Summary
The understanding of the molecular mechanisms of sex differentiation in the mosquito Anopheles gambiae could identify important candidate genes for inducing selective male sterility in transgenic lines or for sex-controlled expression of lethal genes. In many insects doublesex (dsx) is the double-switch gene at the bottom of the somatic sex-determination cascade that determines the differentiation of sexually dimorphic traits. We report here on the identification of the dsx homologue in A. gambiae, and on the characterization of its sex-specific transcripts. Agdsx consists of 7 exons distributed over an 85 kb region on chromosome 2R, which are sex-specifically spliced to produce the female and male AgdsxF and AgdsxM transcripts. AgdsxF contains a 795 bp ORF, coding for a protein of 265 amino acids, while AgdsxM comprises a much longer (1866 bp) ORF, coding for a 622 aa protein. Differences in the exon/intron organization suggest that Agdsx sex-specific splicing results from a different mechanism from Drosophila melanogaster dsx. These findings represent an important step towards the understanding of sex differentiation in Anopheles and will facilitate the use of gene transfer technologies to manipulate sex ratios for vector control programs based on the Sterile Insect Technique.
Keywords: Doublesex, Anopheles gambiae, sex determination, sex-specific transcripts
Introduction
The molecular processes leading to sexual differentiation in Anopheles gambiae have not yet been elucidated despite the importance of these mosquitoes as vectors of human malaria. If identified, genes responsible for the sexual fate of these insects could represent invaluable tools for the development of novel malaria control measures such as those based on the Sterile Insect Technique (SIT). SIT involves the release of large numbers of sexually active but genetically sterile males in order to achieve eradication of wild-type insects. SIT chances of success ultimately rely on the development of efficient genetic sexing strains and on the mating competitiveness of the released sterile males. Limited success or failure of previous mosquito SIT programs has emphasized the limitations of current methods for sex separation and the downsides of irradiation-based sterilization procedures, which have detrimental effects on the fitness of the released insects (Benedict and Robinson, 2003). Unraveling the molecular mechanisms of sex determination in Anopheles would provide crucial information on target genes and sex-specific splicing patterns, which could be used to induce male sterility and develop new molecular sexing methods, thus expanding the tool kit available for SIT programs.
In nature, sexual differentiation is achieved though a variety of mechanisms, which determine morphological, physiological and behavioural traits in most living organisms. Despite the striking differences in the machinery determining sex and in the individual genes involved, a common pattern can be recognised among distinct taxonomic groups: a primary signal, a key gene and a regulatory control gene, lead to a double-switch gene, which selects between alternative sexual programmes. In Drosophila melanogaster, one of the best characterized organisms both genetically and molecularly, the primary signal of somatic sex determination is the ratio of X chromosomes to autosomes (X:A ratio). An X:A ratio of 1.0 (2X:2A) dictates female development while an X:A ratio of 0.5 (1X:2A) determines male development (Cline, 1993). The sole target of the primary signal is the key gene sex-lethal (sxl), which becomes active in females and is inactive in males. SXL then acts as a splicing regulator of transformer (tra) pre-mRNA (Boggs et al., 1987). Although tra is transcribed in both sexes, a splice form of its transcript leading to a complete open reading frame (ORF) occurs only in females. The final double-switch gene in the somatic sex determination cascade is doublesex (dsx). Dsx codes for two sex-specific transcription factors (DSXF in females and DSXM in males) that activate or repress the final genes necessary for the differentiation of sexually dimorphic traits. Sex-specific transcripts of dsx result from differential splicing that depends on the function of tra. In females, TRA, along with the product of the constitutively active gene tra2, act as a splicing regulator by binding to splice enhancer sites (dsxREs) present on the pre-mRNA of dsx, activating a weak female-specific 3’ acceptor site preceding the female specific exon (Baker and Wolfner, 1988; Burtis and Baker, 1989; Hoshijima et al., 1991). In males, where TRA is inactive, the weak 3’ acceptor site is not recognised, causing the splicing of the female-specific exon and, instead, the inclusion of two downstream male-specific exons.
