Abstract
Oligopeptidase B (OpdB) is a serine peptidase broadly distributed among unicellular eukaryotes, gram-negative bacteria, and spirochetes which has emerged as an important virulence factor and potential therapeutic target in infectious diseases. We report here the cloning and expression of the opdB homologue from Salmonella enterica serovar Typhimurium and demonstrate that it exhibits amidolytic activity exclusively against substrates with basic residues in P1. While similar to its eukaryotic homologues in terms of substrate specificity, Salmonella OpdB differs significantly in catalytic power and inhibition and activation properties. In addition to oligopeptide substrates, restricted proteolysis of histone proteins was observed, although no cleavage was seen at or near residues that had been posttranslationally modified or at defined secondary structures. This supports the idea that the catalytic site of OpdB may be accessible only to unstructured oligopeptides, similar to the closely related prolyl oligopeptidase (POP). Salmonella OpdB was employed as a model enzyme to define determinants of substrate specificity that distinguish OpdB from POP, which hydrolyzes substrates exclusively at proline residues. Using site-directed mutagenesis, nine acidic residues that are conserved in OpdBs but absent from POPs were converted to their corresponding residues in POP. In this manner, we identified a pair of glutamic acid residues, Glu576 and Glu578, that define P1 specificity and direct OpdB cleavage C terminal to basic residues. We have also identified a second pair of residues, Asp460 and Asp462, that may be involved in defining P2 specificity and thus direct preferential cleavage by OpdB after pairs of basic residues.
The oligopeptidase B (OpdB; EC 3.4.21.83) subfamily of serine peptidases represents one of two branches of the prolyl oligopeptidase family of serine peptidases (the S9 family, in the nomenclature of Barrett and Rawlings [2]). The archetypical member of this family, prolyl oligopeptidase (POP; EC 3.4.21.26), exclusively hydrolyzes peptide bonds C terminal to proline residues in peptides (31). It has been implicated in the pathophysiology of depression (20) and has attracted pharmaceutical attention, since POP inhibitors have shown potential in the treatment of amnesia (39) and Alzheimer's disease (36). Prolyl oligopeptidase has also served as a model for structural studies of serine oligopeptidases. A 1.4-Å crystal structure analysis of POP recently revealed that an N-terminal regulatory domain, consisting of a seven-bladed β-propeller, regulates substrate access to the C-terminal catalytic domain (7) by a gating filter mechanism (8).
In contrast to the POPs, the OpdB branch of the POP family has received much less attention. These enzymes demonstrate a trypsin-like substrate specificity, hydrolyzing peptide bonds on the C-terminal side of basic amino acid residues of low-molecular-mass (<3 kDa) peptides (17, 30, 37, 41). Of great importance for potential therapeutic applications, OpdB is only found in ancient eukaryotic unicellular organisms, in gram-negative bacteria, and in spirochetes.
Both POP (11) and OpdB (4) have been isolated from the stercorarian trypanosome Trypanosoma cruzi, the etiological agent of Chagas' disease, and both oligopeptidases have been implicated in the pathogenesis of this disease. In infective forms of T. cruzi, OpdB generates a calcium signaling factor which, via an interaction with a receptor at the mammalian cell surface (18), is responsible for the mobilization of Ca2+ from intracellular calcium pools (4). This Ca2+ signaling is a prerequisite for trypanosome invasion. Targeted deletion of the opdB gene in T. cruzi resulted in trypanosomes that were severely impaired with respect to mammalian cell Ca2+ signaling and invasion and were thus attenuated for virulence in a mouse model of infection (5). OpdB has also been isolated from the salivarian trypanosomes Trypanosoma brucei (37) and Trypanosoma congolense (23), and homologues have been cloned from both T. brucei and Leishmania major (25). In the case of T. brucei infections in mice, OpdB is released by dying parasites into the host bloodstream, where it remains stable and catalytically active. OpdB may thus contribute to the pathogenesis of African sleeping sickness through the anomalous degradation of biologically active peptides in the bloodstream of infected hosts (24). Consistent with this view, administration of irreversible OpdB inhibitors to trypanosome-infected mice significantly impaired disease progression (26, 27). Together, these reports point to important roles for this group of enzymes in microbial virulence.
The role of OpdB in the pathogenesis of several parasitic diseases and the possibility that OpdB represents a novel target for antimicrobial chemotherapy prompted an analysis of OpdB homologues from bacterial pathogens. Genes encoding OpdB have been isolated from the gram-negative bacterial pathogens Escherichia coli (17, 32) and Moraxella lacunata (41) and the spirochete Treponema denticola (6). A preliminary analysis of OpdB function in the context of bacterial growth and protein turnover has been undertaken with Salmonella (13). A preliminary kinetic characterization of the E. coli enzyme illustrated that it cleaved substrates at the C terminus of basic residues, exhibited pronounced substrate inhibition and that the enzyme-substrate interaction was disrupted by high ionic strength, implicating ionic interactions in substrate binding (16, 29, 32). However, OpdB homologues from prokaryotes have not been extensively characterized, particularly with respect to substrate recognition properties and roles in bacterial virulence. In this report, we have addressed the former issue, showing that while catalytically similar to its eukaryotic homologues in terms of substrate affinity, OpdB from Salmonella exhibits a diminished catalytic capacity and differs in inhibition and activation properties. We go on to demonstrate that OpdB, in addition to low-molecular-mass peptides, can cleave in a restricted fashion several basic proteins, including human histones H1, H2A, H3, and H4. We have also used Salmonella OpdB as a model enzyme to define structural elements that are responsible for differences in the substrate recognition properties of OpdB and the closely related POP. These elements include two pairs of conserved acidic residues that are likely to direct P1 and P2 substrate specificity (using the nomenclature of Schechter and Berger [35]) for this subfamily of enzymes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium virulent, wild-type strain SL1344 (14) was obtained from Jorge E. Galán (Yale University School of Medicine, New Haven, Conn.) and propogated in Luria-Burtani (LB) broth (1% [mass/vol] NaCl, 1% [mass/vol] tryptone, 0.5% [mass/vol] yeast extract) or on LB agar (LB broth containing 1.5% [mass/vol] agar), supplemented with streptomycin (50 μg · ml−1). E. coli cloning (DH5α, Gibco; TOP10F′, Invitrogen) and expression [BL21(λDE3); Novagen] strains were cultured in LB media containing ampicillin (100 μg · ml−1) or kanamycin (30 μg · ml−1) as appropriate.
Isolation of the Salmonella opdB gene.
