groeneveld E, Schuetz JD, Brouwer C, De Abreu RA, Wijnholds J, Beijnen JH, Borst P. Thiopurine metabolism and identification of the thiopurine metabolites transported by MRP4 and MRP5 overexpressed in human embryonic kidney cells. Mol Pharmacol. 2002;62:1321–31. doi: 10.1124/mol.62.6.1321. [DOI] [PubMed] [Google Scholar]
- 28.Klokouzas A, Wu CP, van Veen HW, Barrand MA, Hladky SB. cGMP and glutathione-conjugate transport in human erythrocytes. Eur J Biochem. 2003;270:3696–708. doi: 10.1046/j.1432-1033.2003.03753.x. [DOI] [PubMed] [Google Scholar]
- 29.McAleer MA, Breen MA, White NL, Matthews N. pABC11 (also known as MOAT-C and MRP5), a member of the ABC family of proteins, has anion transporter activity but does not confer multidrug resistance when overexpressed in human embryonic kidney 293 cells. J Biol Chem. 1999;274:23541–8. doi: 10.1074/jbc.274.33.23541. [DOI] [PubMed] [Google Scholar]
- 30.Sauna ZE, Nandigama K, Ambudkar SV. Multidrug resistance protein 4 (ABCC4)-mediated ATP hydrolysis: effect of transport substrates and characterization of the post-hydrolysis transition state. J Biol Chem. 2004;279:48855–64. doi: 10.1074/jbc.M408849200. [DOI] [PubMed] [Google Scholar]
- 31.Sauna ZE, Müller M, Peng XH, Ambudkar SV. Importance of the conserved Walker B glutamate residues, 556 and 1201, for the completion of the catalytic cycle of ATP hydrolysis by human P-glycoprotein (ABCB1) Biochemistry. 2002;41:13989–4000. doi: 10.1021/bi026626e. [DOI] [PubMed] [Google Scholar]
- 32.Ambudkar SV, Cardarelli CO, Pashinsky I, Stein WD. Relation between the turnover number for vinblastine transport and for vinblastine-stimulated ATP hydrolysis by human P-glycoprotein. J Biol Chem. 1997;272:21160–6. doi: 10.1074/jbc.272.34.21160. [DOI] [PubMed] [Google Scholar]
- 33.Ambudkar S, Dey S, Hrycyna C, Ramachandra M, Pastan I, Gottesman M. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–98. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 34.Kerr KM, Sauna ZE, Ambudkar SV. Correlation between steady-state ATP hydrolysis and vanadate-induced ADP trapping in Human P-glycoprotein. Evidence for ADP release as the rate-limiting step in the catalytic cycle and its modulation by substrates. J Biol Chem. 2001;276:8657–64. doi: 10.1074/jbc.M010044200. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa T. Leukotriene C4 inhibits ATP-dependent transport of glutathione S-conjugate across rat heart sarcolemma. Implication for leukotriene C4 translocation mediated by glutathione S-conjugate carrier. FEBS Lett. 1989;246:177–80. doi: 10.1016/0014-5793(89)80278-9. [DOI] [PubMed] [Google Scholar]
- 36.Klokouzas A, Barrand MA, Hladky SB. Effects of clotrimazole on transport mediated by multidrug resistance associated protein 1 (MRP1) in human erythrocytes and tumour cells. Eur J Biochem. 2001;268:6569–77. doi: 10.1046/j.0014-2956.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- 37.Wijnholds J, Mol CA, van Deemter L, de Haas M, Scheffer GL, Baas F, Beijnen JH, Scheper RJ, Hatse S, De Clercq E, Balzarini J, Borst P. Multidrug-resistance protein 5 is a multispecific organic anion transporter able to transport nucleotide analogs. Proc Natl Acad Sci. 2000;97:7476–81. doi: 10.1073/pnas.120159197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishiyama M, Tominaga H, Shiga M, Sasamoto K, Ohkura Y, Ueno K. A combined assay of cell viability and in vitro cytotoxicity with a highly water-soluble tetrazolium salt, neutral red and crystal violet. Biol Pharm Bull. 1996;11:1518–20. doi: 10.1248/bpb.19.1518. [DOI] [PubMed] [Google Scholar]
- 39.Wonnacott, R. J. a. W., T.H. (1985) Introductory Statistics, 4th edition edn, John Wiley & Son, New York.
- 40.Ambudkar SV. Drug-stimulatable ATPase activity in crude membranes of human MDR1-transfected mammalian cells. Methods Enzymol. 1998;292:504–14. doi: 10.1016/s0076-6879(98)92039-0. [DOI] [PubMed] [Google Scholar]
- 41.Sauna ZE, Ambudkar SV. Characterization of the catalytic cycle of ATP hydrolysis by human P-glycoprotein. The two ATP hydrolysis events in a single catalytic cycle are kinetically similar but affect different functional outcomes. J Biol Chem. 2001;276:11653–61. doi: 10.1074/jbc.M011294200. [DOI] [PubMed] [Google Scholar]