Analysis of the cascade in other insects supports a bottom-up model of evolution for sex determination (Wilkins, 1995). The last gene in the cascade is the most ancient and conserved while upstream regulators have strongly diverged during evolution. Homologues of the key gene sxl have been isolated from Chrysomya rufifacies (Muller-Holtkamp, 1995), Megaselia scalaris (Sievert et al., 2000), Musca domestica (Meise et al., 1998) and Ceratitis capitata (Saccone et al., 1998) where they do not play a role in sex determination. With the exception of C. capitata (Pane et al., 2002), no homologues of tra have been found outside of the genus Drosophila, and the sequence has diverged considerably even within this genus (O'Neil and Belote, 1992). Homologues of the gene tra2 have been found in D. virilis (Chandler et al., 1997), human (Dauwalder et al., 1996; Nayler et al., 1998) and mouse (Segade et al., 1996). On the other hand, homologues of Dmdsx in B. tryoni (Shearman and Frommer, 1998), M. scalaris (Kuhn et al., 2000), C. capitata (Saccone, 1996) and M. domestica (Hediger et al., 2004) are largely conserved in exon-intron structure and sex-specific splicing regulation, suggesting functional conservation. These dsx homologues share most of the features of Dmdsx, which include cis regulatory elements such as the TRA/TRA2 splice enhancer elements dsxREs and a purine-rich enhancer (PRE), a weak 3’ splice acceptor site preceding the female-specific exon and the domains necessary for functional properties related to DSX (DNA-binding domain DBD, oligomerization domains OD1 and OD2). Some features are also conserved in the more distantly related Lepidoptera Bombyx mori, where however the regulatory mechanisms of the sex-specific processing are different (Ohbayashi et al., 2001; Suzuki et al., 2001). Dsx homologues have also been identified in C. elegans (Raymond et al., 1998), humans (Raymond et al., 1999b), mice and chickens (Raymond et al., 1999a).
In this study, we have characterized the sex-specific splicing forms of the A. gambiae dsx gene, and elucidated its chromosomal exon-intron organization. The gene is distributed over an 85 kb region of chromosome 2R and is composed of multiple exons, which are alternatively spliced to produce female-and male-specific transcripts. Based on the sequence conservation, exon-intron organization and transcription pattern, our results strongly indicate a role for Agdsx in determining the sexual fate in A. gambiae.
Materials and methods
Sequencing tools and screening of the cDNA library
To isolate the putative homologue of the dsx gene in A. gambiae, the Ensembl Mosquito Genome (http://www.ensembl.org/Anopheles_gambiae) and NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) browsers were searched. Amino acid sequence alignment of DSX orthologues was performed using the CLUSTAL W programme. Degenerate primers commonfor (5’-GTACTGCAAGTATCGCGCC-3’) and commonrev (5’-GCTCCCATATGCTGCGG-3’) were designed to amplify the dsx common region (DBD/OD1) from A. gambiae Giles sensu stricto genomic DNA (KWA Strain). The cycle conditions included an initial denaturation of 5 min at 95°C, followed by 40 cycles of 95°C for 30s, 54°C for 30s and 72°C for 1 min, with a final elongation step of 72°C for 10 min. This fragment (Probe 5’) was then used to screen a λ ZAP (Stratagene, La Jolla, CA, USA) pupal A. gambiae cDNA library (kindly provided by Peter Billingsley) following the manufacturer's instructions (Gare et al., 2003).