Using the E. coli opdB sequence (17), forward (5′-CCA GAA AGA ACA ATA ACA TGC TAC CAA AAG CC-3′) and reverse (5′-GTA GAT CAG TTT ATC TTG CGA TG-3′) primers were synthesized complementary to the E. coli sequence and used to generate a 668-bp probe using Salmonella genomic DNA, isolated as described in reference 38, as a template. The PCR product was cloned into pCR2.1 (Invitrogen) to create pNA102, from which the opdB fragment could be excised using the flanking EcoRI sites. Radiolabeled DNA probes were prepared using the purified DNA fragment (25 ng) as a template for random prime labeling (Rediprime; Amersham) with 50 μCi of [α-32P]dCTP. Salmonella genomic DNA was subjected to single and double digests with various restriction endonucleases. Digested DNA was resolved on a 0.7% (mass/vol) agarose gel, transferred to a Hybond N+ nylon membrane, and probed with the [α-32P]dCTP-labeled probes as described previously (4). Probes were hybridized with a ±3.5-kb fragment from a SalI/HindIII double digest of Salmonella DNA (not shown). Genomic DNA was double digested with SalI and HindIII and resolved on a 0.7% (mass/vol) low-melting-point agarose gel, and DNA was extracted from a gel slice containing fragments in the 3- to 4-kb size range. Fragments were cloned into the SalI and HindIII sites of pBlueScript II KS(+) (Stratagene) and transformed into E. coli TOP10F′. Colonies could not be screened by hybridization due to annealing of the probe with the genomic DNA-encoded E. coli opdB gene. Colonies were pooled into batches of 50, from which plasmid DNA was screened by PCR. Colonies from PCR-positive batches were screened individually, and a single PCR-positive clone containing a ±3.5-kb fragment (pNA104) was sequenced in both directions from the T3 and T7 pBlueScript II K(+) promoters using T3 (5′-AAT TAA CCC TCA CTA AAG GG-3′) and T7 (5′-GTA ATA CGA CTC ACT ATA GGG C-3′) sequencing primers. This construct contained the full-length Salmonella opdB gene. Nucleotide sequencing was undertaken by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
Expression of recombinant OpdB.
The Salmonella opdB gene was amplified by PCR from pNA104 with PfuTurbo polymerase (Stratagene) using forward (5′-GA AAA CTC GAG ATGTTG CCA AAA GCC AAT CGA ATT CCC-3′) and reverse (5′-TAC GCT CTC GAG CTA TGC GCT ATG TAA GGT TCC CTG CGC-3′) primers (with internal XhoI sites in boldface type; the initiation codon of the forward primer is underlined). The PCR product was cloned into the SmaI site of pBlueScript II KS(+) to create pNA110, and the 2.1-kb XhoI-XhoI fragment was excised from pNA110 and subcloned into the pET19b expression vector (Novagen) to create pNA111. Sequencing from the pET19b T7 promoter using the T7 forward sequencing primer (5′-TAA TAC GAC TCA CTA TAG G-3′) demonstrated that fragment was correctly orientated and the ATG start codon was in frame.
For expression, pNA111 was transformed into E. coli BL21(λDE3). N-terminal polyhistidine-tagged fusion proteins were expressed by induction of a log-phase culture (A600 ≈ 0.6) with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h at 27°C with vigorous (270 rpm) shaking in the presence of ampicillin (100 μg · ml−1). Bacteria from a 500-ml culture were harvested by centrifugation (7,000 × g, 10 min, 4°C); resuspended in 20 mM Tris-HCl, 5 mM imidazole, and 500 mM NaCl (pH 7.9) (25 ml, 4°C); and sonicated on ice with a Branson 250 sonicator (duty cycle = 80%, output control = 8; four cycles were carried out for 2 min each time). Bacterial lysates were clarified by centrifugation (15,000 × g, 15 min, 4°C) and passage through a 0.45-μm syringe filter unit, prior to peptidase purification on Ni2+-agarose (Novagen) according to the manufacturer's specifications. In some instances, the polyhistidine tag was removed with an Enterokinase Cleavage Capture kit (Novagen).
Nucleotide and protein sequence alignments.
An unrooted dendrogram was prepared, comparing full-length amino acid sequences of 18 members of the POP family with the CLUSTAL W software from the MEGALIGN program (DNASTAR, Madison, Wis.), with a PAM250 weight table set with the following parameters: ktuple = 1, gap penalty = 3, and gap window = 5 (1).
Site-directed mutagenesis.
The pNA111 expression construct was employed as a template for the generation of site-directed OpdB mutants. Site-directed mutagenesis was performed using the PCR-based Quickchange system (Stratagene). Mutagenic oligonucleotide primers were synthesized containing single- or multiple-base-pair changes with at least 15 bp of flanking sequence on either side of the base pair change. Base pair changes were confirmed by DNA sequencing from either the pET19b T7 terminator with a T7 terminator primer (5′-GCT AGT TAT TGC TCA GCG G-3′) or from a reverse primer complementary to bp 1648 to 1664 of the opdB gene (5′-CCA TAA CAC AAG CGA CGG-3′).
Kinetic analysis of recombinant OpdB.
Substrate specificity of OpdB was determined using fluorogenic substrates, by preincubation of OpdB (20 to 60 fmol of active enzyme, 37°C, 5 min) in 50 mM Tris-HCl, pH 8, followed by addition of substrate. The initial steady-state velocity (v0) was determined by continuous assay for a range of substrate concentrations (50 nM to 100 μM). Km and Vmax values were determined by hyperbolic regression of the kinetic data using the software package Hyper 1.01 (obtained from J. S. Easterby, University of Liverpool, Liverpool, United Kingdom). The kcat was determined from the expression kcat = Vmax/[E]0, where [E]0 represents the active enzyme concentration, determined by active-site titration with 4-methylumbelliferyl-p-guanidinobenzoate as described previously (25). Fluorogenic leaving groups were detected with a Hitachi F-2000 spectrofluorimeter at excitation and emission wavelengths of 370 and 460 nm, respectively, for 7-amino-4-methylcoumarin (AMC) (25) and 337 and 420 nm for β-naphthylamide (βNA) (9). Hydrolysis of para-nitroanilide (pNA) substrates was monitored spectrophotometrically at 405 nm with a Pharmacia Biotech Ultraspec 2000 spectrophotometer (29).
The effect of reducing agents on OpdB activity was investigated by preincubating OpdB in assay buffer containing l-cysteine, β-mercaptoethanol, dithiothreitol, or reduced glutathione (1 to 25 mM, 37°C, 5 min) prior to the addition of Carbobenzyloxy (Cbz)-Arg-Arg-AMC (to 5 μM).
For reversible competitive inhibitors, the Ki value was determined as described in reference 33. The enzyme-catalyzed hydrolysis of Cbz-Arg-Arg-AMC was monitored continuously to establish an uninhibited rate of substrate hydrolysis (v0), after which a 20-fold molar excess of inhibitor over enzyme was added (in less than 5% of the total assay volume), and the new steady-state velocity in the presence of the inhibitor (vi) determined. The apparent inhibition constant in the presence of substrate [Ki(app)] was given by the expression v0/vi = 1 + [I]/Ki(app). The true Ki was calculated for competitive inhibitors (I), catering for the presence of substrate, from the relationship Ki = Ki(app)/1 + [S]/Km, where [S] indicates substrate concentration.
The effects of irreversible peptidase inhibitors were investigated as described in reference 33 by adding an aliquot of inhibitor (10 μl) to a buffered enzyme solution (140 μl, containing 90 to 150 ng of OpdB in 50 mM Tris-HCl [pH 8.0], 37°C) to initiate the inactivation. Aliquots were removed at timed intervals and residual activity (vt) was determined against Cbz-Arg-Arg-AMC as described above. Pseudo-first-order inhibition rate constants (kobs) were obtained from plots of ln vt/v0 versus time, where v0 represents the activity prior to addition of inhibitor. Second-order inhibition rate constants (kass) were obtained from the expression kass = kobs/[I], where [I] represents inhibitor concentration.
Histone degradation by OpdB.
