Abstract
Transcription antitermination by the bacteriophage λ N protein is stimulated in vitro by the Escherichia coli NusG protein. Earlier work suggested that NusG was not required for N activity in vivo. Here we present evidence that NusG also stimulates N-mediated transcription antitermination in intact cells.
NusG, a 21-kDa essential protein in Escherichia coli (8, 20), is an abundant transcription elongation factor that modulates Rho-dependent transcription termination (22, 24, 25). Rho-dependent termination is inhibited by depletion or overexpression of NusG (2, 24). Affinity chromatography experiments showed that NusG associates with Rho or core RNA polymerase (19). It was proposed that NusG facilitates termination by aiding in the recruitment of Rho to the transcription complex (1, 18, 21, 22, 24).
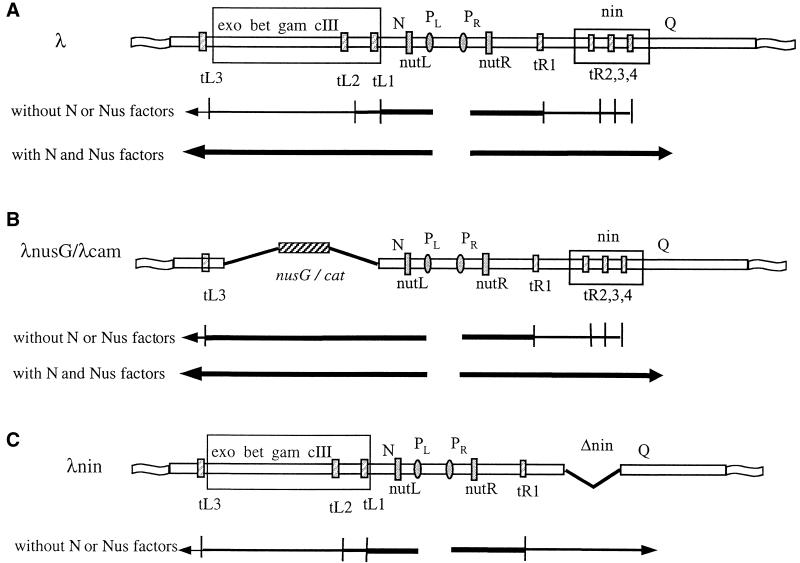
In vitro studies identified NusG as an E. coli protein that influences transcription antitermination mediated by the N protein of phage λ (18, 20). Transcription of the λ delayed early genes requires RNA polymerase to read through a number of transcription terminators. N, in complex with the E. coli Nus factors, modifies RNA polymerase at specific RNA sites, NUT, to a form that overrides downstream transcription terminators (Fig. 1) (7, 11, 12, 16, 23, 27).
FIG. 1.
Map of relevant region of the λ genome showing location of promoters (P), terminators (t), nut sites, and representative genes. Also shown are the regions of the nin deletion and the gene replacements. The λ recombination system (28) was used to replace λ sequences from nucleotides 31,351 to 34,700 in the standard λ sequence (6) with either the nusG or cat gene. (A) λ wild-type; (B) λnusG and λcam; (C) λnin. Below each genetic map are the patterns of transcription in both the presence and absence of N or Nus factors (A and B) or only in the absence of N or Nus factors (C).
Nus factors were first identified by mutations in E. coli that influence N action. Mutations in nusA (nusA1) (9), nusB (a number of mutations) (10, 17), and rpsJ (nusE71) (14) reduce N activity, particularly at higher temperatures. In contrast, the nusB101 mutation suppresses this effect of the nusA1 and nusE71 alleles on N action (26).
The only genetic evidence for a role of NusG in N antitermination was the isolation of nusG4, which has a suppressor phenotype identical to that of the nusB101 allele (25). However, selection for nus mutations that reduce N action did not yield nusG mutants. Furthermore, in contrast to its function in vitro, in vivo studies provided evidence that NusG was not required for effective N-mediated transcription antitermination. The in vivo studies employed a strain bearing a lacZ reporter fused downstream of the λ pL promoter, nutL, and transcription terminators. Because of the terminators, expression of the lacZ reporter is N dependent. NusG was expressed from a plasmid with a temperature-sensitive replicon. NusG depletion was accomplished by shifting the strain from 32 to 42°C. N was expressed from a plasmid under the control of the lac promoter. The experimental design was to supply N after depleting the bacteria of NusG (see below for the method of depletion) by treating the bacteria with IPTG (isopropyl-β-d-thiogalactopyranoside). Depleting this strain of NusG did not reduce lacZ expression, suggesting that N-mediated antitermination did not require NusG (24). However, more recent experiments show that this result can be explained on the basis of leaky expression of N prior to the depletion of NusG (data not shown). We were concerned because the plasmid supplying N was present during the entire time of growth at 42°C. Since the pL-nutL-tL1-lacZ fusion used to measure N-mediated transcription antitermination is controlled by the temperature-sensitive λ cI857 repressor, transcription from pL would be active during NusG depletion. Thus, a leaky level of N expression during depletion supports read-through of the terminator and permits expression of lacZ. We avoided this problem by devising a protocol in which N was supplied only after NusG depletion had been completed.