RT-PCR and Northern Blot analyses
Total RNA was extracted from adult male and female A. gambiae mosquitoes, using Tri Reagent (Helena Biosciences, Sunderland, UK) according to manufacturer's instructions. Pupae were sexed by comparing their terminalia under a dissecting microscope. The Superscript First-Strand Synthesis System (Gibco, Gaithersburg MD, USA) was used for first-strand cDNA synthesis using oligo d(T) primers and total RNA, according to manufacturer's instructions. PCR was performed essentially as described above except the annealing temperatures were adjusted to individual primers. Standardization was performed with primers S7for (5’-GGCGATCATCATCTACGTGC-3’) and S7rev (5’-GTAGCTGCTGCAAACTTCGG-3’), which amplify the housekeeping S7 ribosomal gene. Other primers include: dsx1f (5’-AAAGCACACCAGCGGATCG-3’), dsx2f (5’-TCTACAATCAATCAATCCGTG-3’), dsx3f (5’-ACCATCGTTCAACCAATACC-3’), dsx1r (5’-CACCGAGATGTTCTCGTCC-3’), dsx2r (5’-TCCACTCTGACGGGTGGTATTGCG), dsx3r (5’-GATTGATTGATTGTAGAGTGG-3’), dsxef (5’-TTTCGATCGTGCAACGAAGG-3’) and dsxer (5’-TTTGGTGGGAAATTGGGCG-3’).
For Northern blot analysis, 10μg of total RNA from male and female adults was isolated as described above and hybridized with a 32P-labeled Probe 5’ (described above) and Probe 3’, a 446 bp fragment amplified from genomic DNA with dsx3’f (5’-GAAGTCATCGCTCGATCCG-3’) and dsx3’r (5’-CCAGGCTCTCGTACACG-3’), following well established protocols (Sambrook, 1989).
Southern blot analysis
For Southern blot analysis, a total of 5μg of genomic DNA from male and female A. gambiae adults were digested with BamHI, HindIII, or XhoI and hybridized with Probe 5’ as previously described (Catteruccia et al., 2000).
Sequencing of Splice Acceptor Sites
In order to obtain the sequence of the 3’ splice acceptors within introns 2, 3, 4 and 5, PCR amplification from genomic DNA was performed as described above. Primers intron2f (5’-CTCCGAGGTGAACAATCGG-3’) and intron2r (5’-AGGAGATTTACAGGTTCTGG-3’) amplified a fragment of intron 2; intron3f (5’-GCTGCCACGATTGACACC-3’) and intron3r (5’-TTCAGTATGACGTACATCAGG-3’) amplified a fragment of intron 3; intron4f (5’-TAAAGAGCGCCGATGGCG-3’) and dsx2r (above) amplified intron 4; and intron5f (5’-ACAATCAATCAATCCGTGCAG-3’) and dsx1r amplified intron 5. Amplification products were cloned into the pGEM T-easy vector according to manufacturer's instructions. Sequencing reactions were performed using the ABI Prism fluorescent sequencing kit. Sequencing of the inserts was performed using primers annealing on the vector, M13left (5’-GTAAAACGACGGCCAGT-3’) or M13rev (5’-GGAAACAGCTATGACCATG-3’).