Human histones H1, H2A, H3, and H4 (Roche Molecular Biochemicals) were prepared as aqueous 1-mg · ml−1 solutions. Reaction mixtures, composed of histone (10 μg) and OpdB (0.1 to 10 μg; giving enzyme/substrate ratios of 1:100, 1:10, and 1:1) in 50 mM Tris-HCl, pH 8, were incubated at 37°C. Samples, incubated for 20 min, 1 h, 4 h, and 12 h, were resolved by reducing Tris-Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 16% polyacrylamide gels containing 6 M urea (34), and protein was detected with Coomassie blue. For N-terminal analysis, proteins were electroblotted onto polyvinylene difluoride membranes as described in reference 21 and N-terminal analysis was undertaken by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
Fluorescence measurements.
Steady-state fluorescence intensities and emission spectra were recorded with a Hitachi F-2000 spectrofluorimeter with excitation and emission slit widths of 2 nm and an excitation wavelength of 278 nm. To monitor the unfolding transition of OpdB and its site-mutated variants as a function of guanidine hydrochloride (GuHCl) concentration, aliquots of GuHCl (50 to 1,000 μl) were added from an 8 M stock to the sample (final concentration, 25 μg · ml−1 in 50 mM Tris-HCl, pH 8; final volume, 2 ml). Samples were equilibrated at 25°C before the emission spectrum was recorded.
Nucleotide sequence accession number.
The nucleotide sequence of S. enterica serovar Typhimurium opdB is available under GenBank/EBI database accession number AF237705.
RESULTS
Analysis and expression of the Salmonella opdB gene.
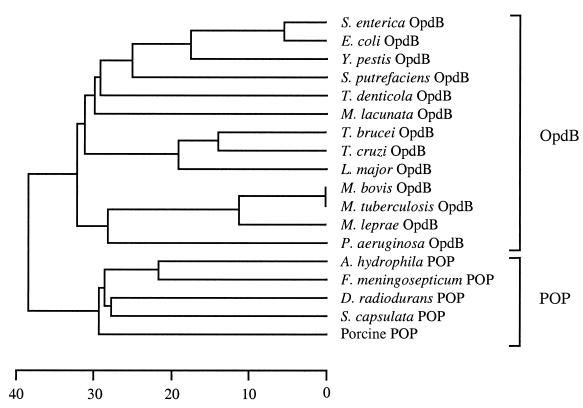
The opdB gene isolated from S. enterica serovar Typhimurium strain SL1344 contained an open reading frame of 2,052 bp, encoding a polypeptide of 684 amino acids with a predicted molecular mass of 78.969 kDa. The polypeptide contained a POP family consensus sequence (GXSXGGZZ, where X = any residue and Z = a hydrophobic residue [3]) at residues 530 to 537, which harbors the active-site serine residue. The opdB gene from this highly virulent Salmonella strain (SL1344) contained a single thymine→cytosine base pair change (at nucleotide 182) when compared with the opdB gene found in the genome sequence of nonvirulent S. enterica serovar Typhimurium strain LT2 (22), resulting in a Thr61→Ile61 replacement. The Salmonella opdB gene exhibited 75% identity with its E. coli homologue (17), and their encoded proteins share 89% identity. Salmonella OpdB retained a high degree of identity to OpdB from several other members of the gamma subdivision of the Proteobacteria (Fig. 1), including Yersinia pestis (63%) and Schewanella putrefaciens (44%). Interestingly, Salmonella OpdB exhibited higher identity to the deduced amino acid sequence of several eukaryotic opdB homologues, including those from the eukaryotes T. brucei (33%) and L. major (33%), than it did to OpdB from some prokaryotes, for example, three Mycobacterium species (31 to 32%). The wild-type opdB gene and its site-mutated variants were expressed in E. coli as soluble, catalytically active polyhistidine affinity-tagged enzymes with protein yields of between 10 to 15 mg per liter of bacterial culture.
FIG. 1.
Phylogenetic relationship of members of the POP family of serine peptidases. An unrooted dendrogram was prepared by comparing the full-length amino acid sequences of 18 members of the POP family using the CLUSTAL W alignment software of the MEGALIGN program (DNASTAR) with a PAM250 weight table set with the following parameters: ktuple = 1, gap penalty = 3, gap window = 5. The scale at the bottom measures the distance between sequences. The units indicate the number of substitution events (as a percentage). Sequences were obtained from the GenBank/EBI database under the following accession numbers: D10976 (E. coli OpdB), AF078916 (T. brucei OpdB), U69897 (T. cruzi OpdB), AF109875 (L. major OpdB), D38405 (M. lacunata OpdB), Z80226 (Mycobacterium tuberculosis OpdB), Z95151 (Mycobacterium leprae OpdB), AAK39550 (T. denticola OpdB), D14005 (Aeromonas hydrophila POP), D10980 (Flavobacterium meningosepticum POP), U08343 (Deinococcus radiodurans POP), AB010298 (Sphingomonas capsulata POP) and M64227 (porcine brain POP). The OpdB sequences for Y. pestis and Mycobacterium bovis were extracted from their partially sequenced genomes at the Sequencing Group at the Sanger Centre (available at www.sanger.ac.uk). The OpdB sequences from S. putrefaciens and Vibrio cholerae were extracted from their partially sequenced genomes at The Institute for Genome Research (available at www.tigr.org). The Pseudomonas OpdB sequence was extracted from the partially sequenced Pseudomonas genome at the Pseudomonas Genome Project (available at www.pseudomonas.com).
Amidolytic activity of recombinant Salmonella OpdB.
Recombinant OpdB exhibited hydrolytic activity against several fluorogenic substrates (Table 1). The best substrate for the peptidase was Cbz-Arg-Arg-AMC with a kcat/Km = 36 s−1 μM−1, comparing well with the 63 s−1 μM−1 reported for the E. coli homologue (30). Removal of the N-terminal polyhistidine affinity tag did not alter the kinetic profile (Table 1), illustrating that the tag did not interfere with catalysis. This is consistent with our previous report that a polyhistidine affinity-tagged recombinant OpdB from T. brucei behaved identically to the native enzyme isolated directly from trypanosomes (25). OpdB only hydrolyzed peptide substrates with basic amino acid residues (arginine or lysine) in the P1 position (using the nomenclature of Schechter and Berger [35]; Table 1), although no activity was observed against the aminopeptidase substrates H-Arg-AMC and H-Lys-AMC. Hydrolytic activity was observed when the N terminus of H-Arg-AMC was blocked with a Cbz group (i.e., Cbz-Arg-AMC; kcat/Km = 2 s−1 μM−1). Further lengthening of the peptide (Cbz-Arg-Arg-AMC; kcat/Km = 36 s−1 μM−1) considerably (18-fold) improved activity, suggesting that substrate hydrolysis is more efficient when multiple substrate-binding sites are occupied. Arginine appeared to be the preferred P1 residue, since replacement of Arg in Cbz-Arg-AMC with Lys was accompanied by a 50% reduction in kcat/Km (Table 1). Many different residues could be accommodated in P2, including aromatic (Phe), hydrophobic (Val), hydrophilic (Thr), small uncharged (Gly) residues and proline. However, basic residues (Arg and Lys) were also favored in P2. Both arginine and lysine appeared equally acceptable in P2 (since replacement of the P2-Arg with Lys in Boc-Leu-Arg-Arg-AMC and Boc-Gly-Arg-Arg-AMC (the replacement site is shown in boldface) did not appreciably [9%] alter the Km).