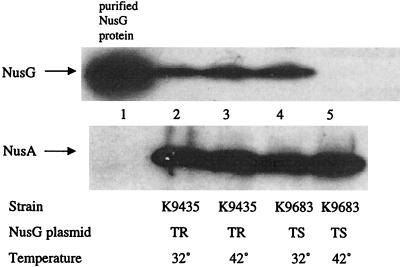
The method of Sullivan and Gottesman was used to produce bacteria depleted of NusG (24). Briefly, a conditionally lethal E. coli strain, K9683, was constructed by disrupting the chromosomal nusG allele with an insertion of a gene conferring resistance to kanamycin, kanR. The nusG defect is complemented with an expressible wild-type nusG gene on pSS119, a plasmid that has a thermosensitive defect in replication. As shown previously, the NusG protein in the E. coli cells can be depleted by growing the cells at 42°C in Luria-Bertani broth (15) for 6 h. The cultures were diluted every hour into fresh prewarmed Luria-Bertani broth to maintain logarithmic growth. Because the plasmid fails to replicate under these conditions, cell division segregates cells that lack the nusG plasmid and eventually the NusG protein. The control strain, K9435, carries the disrupted chromosomal nusG allele, but the nusG plasmid, pSS120, is replication proficient at high temperature. Depletion was confirmed by Western blotting. As shown in Fig. 2, K9683 and K9435 grown at 32°C contained similar quantities of NusG protein. However, when the bacteria were grown at 42°C, NusG was undetectable in strain K9683, which carries the temperature-sensitive nusG plasmid, whereas no decrease in NusG concentration was observed in the control strain, K9435.
FIG. 2.
Western blot to assess levels of NusG. Samples from logarithmically grown bacteria were isolated, prepared, and analyzed as described previously (5). The upper panels were blotted with anti-NusG, and the lower panels were blotted with anti-NusA. Lane 1 was loaded with purified NusG (24). Other lanes were loaded with extracts prepared from the indicated strains. TR, the plasmid in the strain replicates at high temperature; TS, the plasmid in the strain fails to replicate at high temperature. The last line shows the temperature at which the bacterial cultures were grown prior to preparation of extracts.
We assessed the role of NusG in N antitermination by infecting K9435 and K9863 with four λ variants. λ, λnusG, and λcam all require N; they fail to grow in E. coli carrying the nusA1, nusB5, or nusE71 mutation at the nonpermissive temperature (reference 14 and data not shown). λnin has a deletion of a critical set of transcription terminators (Fig. 1) and thus can grow independently of N (3, 4). As expected, λnin grows in the three nus mutant strains (reference 14 and data not shown). λnusG is identical to λcam but carries a nusG gene instead of cat. λnusG is expected to grow in an E. coli strain depleted of NusG even if NusG is required for N action. λcam serves as a control for λnusG (see the legend to Fig. 1). Phage growth was determined by measuring burst size (phage produced per infected bacterium) 90 min after infection (13). The results of these experiments are shown in Table 1. The burst sizes of λ and λnin at 42°C are the same in the control strain, K9435, where NusG is present. However, the burst size of λ is ∼30-fold lower than that of λnin in strain K9683, which is depleted of NusG at 42°C. Note that the burst size of λnin is reduced in K9683 relative to that produced in K9435, likely reflecting the unfavorable environment provided by a bacterium growing in the near absence of NusG. λnin, then, provides a way to assess the effectiveness of bacteria depleted of NusG in supporting growth of λ independently of the requirement for N-mediated antitermination; i.e., the nin phage is the positive control. In summary, these data suggest that poorer growth of λ in NusG-depleted E. coli is due to a failure of N-mediated transcription antitermination over and above that caused by the failing physiological environment.
TABLE 1.
Growth of λ derivatives in the absence of NusG
| Phagec | Burst sizea at 42°C
|
|
|---|---|---|
| NusG present (K9435) | NusG absentb (K9683) | |
| λ | 22 ± 7 | 0.1 ± 0.1 |
| λnin | 22 ± 9 | 3 ± 2 |
| λnusG | 72 ± 17 | 6 ± 4 |
| λcam | 68 ± 35 | 0.2 ± 0.1 |
Burst size is calculated as the number of phage produced per infected bacterium.
NusG was depleted as described in the text.
All of these phages have the cI857 allele, which encodes a temperature-sensitive repressor. Since the phenotype imposed by this mutation has no bearing on the results of our studies, we have not included this genotype in the nomenclature of the various phages in the text.
To show that N would be effective if NusG were present in a NusG-depleted bacterium, we compared growth of λnusG and λcam in the absence of NusG. Since these phages are essentially identical except for the ability of the former to express NusG, support of growth of λnusG but not λcam would be evidence that NusG is required for λ growth. The bursts of these two phages were the same when grown in the host K9435, which expresses NusG at 42°C. However, the burst of λcam is ∼30-fold lower than the burst of λnusG in K9683 depleted of NusG by prior growth at 42°C.