Results
Female and male specific transcripts of Agdsx
The A. gambiae EST database was searched in order to identify the putative orthologue of the D. melanogaster dsx gene. A clone P4060 (Genbank accession number EAA05330) was identified with significant homology to Dmdsx. Analysis of the available A. gambiae genomic sequence could identify the DBD/OD1 and OD2 regions as well as a region with high similarity to the female-specific coding sequence from other insects. However, sequence analysis and comparison to D. melanogaster did not provide any clue about the nature of the male specific sequence and about the exon/intron organization of the Agdsx locus. Degenerate primers were designed using those parts of the translated sequence of the P4060 clone displaying high homology to the DBD/OD1 domains of Dmdsx. A 355 bp fragment was amplified from genomic DNA, which exhibited 82% identity at the nucleotide level and 80% identity at the protein level with Dmdsx. To search for the sequence of the female and male specific transcripts of Agdsx, a mixed pupal A. gambiae cDNA library was screened with this fragment (Probe 5’). A total of fourteen clones were isolated from the screening of the cDNA library, some of which encoded truncated proteins, because of incomplete splicing, and were disregarded in the analysis (Fig. 1). Three of the clones contained the highly conserved female-specific region and were therefore classified as female (AgdsxF), while one clone lacking this region was classified as male (AgdsxM). The three putative AgdsxF clones F1, F2 and F3 were 3.1 kb, 4.5 kb and 5.8 kb long, respectively (Genbank accession number DQ137802). Clones F1 and F3 shared similar 5’ regions, except F1 was 230 bp longer at the 5’ end, while F2 and F3 had identical 3’ ends, that extended for a further 3 kb downstream from the 3’ end of F1 (Fig. 1). The putative AgdsxM clone M1 was 6.0 kb in length, and did not contain the highly conserved female-specific coding sequence and 1.6 kb of 3’ UTR immediately downstream of it (Genbank accession number DQ137801). This suggests that this region (1.7 kb in total) is spliced out in male transcripts (Fig. 1), resulting in the translation of a downstream 387 amino acid region, likely to correspond to the male-specific part of Agdsx. Clone M1 extended for a further 2.6 kb downstream from the 3’ end of female clones F2 and F3. This region contained a series of polyadenylation signal sequences, which were absent in the female clones (Fig. 1).
Fig. 1.
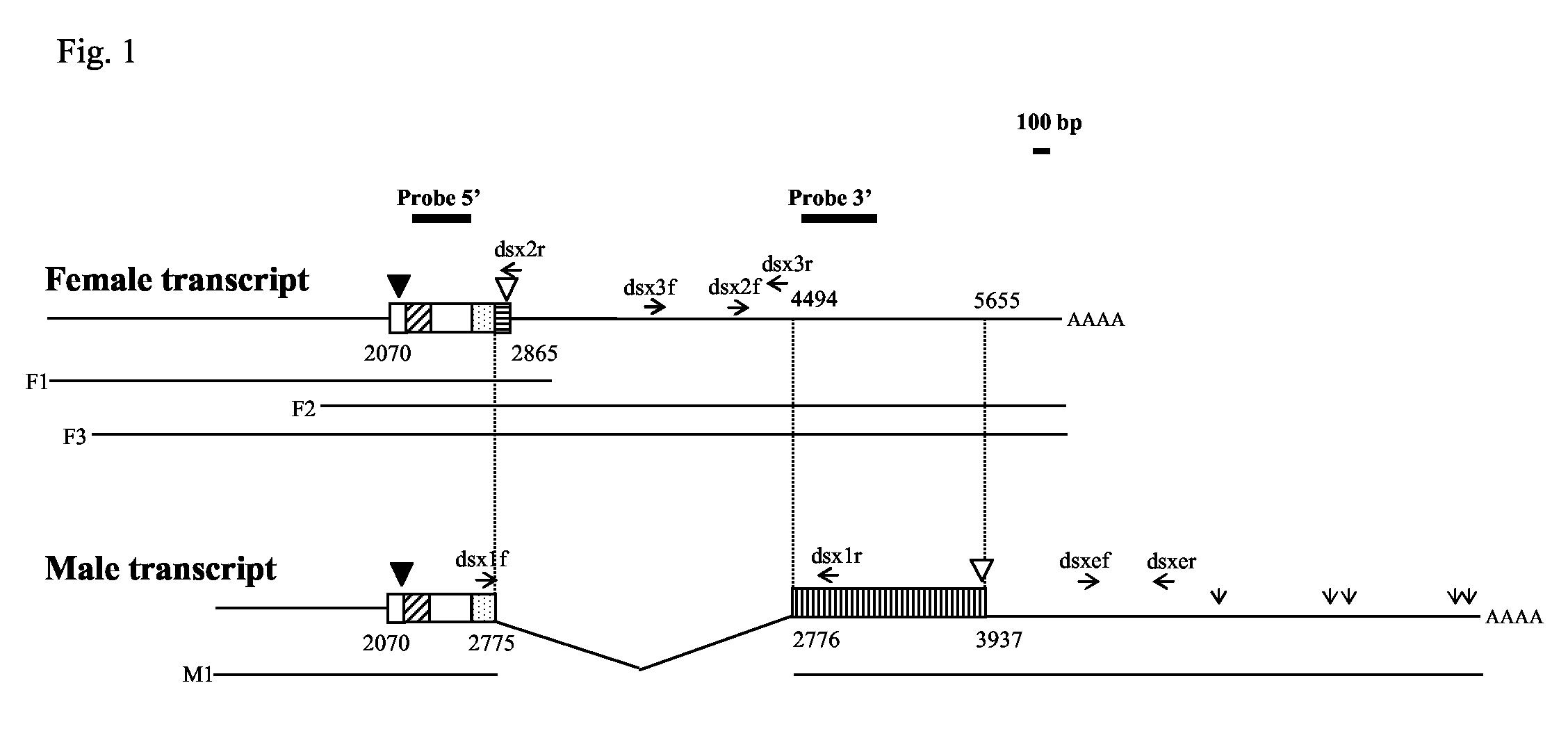