TABLE 1.
Substrate specificity of Salmonella OpdB
| Substratea,b | Kinetics
|
||
|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | |
| Cbz-Arg-Arg-AMC | 0.8 | 29 | 36 |
| Boc-Leu-Lys-Arg-AMC | 1.0 | 28 | 28 |
| Boc-Leu-Arg-Arg-AMC | 1.1 | 26 | 24 |
| Boc-Gly-Arg-Arg-AMC | 1.9 | 23 | 12 |
| Cbz-Gly-Gly-Arg-AMC | 2.3 | 25 | 11 |
| Boc-Gly-Lys-Arg-AMC | 1.9 | 20 | 11 |
| Cbz-Phe-Arg-AMC | 2.1 | 21 | 10 |
| Boc-Leu-Thr-Arg-AMC | 2.2 | 18 | 8 |
| Boc-Leu-Gly-Arg-AMC | 2.6 | 20 | 8 |
| Boc-Leu-Gly-Lys-AMC | 2.4 | 16 | 7 |
| Boc-Gln-Gly-Arg-AMC | 2.8 | 17 | 6 |
| Cbz-Val-Arg-AMC | 2.6 | 15 | 6 |
| Cbz-Gly-Arg-AMC | 3.4 | 15 | 4 |
| Tos-Gly-Pro-Lys-AMC | 8.1 | 18 | 2 |
| Cbz-Arg-AMC | 4.4 | 9 | 2 |
| Tos-Gly-Pro-Arg-AMC | 9.9 | 13 | 1 |
| Cbz-Lys-AMC | 5.4 | 7 | 1 |
No activity was observed against H-Arg-AMC, H-Leu-AMC, H-Lys-AMC, Cbz-Leu-Leu-Glu-βNA, Cbz-Gly-Gly-Leu-pNA, Cbz-Ala-Ala-Phe-AMC, Suc-Ile-Ala-AMC, Suc-Gly-Pro-AMC, Ac-Tyr-Val-Ala-Asp-pNA, and Suc-Ala-Ala-Pro-Phe-AMC. Standard errors of Km and kcat values were within 7% of their respective means.
Kinetic constants for Salmonella OpdB from which the polyhistidine affinity tag had been removed were as follows: for Cbz-Arg-Arg-AMC, Km = 0.88 μM, kcat = 34 s−1, and kcat/Km = 38.64 s−1 μM−1; for Cbz-Arg-AMC, Km = 5.1 μM, kcat = 10 s−1, and kcat/Km = 1.96 s−1 μM−1.
In contrast to its eukaryotic homologues, Salmonella OpdB did not demonstrate reductive activation, since preincubation with l-cysteine, β-mercaptoethanol, dithiothreitol, and reduced glutathione had no affect on activity against Cbz-Arg-Arg-AMC (not shown). Similarly, OpdB activity was not influenced by thiol-alkylating agents, including iodoacetamide, iodoacetate and N-ethylmaleimide (not shown), although these compounds are potent inhibitors of eukaryotic OpdB. Salmonella OpdB was rapidly inactivated by irreversible serine peptidase inhibitors, 4-(2-aminoethyl)benzenesulfonyl fluoride, 2,3-dichloroisocoumarin, and phenylmethane sulfonylfluoride at rates comparable to those for eukaryotic OpdB (25). Salmonella OpdB was potently inhibited by the tripeptide aldehydes antipain and leupeptin (Table 2) although the arginine analogues benzamidine and para-aminobenzamidine were comparatively poor inhibitors (Table 2), supporting our earlier suggestion that substrate (or inhibitor) binding is more effective when multiple substrate-binding sites are occupied.
TABLE 2.
Inhibition of Salmonella OpdB
| Inhibitora | Inhibition constant
|
|
|---|---|---|
| Ki (μM) | kass (M−1 s−1) | |
| Reversible | ||
| para-Aminobenzamidine | 117.1 | |
| Benzamidine | 186.7 | |
| Leupeptin | 0.4 | |
| Antipain | 0.0081 | |
| Irreversible inhibitors | ||
| 3,4-Dichloroisocoumarin | 129.0 ± 12 | |
| 4-(2-Aminoethyl)benzenesulfonyl fluoride | 31.1 ± 8 | |
| Phenylmethane sulfonylfluoride | 0.2 ± 0.04 | |
No inhibition of activity of the wild-type enzyme was observed with iodoacetamide (1 mM), iodoacetate (1 mM), N-ethylmaleimide (1 mM), EDTA (1 mM), EGTA (1 mM), bestatin (10 μM), amastatin (10 μM), arphamenine A (10 μM), elastinal (10 μM), dynorphin A (10 μM), or dynorphin B (10 μM). The standard error of the Ki values was within 5% of the mean.
Histone hydrolysis by recombinant Salmonella OpdB.
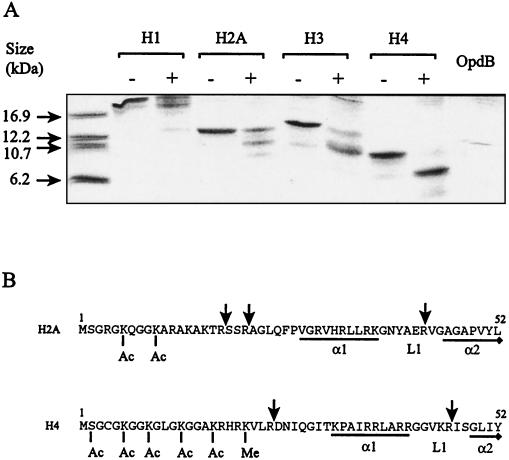
Several histone degradation products were observed upon coincubation with recombinant polyhistidine affinity-tagged OpdB at an enzyme/substrate ratio of 1:10 for 4 h (Fig. 2A). Identical data were obtained using recombinant OpdB from which the polyhistidine affinity tag had been removed (data not shown). Proteolysis was limited, since identical digestion patterns were observed with three different enzyme/substrate ratios (including 1:100, 1:10, 1:1), and in the case of H2A and H4, a enzyme/substrate ratio of 10:1 was also attempted (data not shown). Upon 4 h of coincubation with OpdB, histones H2A and H4 each yielded two major degradation products (enzyme/substrate ratio, 1:10) (Fig. 2A). These degradation products appear to be generated simultaneously, since they appear at similar relative intensities at 1, 4, and 12 h and are not evident at 20 min (not shown). Thus, cleavage at the N-terminal proximal site and the N-terminal distal site probably takes place independently, not sequentially. Several cleavage points were identified in H2A and H4 by N-terminal analysis of the degradation products. Hydrolysis was always observed C terminal to an Arg residue and occurred only after residues that were not posttranslationally modified, or locked into defined secondary structures such as α-helices (Fig. 2B). The major OpdB cleavage sites in the H2A molecule were identified by N-terminal analysis as Arg18↓Ser19, Arg21↓Ala22 and Arg43↓Val44, and in the H4 molecule as Arg24↓Asp25 and Arg46↓Ile47. Histone hydrolysis by OpdB was abrogated in the presence of anti-OpdB antibodies (not shown). Since anti-OpdB antibodies are potent and selective inhibitors of OpdB (24), we are confident that it was OpdB, and not residual contaminating E. coli peptidases, that is responsible for the observed hydrolysis of histone proteins.