Thus, the defect in N activity in K9683 can be corrected by expressing NusG. The 10-fold-lower burst of λnusG in K9683 relative to K9435 is not unexpected, since NusG is supplied to the depleted bacterium only during the short time of infection.
Taken together, these observations indicate that NusG is required for N action. First, the low burst of λ relative to λnin in K9683 provides evidence that N-mediated transcription antitermination is reduced in vivo in the absence of NusG. Second, the ability of λnusG to grow in K9683 shows that the absence of NusG is responsible for the failure of N antitermination. We conclude, therefore, that NusG is required for N-mediated antitermination in vivo as it is in vitro.
Acknowledgments
Jack Greenblatt is thanked for anti-NusG antiserum.
The work at the University of Michigan was supported by Public Health Research grant A111459-10. M.E.G. is the recipient of NIH grant GM37219.
REFERENCES
- 1.Burns, C. M., and J. P. Richardson. 1995. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc. Natl. Acad. Sci. USA 92:4738-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burova, E., and M. E. Gottesman. 1995. NusG overexpression inhibits Rho-dependent termination in Escherichia coli. Mol. Microbiol. 17:633-641. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, S. C., D. L. Court, and D. I. Friedman. 1995. Transcription termination signals in the nin region of bacteriophage lambda: Identification of Rho-dependent termination regions. Genetics 140:875-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Court, D., and K. Sato. 1969. Studies of novel transducing variants of lambda: dispensability of genes N and Q. Virology 39:348-352. [DOI] [PubMed] [Google Scholar]
- 5.Craven, M. G., and D. I. Friedman. 1991. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J. Bacteriol. 173:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels, D. L., J. L. Schroeder, W. Szybalski, F. Sanger, and F. R. Blattner. 1983. A molecular map of coliphage lambda, p. 469-676. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 7.Das, A. 1992. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J. Bacteriol. 174:6711-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downing, W. L., S. L. Sullivan, M. E. Gottesman, and P. P. Dennis. 1990. Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J. Bacteriol. 172:1621-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman, D. I., and L. S. Baron. 1974. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology 58:141-148. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, D. I., M. Baumann, and L. S. Baron. 1976. Cooperative effects of bacterial mutations affecting lambda N gene expression. I. Isolation and characterization of a nusB mutant. Virology 73:119-127. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, D. I., and D. L. Court. 1995. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 18:191-200. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, D. I., and D. L. Court. 2001. Bacteriophage lambda: alive and well and still doing its thing. Curr. Opin. Microbiol. 4:201-207. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, D. I., C. T. Jolly, and R. J. Mural. 1973. Interference with the expression of the N gene function of phage lambda in a mutant of Escherichia coli. Virology 51:216-226. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, D. I., A. T. Schauer, M. R. Baumann, L. S. Baron, and S. L. Adhya. 1981. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc. Natl. Acad. Sci. USA 78:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman, D. I., and M. B. Yarmolinsky. 1972. Prevention of the lethality of induced lambda prophage by an isogenic lambda plasmid. Virology 50:472-481. [DOI] [PubMed] [Google Scholar]
- 16.Gottesman, M. E., and R. A. Weisberg. 1995. Termination and antitermination of transcription in temperate bacteriophage. Semin. Virol. 6:35-42. [Google Scholar]
- 17.Keppel, F., C. P. Georgopoulos, and H. Eisen. 1974. Host interference with expression of the lambda N gene product. Biochimie 56:1505-1509. [DOI] [PubMed] [Google Scholar]
- 18.Li, J., R. Horwitz, S. McCracken, and J. Greenblatt. 1992. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J. Biol. Chem. 267:6012-6019. [PubMed] [Google Scholar]
- 19.Li, J., S. W. Mason, and J. Greenblatt. 1993. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 7:161-172. [DOI] [PubMed] [Google Scholar]
- 20.Mason, S. W., and J. Greenblatt. 1991. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5:1504-1512. [DOI] [PubMed] [Google Scholar]
- 21.Nehrke, K. W., and T. Platt. 1994. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J. Mol. Biol. 243:830-839. [DOI] [PubMed] [Google Scholar]
- 22.Pasman, Z., and P. H. von Hippel. 2000. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry 39:5573-5585. [DOI] [PubMed] [Google Scholar]
- 23.Richardson, J. P., and J. Greenblatt. 1996. Control of RNA chain elongation and termination, p. 822-848. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 24.Sullivan, S. L., and M. E. Gottesman. 1992. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 68:989-994. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan, S. L., D. F. Ward, and M. E. Gottesman. 1992. Effect of Escherichia coli nusG function on lambda N-mediated transcription antitermination. J. Bacteriol. 174:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward, D. F., A. DeLong, and M. E. Gottesman. 1983. Escherichia coli nusB mutations that suppress nusA1 exhibit lambda N specificity. J. Mol. Biol. 168:73-85. [DOI] [PubMed] [Google Scholar]
- 27.Weisberg, R. A., and M. E. Gottesman. 1999. Processive antitermination. J. Bacteriol. 181:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]