Schematic representation of the putative female (F1, F2 and F3) and male (M1) Agdsx transcripts isolated from a mixed pupal A. gambiae cDNA library. The sequence of the putative female transcript was deduced by combining the information provided by clones F1, F2 and F3, while the sequence of the putative male transcript was based on clone M1. The diagonally hatched box represents the DBD/OD1 domain, and the stippled box represents the non-sex-specific region of the OD2 domain. The putative female- and male-specific regions are indicated by a horizontal and a vertical hatched box, respectively. Black and white triangles indicate the position of the initiation and stop codon, respectively. Open vertical arrows in the male transcript indicate polyadenylation signal sequences. Numbers indicate the distance in bp from the start of clone F1, which was arbitrarily considered as the start for both female and male transcripts. The location of Probe 5’ and Probe 3’ is indicated by solid lines. Primers used in RT-PCR experiments are indicated as horizontal arrows. The bar (100 bp) indicates the scale of the figure.
Analysis of the Agdsx sex-specific transcripts
RT-PCR analyses were performed to confirm the structure of the male and female transcripts (Fig. 2). A first set of primers was designed to flank the 1.7 kb region present in female clones F2 and F3 but absent in male clone M1 (for primer locations see Fig. 1). RT-PCRs using primers dsx1f and dsx1r amplified a 125 bp product in male adults, consistent with excision of this region in the mature transcript as predicted by the analysis of the cDNA (Fig. 2A). It was not possible to amplify the expected 1.7 kb fragment in females, possibly due to the length of the transcript. However, RT-PCR reactions performed with primers dsx1f and dsx1r, in combination with primers dsx2r and dsx2f respectively, produced amplification products in females that were consistent with the structure of the putative female transcripts. These combinations of primers did not generate any product in males (Fig. 2B,C). Similarly, RT-PCR experiments carried out using primers dsx3f and dsx3r, which anneal within the 1.7 kb region, amplified a 475 bp band in female, but not male, adults (Fig. 2D). All PCR products were cloned and their sequences were shown to match the results obtained from female and male cDNA clones. No amplification was detected in the absence of reverse transcription (data not shown). These results strongly indicate that the 1.7 kb region containing the female-specific exon is spliced out in the mature male transcript, a situation observed in other dsx homologues (Hediger et al., 2004; Kuhn et al., 2000; Ohbayashi et al., 2001; Saccone, 1996; Shearman and Frommer, 1998). An additional RT-PCR reaction was performed with primers dsxef and dsxer (Fig. 1), annealing on the 3’ extension present in the cDNA clone M1 but not in the female clones. Interestingly, these primers amplified a band of 379 bp from both female and male adult cDNA, indicating that the mature transcripts in both sexes share similar 3’ ends (Fig. 2E). This suggests the AgdsxF clones isolated from the cDNA library were not full-length at the 3’ end, in agreement with the observation that they lacked polyadenylation signals.