FIG. 2.
Degradation of histone proteins by OpdB. Reactions consisted of histone proteins (10 μg) incubated alone (indicated by −) or together with recombinant polyhistidine affinity-tagged OpdB (1 μg; indicated by +) in 50 mM Tris-HCl, pH 8, for 4 h. As a control, recombinant polyhistidine affinity-tagged OpdB (1 μg) was incubated without histone proteins (OpdB) under the same conditions. (A) Samples were resolved by reducing Tris-Tricine SDS-PAGE on 16% polyacrylamide gels containing 6 M urea. (B) Selected degradation products were blotted onto polyvinylidene difluoride membranes, and cleavage points were identified by N-terminal amino acid sequence analysis. Cleavage points in the H2A and H4 N-terminal regions are indicated by an arrow (↓). Residues that are acetylated (Ac) and methylated (Me) in vivo are indicated. The first two α-helices of the core histone domains and their connecting random loops are indicated by α1, α2, and L1, respectively. Residues forming α-helices are underlined. Residue numbering is shown above the first and last residues in each sequence.
Alignment of catalytic domains in POP and OpdB.
Amino acid sequence alignment of the C-terminal catalytic domain of five OpdB and three POP sequences (Fig. 3) revealed several stretches of sequence or individual residues that were conserved within respective branches of the phylogenetic tree (Fig. 1) but were consistently different between the two branches. We proposed that residues contained within these differentially conserved stretches of sequence were likely to be responsible for substrate specificity. In particular, aspartic or glutamic acid residues conserved in the OpdB group of enzymes were of interest, since they would facilitate electrostatic interaction with the basic amino acid side chains of arginine and lysine residues in substrates. Nine such residues, Asp460, Asp462, Glu494, Asp567, Glu576, Glu578, Asp599, Glu624, and Asp638, were targeted for mutagenesis studies. Site-mutated variants were expressed equally well as the wild-type enzyme in E. coli and, after two passages over Ni2+-agarose, were almost 100% pure, as judged by a Coomassie blue stained SDS-PAGE gel (not shown).
FIG. 3.
Multiple sequence alignment of the amino acid sequences of the catalytic domains of OpdB from representatives of the OpdB and POP branches of the prolyl endopeptidase family of serine peptidases. Sources of sequences are given in the legend to Fig. 1. The active-site serine, histidine, and aspartic acid residues that constitute the catalytic triad are indicated by asterisks. Residues that were mutated in this study are grouped in boxes.
Fluorescence emission spectra of recombinant S. enterica serovar Typhimurium OpdB.
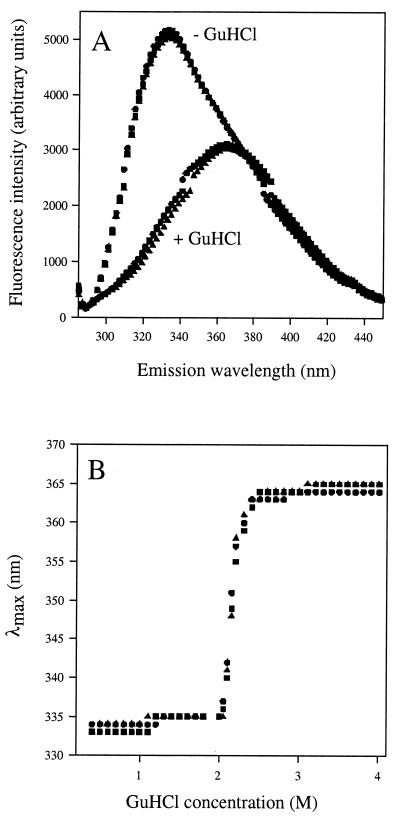
Emission spectra for all the native forms of all OpdB variants peaked at 336 nm (λmax) and showed a bathochromic (red) shift in λmax to 366 nm in the presence of 4 M GuHCl. Fluorescence emission spectra of native and GuHCl-denatured wild-type OpdB and the D460T-D462N and E576A-E578A site-mutated OpdB variants are illustrated in Fig. 4A. All mutants also exhibited identical unfolding transitions in the presence of increasing GuHCl concentrations (Fig. 4B). In the interests of clarity, only spectra for the wild-type, D460T-D462N, and E576A-E578A site-mutated OpdB variants are illustrated. The remaining nine site-mutated OpdB variants exhibited superimposable native and GuHCl-denatured emission spectra. These data strongly suggest that differences in the observed kinetic properties of the wild-type and active-site mutated variants of OpdB are not due to the disruption of the structural integrity of the enzyme (although it remains possible that very subtle structural changes were not detected in the emission spectra).
FIG. 4.
Equilibrium unfolding transitions by GuHCl. (A) Fluorescence emission spectra for wild-type (•), D460T-D462N (▴), and E576A-E578A (▪) OpdB mutant proteins. Each enzyme was 25 μg · ml−1 in 50 mM Tris-HCl, pH 8. Spectra were recorded using an excitation wavelength of 278 nm, in the presence (+ GuHCl) and absence (− GuHCl) of 4 M GuHCl. (B) Changes in the λmax of the emission spectra of OpdB wild-type and mutant proteins as a function of denaturant concentration.
Effect of point mutations on P1 substrate specificity.
The substrate Cbz-Arg-AMC was employed to monitor changes in P1 substrate specificity of site-mutated OpdB variants. Only two mutations, those in Glu576 and Glu578, resulted in significant changes in the hydrolysis of Cbz-Arg-AMC by OpdB (Table 3). Replacement of Glu576 with alanine (E576A) caused a 7.6-fold elevation in the Km and a 4.5-fold reduction in the kcat, in comparison to the wild-type enzyme (Table 3). Similarly, although less dramatic, replacement of Glu578 with alanine (E578A) resulted in a 3.7-fold elevation in Km and a 2.3-fold reduction in kcat. Simultaneous replacement of both Glu576 and Glu578 with tryptophan and threonine, respectively, (E576W:E578T), or with alanines (E576A:E578A) or arginines (E576R:E578R), abolished OpdB activity against Cbz-Arg-AMC (Table 3).
TABLE 3.
Affect of amino acid substitutions on P1 specificity of OpdB using Cbz-Arg-AMCa
| Mutant | Kinetics
|
||
|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | |
| Wild type | 4.4 | 9 | 2 |
| D460T | 4.6 | 10 | 2 |
| D462N | 4.7 | 10 | 2 |
| D460T-D462N | 5.2 | 7 | 1 |
| E494H | 4.6 | 9 | 2 |
| D567H | 5.0 | 9 | 2 |
| E576A | 33.4 | 2 | 0.06 |
| E578A | 16.4 | 4 | 0.2 |
| E576A-E578A | ND | ND | ND |
| E576W-E578T | ND | ND | ND |
| E576R-E578R | ND | ND | ND |
| D599H | 4.5 | 9 | 2 |
| E624H | 5.4 | 10 | 2 |
| D638Q | 4.0 | 11 | 3 |
No activity was observed against H-Arg-AMC, H-Leu-AMC, H-Lys-AMC, Cbz-Leu-Leu-Glu-βNA, Cbz-Ala-Ala-Phe-AMC, Cbz-Gly-Gly-Leu-pNA, Suc-Ile-Ala-AMC, Suc-Gly-Pro-AMC, Ac-Tyr-Val-Ala-Asp-pNA, and Suc-Ala-Ala-Pro-Phe-AMC. ND, no detectable activity. The standard errors of the Km and kcat values were within 10% of their respective means.