Fig. 2.

RT-PCR and Northern blot analyses of total A. gambiae RNA. A-F): RT-PCR products were amplified from female and male adult A. gambiae total RNA using the following combinations of primers: A) dsx1f and dsx1r; B) dsx2f and dsx1r; C) dsx1f and dsx2r; D) dsx3f and dsx3r; E) dsxef and dsxer; F) S7for and S7rev (which amplify the housekeeping ribosomal S7 gene as a control). For primer locations see Fig. 1. G) Northern blot analysis was performed on total RNA (10μg) extracted from male and female A. gambiae adults. Lane 1: female adult; lane 2: male adult. Upper panel: hybridization performed with Probe 5’; lower panel: standardization with probe S7. The molecular size is shown in kb.
To determine whether sex-specific transcripts of dsx were present in A. gambiae, Northern blot hybridisation was performed. Probe 5’ was hybridized to total RNA from male and female adults (Fig. 2G). In female adults, two transcripts of approximately 9 kb and 8.2 kb were detected, which possibly corresponded to identical transcripts resulting from the use of polyadenylation sites downstream of the 3’ end of the isolated female cDNA clones. In male adults, a single transcript of approximately 6.5 kb was detected, consistent with the size of the cDNA clone M1. Hybridization experiments performed with Probe 3’ (Fig. 1), which encompasses a 446 bp sequence downstream of the female-specific region, showed the same banding pattern observed with Probe 5’, confirming the presence of this region in both female and male transcripts (data not shown).
Genomic organization of Agdsx
Transcript analysis and genomic sequence allowed us to draft the structural organization of Agdsx, define exon-intron boundaries and characterize alternative sex-specific splicing. In Drosophila, the Dmdsx gene is spread over a 45 kb region on chromosome 3R and consists of three common exons, followed by a female-specific and two male-specific exons (Fig. 3). DmdsxF translation initiates at the AUG within exon 2 and terminates within the female-specific exon 4, while in the case of DmdsxM, translation begins at the same AUG and terminates within the first male-specific exon 5. Similarly, the Agdsx gene is spread over an 85 kb region of chromosome 2R and consists of 7 exons, the first 4 of which code for the common region of the protein (705 bp) (Fig. 3). Exon 5 (1.7 kb) is female-specific and exon 6 (1.2 kb) represents the male-specific coding sequence and flanking UTR, which is transcribed completely as UTR in the female. Finally, exon 7 (2.6 kb) is a non-coding exon in the UTR of both female and male transcripts. The complete female-specific transcript contains a 795 bp ORF, coding for a protein of 265 amino acids. By sequence comparison with Dmdsx, it can be assumed that translation initiates at an AUG within exon 2 and terminates in the female-specific exon 5 with two successive stop codons, opal and ochre (UGAUAA), a feature conserved in other Diptera (Kuhn et al., 2000). The male-specific transcript contains a much longer (1866 bp) ORF, coding for a 622 aa protein. The ORF starts at the same position in exon 2 and terminates within exon 6 (Fig. 3).
Fig. 3.