Effect of point mutations on P2 substrate specificity.
The substrates Cbz-Arg-Arg-AMC and Cbz-Phe-Arg-AMC were employed to monitor changes in P2 substrate specificity of site-mutated OpdB variants. Only two mutations, those in Asp460 and Asp462, altered the hydrolytic activity of OpdB against Cbz-Arg-Arg-AMC (Table 4) apart from Glu576 and Glu578. Mutation of Asp460 to threonine (D460T) caused a 2.6-fold elevation in Km for Cbz-Arg-Arg-AMC hydrolysis and a 24% reduction in kcat. Similarly, replacement of Asp462 with Asn (D462N) caused a threefold elevation in Km and a 21% reduction in kcat for Cbz-Arg-Arg-AMC hydrolysis. Simultaneous replacement of Asp460 and Asp462 with threonine and asparagine, respectively (D460T-D462N) had a more pronounced affect than the single mutations, yielding an OpdB variant with a 3.6-fold increase in Km and a 38% reduction in kcat for Cbz-Arg-Arg-AMC hydrolysis when compared with the wild-type enzyme. The affects of these mutations on Cbz-Phe-Arg-AMC hydrolysis by OpdB, where phenylalanine is the P2 residue, were less severe (Table 4), with the D460T-D462N OpdB variant exhibiting only a 28% increase in Km and a 19% reduction in kcat for Cbz-Phe-Arg-AMC.
TABLE 4.
Affect of amino acid substitutions on P2 specificity of OpdBa
| Mutant | Amino acid substitution and kinetics
|
|||||
|---|---|---|---|---|---|---|
| Cbz-Arg-Arg-AMC
|
Cbz-Phe-Arg-AMC
|
|||||
| Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | |
| Wild type | 0.8 | 29 | 36 | 2.1 | 21 | 10 |
| D460T | 2.1 | 22 | 10 | 2.9 | 18 | 6 |
| D462N | 2.3 | 23 | 10 | 3.0 | 19 | 6 |
| D460T-D462N | 2.9 | 18 | 6 | 2.9 | 17 | 6 |
| E494H | 1.0 | 33 | 33 | 2.5 | 18 | 7 |
| D567H | 0.8 | 29 | 36 | 2.0 | 22 | 10 |
| E576A | 4.5 | 9 | 2 | 12.0 | 7 | 0.6 |
| E578A | 3.1 | 9 | 3 | 5.9 | 9 | 2 |
| D599H | 1.0 | 30 | 30 | 2.1 | 20 | 10 |
| E624H | 1.0 | 33 | 33 | 2.6 | 22 | 8 |
| D638Q | 1.1 | 31 | 28 | 2.8 | 20 | 7 |
No activity was observed against H-Arg-AMC, H-Leu-AMC, H-Lys-AMC, Cbz-Leu-Leu-Glu-βNA, Cbz-Ala-Ala-Phe-AMC, Suc-Ile-Ala-AMC, Suc-Gly-Pro-AMC, Ac-Tyr-Val-Ala-Asp-pNA, and Suc-Ala-Ala-Pro-Phe-AMC. The standard errors of the values for Km and kcat were within 10% of their respective means.
DISCUSSION
Salmonella OpdB amidolytic activity was restricted to substrates with basic residues in the P1 position, while basic, aromatic and hydrophobic residues were accommodated in P2. While this substrate specificity of was comparable to that of eukaryotic OpdBs, the catalytic power of the enzyme (kcat) was up to fivefold less than that observed for T. brucei OpdB (25).
Salmonella OpdB neither demonstrated reductive activation nor was inhibited by thiol-alkylating agents, in contrast to its eukaryotic relatives (both POP and OpdB) which demonstrate sensitivity to reducing agents, enhancing activity (23, 25), and are potently inhibited by thiol-alkylating agents (7, 25). In porcine POP, this phenomenon is attributed to the reduction or alkylation of a cysteine residue, Cys255, which is brought into close proximity to the S1 and S3 substrate-binding sites by the folding of the enzyme (7). There is no comparable cysteine residue in the Salmonella OpdB sequence we report here, perhaps explaining this observation. Interestingly, a post-proline-cleaving member of the POP family from the prokaryote Flavobacterium also does not demonstrate sensitivity to reducing and thiol-alkylating agents either (40). Thus, insensitivity to these agents appears to be a property of members of both branches of the prokaryotic POP family.
Histone proteins were employed as prospective OpdB substrates because they are highly basic proteins (pI > 11) (19) and contain a high proportion of basic residues, thus presenting many potential OpdB cleavage sites. Salmonella OpdB degraded human histones H1, H2A, H3, and H4. Cleavage sites in H2A and H4 were located in the N-terminal tail, which projects out from the globular structure of the histone core (19). These tails may thus gain access to the OpdB active site, which by analogy with POP ordinarily excludes proteins with globular tertiary structures via the regulatory β-propeller domain (7). Cleavage was not observed after pairs of basic residues in H2A and H4, possibly because basic amino acid pairs in H2A and H4 are either chemically modified by acetylation or methylation [e.g., Arg21Lys(Me)22 in H4] or locked into α-helices (e.g., Arg36Arg37 in H4) (Fig. 2B) (19). Together, these data suggest that the substrate specificity of OpdB is not as restricted as originally theorized and that proteins, in addition to oligopeptides, are modified by OpdB. However, sites for OpdB hydrolysis in proteins are likely to be confined regions that are not organized into defined, rigid, secondary structures or posttranslationally modified at the P1 or P2 residues. These data add to a growing body of evidence that these oligopeptidases hydrolyze protein substrates, albeit in a restricted fashion (for example, the degradation of p40-phox by POP [12]), and lend support to the theory that OpdB is a specialized protein processing enzyme in prokaryotes and lower eukaryotes (30, 32). This theory is also supported by a report of the limited hydrolysis of aspartokinase I-homoserine dehydrogenase by E. coli OpdB (28).
To further explore substrate recognition properties of OpdB at a molecular level, we attempted to identify residues that may be determinants of substrate specificity. The POP family of serine peptidases splits into two branches, in terms of substrate specificity (Fig. 1). The archetypical member of this family (POP) hydrolyzes peptide bonds exclusively on the C-terminal side of proline residues in oligopeptides. In contrast, the OpdB branch hydrolyzes peptide bonds exclusively on the C-terminal side of basic residues. An alignment of the catalytic domains of both branches of the POP family (Fig. 3) revealed stretches of sequence that were conserved within respective branches of the phylogenetic tree (Fig. 1) but were consistently different between the two branches. We reasoned that residues contained in these differentially conserved regions were likely to be responsible for substrate specificity, since with the exception of substrate specificity, the physicochemical properties POP and OpdB are similar. Given the specificity of OpdB for cleavage after basic residues, we reasoned that differentially conserved acidic residues were likely determinants of specificity, since they may electrostatically interact with the basic side chains of arginine and lysine residues in substrates. In the present study, we examined the role of nine differentially conserved acidic residues located in the catalytic domain of Salmonella OpdB: Asp460, Asp462, Glu494, Asp567, Glu576, Glu578, Asp599, Glu624, and Asp638 (Fig. 3). Mutations in Glu494, Asp567, Asp599, Glu624, and Asp638 yielded OpdB variants with substrate hydrolysis profiles similar to those of the wild-type enzyme. We thus concluded that these residues were not determinants of P1 and P2 specificity.
Mutations in Glu576 and Glu578 significantly altered the hydrolysis of Cbz-Arg-AMC by OpdB, since simultaneous replacement of both Glu576 and Glu578 with alanine abolished this activity. Thus, Glu576 and Glu578 are critically involved in the interaction of OpdB with the P1 substrate residue. We propose that the carboxyl groups of Glu576 and Glu578 direct OpdB substrate specificity by electrostatic interaction with the charged side chains of P1 arginine and lysine residues in substrates. This idea is reinforced by the observations that high ionic strength inhibits the hydrolysis of basic substrates by E. coli OpdB by elevating the Km (15, 30), which is most likely attributable to the disruption of electrostatic interactions between substrate arginyl residues and carboxyl groups of the enzyme. Given the close proximity of Glu576 and Glu578 to one another (they are separated by a single tryptophan residue in the primary sequence (Fig. 3), we speculate that the side chains of Glu576 and Glu578 form a carboxyl dyad that electrostatically binds the basic side chains of arginine and lysine residues in substrates. In support of this idea, Glu576 of Salmonella OpdB corresponds to Trp595 of porcine POP, which forms the base of the S1 specificity pocket of POP and directs P1 specificity of POP for proline residues, by ring stacking between the tryptophan indole ring and the proline residue (7).
We attempted to change the P1 specificity of OpdB by mutating Glu576 and Glu578 to hydrophobic residues (a E576A-E578A double mutant, thus creating a hydrophobic S1 pocket) or to basic residues (a E575R-E578R double mutant, thus creating a basic charge environment in the S1 pocket that could accommodate acidic P1 side chains). However, both double mutants, which were devoid of activity against the OpdB substrate Cbz-Arg-AMC, also lacked activity against substrates with P1 hydrophobic residues (bold) (Cbz-Ala-Ala-Phe-AMC, Suc-Ile-Ala-AMC and Cbz-Gly-Gly-Leu-pNA) and P1 acidic residues (bold) (Cbz-Leu-Leu-Glu-βNA and Ac-Tyr-Val-Ala-Asp-pNA). Attempts to convert OpdB specificity to POP specificity by replacing both Glu576 and Glu578 with their corresponding residues in POP (Glu576→Trp and Glu578→Thr) (Fig. 3) yielded an OpdB double mutant (E576W-E578T) that did not exhibit any activity against the OpdB substrate Cbz-Arg-AMC or the POP substrate Suc-Gly-Pro-AMC. Thus, either these variants were misfolded and catalytically incompetent or other residues were involved in directing P1 specificity. The former possibility seems unlikely, since site-mutated OpdB variants exhibited fluorescence emission spectra identical to that of the wild-type enzyme. More likely, additional residues act in concert with Glu576 and Glu578 to define P1 specificity. Along these lines, differentially conserved sequences (for example, Ser569 Ile570 Pro571 Leu572 Thr573 Thr574) (Fig. 3) that lack acidic residues are observed in close proximity to Glu576 and Glu578 in all OpdB homologues.
Mutations in a second pair of residues, Asp460 and Asp462, altered the hydrolytic activity of OpdB against Cbz-Arg-Arg-AMC but had considerably less affect when a hydrophobic residue was in P2 (phenylalanine, in the case of Cbz-Phe-Arg-AMC). Since these mutations did not affect the hydrolysis of Cbz-Arg-AMC by OpdB, Asp460 and Asp462 are not involved in the S1-P1 interaction but rather appear to be involved in the recognition of the basic, charged side chain of the P2 Arg in Cbz-Arg-Arg-AMC and thus are involved in directing the specificity of OpdB for cleavage after pairs of basic amino acid residues.
Our kinetic data are nicely supported by the recently published three-dimensional model of OpdB built by Gérczei and coworkers (10). These investigators prepared an homology model of E. coli OpdB based on the crystal structure of the closely related porcine prolyl oligopeptidase (7) and docked two substrates, Cbz-Arg-OH and Cbz-Arg-Arg-OH into the OpdB active-site using the Monte Carlo docking method. In this model, the P2 Arg of Cbz-Arg-Arg-OH was bound to the carboxyl groups of Asp460 and Asp462, implicating both Asp460 and Asp462 in directing P2 specificity, which we have demonstrated experimentally in this study using site-directed mutagenesis. We thus propose in this study the existence of two carboxyl dyads: one formed by the side chains of Asp460 and Asp462, which together direct the OpdB P2 substrate specificity, and another formed by the side chains of Glu576 and Glu578, which together direct P1 substrate specificity.
Acknowledgments
We are very grateful to László Polgár and members of his laboratory for fruitful discussions and for providing prepublication copies of manuscripts. We also thank Elisabet V. Caler, Jörn Coers, Jorge E. Galán, David R. Liston, and Craig R. Roy for expert advice and John D. Lonsdale-Eccles for critically reading the manuscript.
This work was supported by grants from the National Institutes of Health to N.W.A. and by a Collaborative Research Grant from the Human Frontier Science Program (grant RG0043/2000-M) to V.F. and N.W.A.