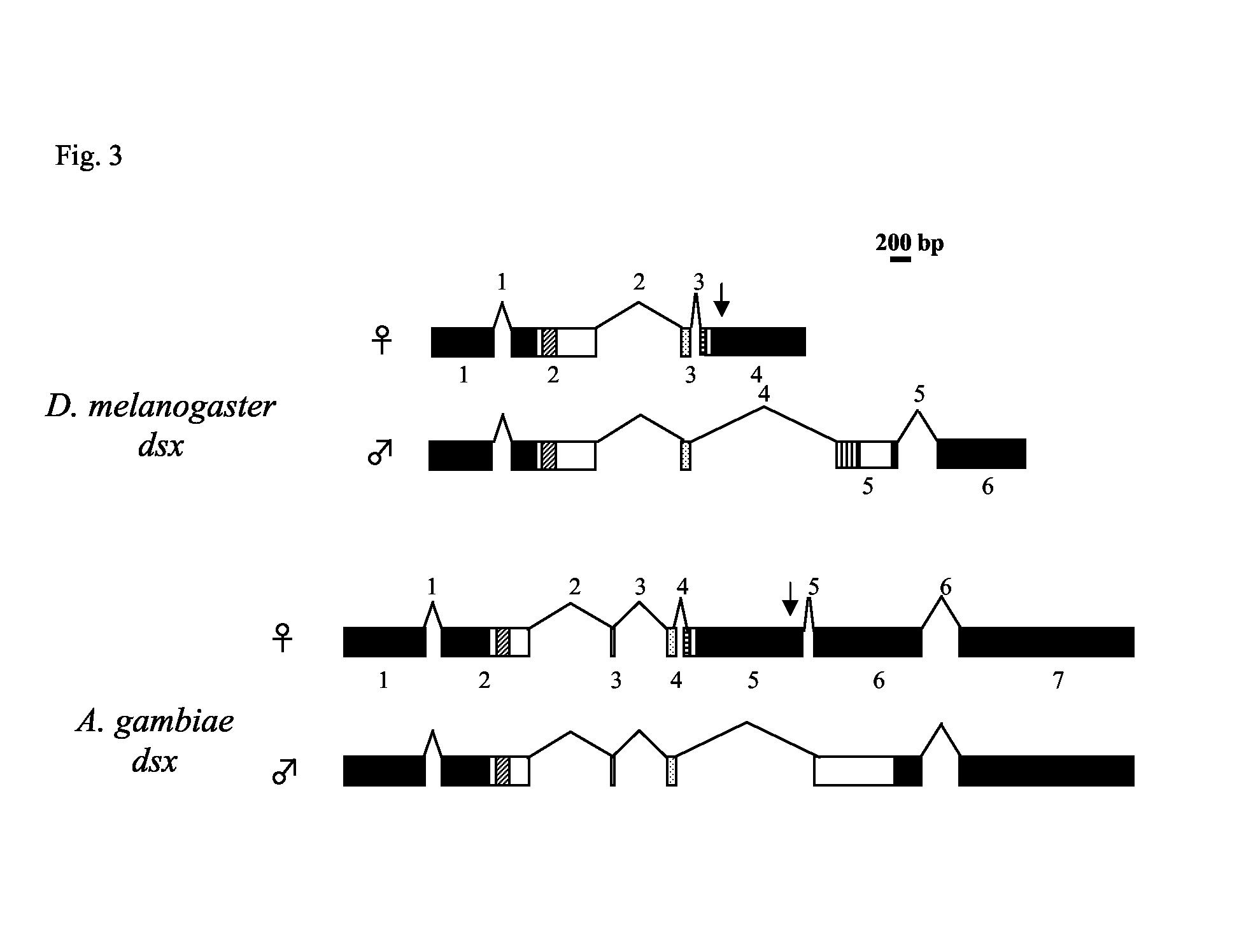
Exon/intron organization of AgdsxF and AgdsxM and comparison with the female and male transcripts of Dmdsx. Exons are shown as boxes where black boxes represent untranslated regions and open boxes represent coding regions. The OD1 domain is indicated by diagonal hatched boxes and the non-sex-specific region of the OD2 domain by stippled boxes. Male- and female-specific regions of the OD2 domains are indicated by vertical and lateral hatched boxes, respectively. In Agdsx, a male-specific region of the OD2 could not be identified. Introns and exons are numbered according to the sequence of the female transcripts. The exon/intron boundary between the last common exon and the sex-specific regions is conserved between Dmdsx and Agdsx Vertical arrows indicate the region where the dsxREs and PREs are found. The bar (200 bp) indicates the scale of the figure. Introns are shown not in scale.