REFERENCES
- 1.Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, A. J., and N. D. Rawlings. 1995. Families and clans of serine peptidases. Arch. Biochem. Biophys. 318:247-250. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, A. J., and N. D. Rawlings. 1992. Oligopeptidases, and the emergence of the prolyl oligopeptidase family. Biol. Chem. Hoppe-Seyler 373:353-360. [DOI] [PubMed] [Google Scholar]
- 4.Burleigh, B. A., E. V. Caler, P. Webster, and N. W. Andrews. 1997. A cytosolic serine endopeptidase from Trypanosoma cruzi is required for the generation of Ca2+-signaling in mammalian cells. J. Cell Biol. 136:609-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caler, E. V., S. Vaena, P. A. Haynes, N. W. Andrews, and B. A. Burleigh. 1998. Oligopeptidase B-dependent signaling mediates host cell invasion by Trypanosoma cruzi. EMBO J. 17:4975-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenno, J. C., S. Young Lee, C. H. Bayer, and Y. Ning. 2001. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect. Immun. 69:6193-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fülöp, V., Z. Böcskei, and L. Polgár. 1998. Prolyl oligopeptidase: an unusual β-propeller domain regulates proteolysis. Cell 94:161-170. [DOI] [PubMed] [Google Scholar]
- 8.Fülöp, V., Z. Szeltner, and L. Polgár. 2000. Catalysis of serine oligopeptidases is controlled by a gating filter mechanism. EMBO Rep. 1:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fülöp, V., Z. Szeltner, V. Renner, and L. Polgár. 2001. Structures of prolyl oligopeptidase substrate/inhibitor complexes: use of inhibitor binding for titration of the catalytic histidine residue. J. Biol. Chem. 276:1262-1266. [DOI] [PubMed] [Google Scholar]
- 10.Gérczei, T., G. M. Keserü, and G. Náray-Szabó. 2000. Construction of a 3D model of oligopeptidase B, a potential processing enzyme in prokaryotes. J. Mol. Graph. Model. 18:7-17. [DOI] [PubMed] [Google Scholar]
- 11.Grellier, P., S. Vendeville, R. Joyeau, I. M. Bastos, H. Drobecq, F. Frappier, A. R. Teixeira, J. Schrével, E. Davioud-Charvet, C. Sergheraert, and J. M. Santana. 2001. Trypanosoma cruzi prolyl oligopeptidase Tc80 is involved in nonphagocytic mammalian cell invasion by trypomastigotes. J. Biol. Chem. 276:47078-47086. [DOI] [PubMed] [Google Scholar]
- 12.Hasebe, H., J. Hua, A. Someya, P. Morain, F. Checler, and I. Nagaoka. 2001. Involvement of cytosolic prolyl endopeptidase in degradation of p40-phox splice variant protein in myeloid cells. J. Leukoc. Biol. 69:963-968. [PubMed] [Google Scholar]
- 13.Heiman, C., and C. G. Miller. 1978. Salmonella typhimurium mutants lacking protease II. J. Bacteriol. 135:588-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 15.Juhász, T., Z. Szeltner, V. Renner, and L. Polgár. 2002. Role of the oxyanion binding site and subsites S1 and S2 in the catalysis of oligopeptidase B, a novel target for antimicrobial chemotherapy. Biochemistry 41:4096-4106. [DOI] [PubMed] [Google Scholar]
- 16.Kahyaoglu, A., K. Haghjoo, F. Guo, F. Jordan, C. Kettner, F. Felföldi, and L. Polgár. 1997. Low barrier hydrogen bond is absent in the catalytic triads in the ground state but is present in a transition state complex in the prolyl oligopeptidase family of serine peptidases. J. Biol. Chem. 272:25547-25554. [DOI] [PubMed] [Google Scholar]
- 17.Kanatani, A., T. Masuda, T. Shimoda, F. Misoka, X. S. Lin, T. Yoshimoto, and D. Tsuru. 1991. Protease II from Escherichia coli: sequencing and expression of the enzyme gene and characterization of the expressed enzyme. J. Biochem. 110:315-320. [DOI] [PubMed] [Google Scholar]
- 18.Leite, M. F., M. S. Moyer, and N. W. Andrews. 1998. Expression of the mammalian calcium signaling response to Trypanosoma cruzi in Xenopus laevis oocytes. Mol. Biochem. Parasitol. 92:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Luger, K., A. W. Mäder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 20.Maes, M., F. Goossens, S. Scharpé, H. Y. Meltzer, P. D'Hondt, and P. Cosyns. 1994. Lower serum prolyl endopeptidase activity in major depression: further evidence that peptidases play a role in the pathophysiology of depression. Biol. Psychiatry 35:545-552. [DOI] [PubMed] [Google Scholar]
- 21.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 22.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterton, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 23.Morty, R. E., E. Authié, L. Troeberg, J. D. Lonsdale-Eccles, and T. H. T. Coetzer. 1999. Purification and characterisation of a trypsin-like serine oligopeptidase from Trypanosoma congolense. Mol. Biochem. Parasitol. 102:145-155. [DOI] [PubMed] [Google Scholar]
- 24.Morty, R. E., J. D. Lonsdale-Eccles, R. Mentele, E. A. Auerswald, and T. H. T. Coetzer. 2001. Trypanosome-derived oligopeptidase B is released into the plasma of infected rodents, where it persists and retains full catalytic activity. Infect. Immun. 69:2757-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morty, R. E., J. D. Lonsdale-Eccles, J. Morehead, E. V. Caler, R. Mentele, E. A. Auerswald, T. H. T. Coetzer, N. W. Andrews, and B. A. Burleigh. 1999. Oligopeptidase B from Trypanosoma brucei: a new member of an emerging subgroup of serine oligopeptidases. J. Biol. Chem. 274:26149-26156. [DOI] [PubMed] [Google Scholar]
- 26.Morty, R. E., L. Troeberg, R. N. Pike, R. Jones, P. Nickel, J. D. Lonsdale-Eccles, and T. H. T. Coetzer. 1998. A trypanosome oligopeptidase as a target for the trypanocidal agents pentamidine, diminazene and suramin. FEBS Lett. 433:251-256. [DOI] [PubMed] [Google Scholar]
- 27.Morty, R. E., L. Troeberg, J. C. Powers, S. Ono, J. D. Lonsdale-Eccles, and T. H. T. Coetzer. 2000. Characterisation of the antitrypanosomal activity of peptidyl α-aminoalkyl phosphonate diphenyl esters. Biochem. Pharmacol. 60:1497-1506. [DOI] [PubMed] [Google Scholar]
- 28.Pacaud, M., and C. Richaud. 1975. Protease II from Escherichia coli. J. Biol. Chem. 250:7771-7779. [PubMed] [Google Scholar]
- 29.Polgár, L. 1999. Oligopeptidase B: a new type of serine peptidase with a unique substrate-dependent temperature sensitivity. Biochemistry 38:15548-15555. [DOI] [PubMed] [Google Scholar]
- 30.Polgár, L. 1997. A potential processing enzyme in prokaryotes: oligopeptidase B, a new type of serine peptidase. Proteins 28:375-379. [PubMed] [Google Scholar]
- 31.Polgár, L. 1994. Prolyl oligopeptidases. Methods Enzymol. 244:188-200. [DOI] [PubMed] [Google Scholar]
- 32.Polgár, L., and F. Felföldi. 1998. Oligopeptidase B: cloning and probing stability under nonequilibrium conditions. Proteins 30:424-434. [PubMed] [Google Scholar]
- 33.Salvesen, G., and H. Nagase. 1989. Inhibition of proteolytic enzymes, p. 83-104. In R. J. Benyon and J. S. Bond (ed.), Proteolytic enzymes: a practical approach. IRL Press, Oxford, United Kingdom.
- 34.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 35.Schechter, I., and A. Berger. 1967. On the size of the active-site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 32:898-902. [DOI] [PubMed] [Google Scholar]
- 36.Toide, K., M. Shinoda, and A. Miyazaki. 1998. A novel prolyl endopeptidase inhibitor, JTP-4819—its behavioural and neurochemical properties for the treatment of Alzheimer's disease. Rev. Neurosci. 9:17-29. [DOI] [PubMed] [Google Scholar]
- 37.Troeberg, L., R. N. Pike, R. E. Morty, R. K. Berry, T. H. T. Coetzer, and J. D. Lonsdale-Eccles. 1996. Proteinases from Trypanosoma brucei brucei: purification, characterisation and interactions with host regulatory molecules. Eur. J. Biochem. 238:728-736. [DOI] [PubMed] [Google Scholar]
- 38.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 39.Yoshimoto, T., K. Kado, F. Matsubara, N. Koriyama, H. Kaneto, and D. Tsuru. 1987. Specific inhibitors for prolyl oligopeptidase and their anti-amnesic effect. J. Pharmacobiodyn. 10:730-735. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimoto, T., A. Kanatani, T. Shimoda, T. Inaoka, T. Kokubo, and D. Tsuru. 1991. Prolyl endopeptidase from Flavobacterium meningosepticum: cloning and sequencing of the enzyme gene. J. Biochem. 110:873-878. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimoto, T., J. Tabira, T. Kabashima, S. Inoue, and K. Ito. 1995. Protease II from Moraxella lacunata: cloning, sequencing, and expression of the enzyme gene, and crystallization of the expressed enzyme. J. Biochem. 117:654-660. [DOI] [PubMed] [Google Scholar]