Multiple sequence alignment of A. gambiae DSXM and DSXF with the DSX proteins from D. melanogaster and other Diptera shows a high degree of sequence conservation in the N-termini up to the unique zinc finger-like DBD/OD1 domain and the common part of the OD2 domain, whose end marks the beginning of the sex-specific regions (Fig. 4). Furthermore, six residues (Cys, His, His, Cys, Cys, and Arg) within the DBD/OD1 that have been shown to be essential for DNA-binding activity in D. melanogaster are conserved in Agdsx (Erdman and Burtis, 1993). The C-terminal regions of the DSXF proteins show very high sequence conservation among the different insects. A sequence corresponding to the OD2 domain in the female region is very highly conserved and the stop signal (UGAUAA) is identical among the different species. On the other hand, multiple alignments show very little similarity in the DSXM regions. One common feature that can be observed is the greater length of the male-specific regions when compared to the female-specific parts. Several of the DSXM orthologues contain strings of repeated amino acids and AgDSXM contains regions rich in proline, serine, glutamic acid and threonine (PEST regions), a characteristic observed in DmDSXM (Fig. 4).
Fig. 4.

Multiple sequence alignment of DSX homologues from D. melanogaster, B. tryoni, M. domestica, M. scalaris, B. mori and A. gambiae. The sequence is divided into: (A) the region that is common to DSXF and DSXM; (B) the conserved female-specific parts; (C) the highly divergent male-specific regions. The DBD/OD1 and OD2 domains are indicated. Black boxes represent areas of amino acid identity; shaded boxes represent areas of amino acid similarity; the asterisks * indicate 6 amino acids whose replacement has been shown to abolish DNA-binding activity in D. melanogaster.
Agdsx is a single copy gene
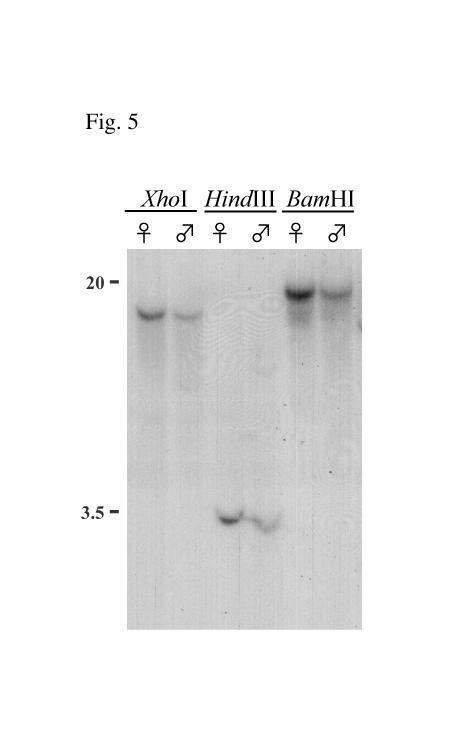
Southern blot hybridisation was performed in order to determine whether sex-specific transcripts of Agdsx are the result of alternative splicing of a single gene or are transcribed from two similar genes. The 5’ probe (see Fig. 1 for location) was hybridized to genomic DNA of male and female A. gambiae adults digested with 3 restriction enzymes that do not cut within the probe sequence. Only one genomic band was observed in each lane and no size differences were observed in the sexes, suggesting that Agdsx is most likely a single-copy gene (Fig. 5). Therefore, the sex-specific Agdsx transcripts are the result of differential splicing of the same primary transcript.
Fig. 5.
Determination of Agdsx gene copy number in male and female genomic DNA. Genomic DNA (5 μg) extracted from male and female A. gambiae was digested with (a) XhoI (b) HindIII and (c) BamHI and electrophoresed on a 1% agarose gel. Hybridization was performed with a radiolabelled Agdsx Probe 5’ that has no restriction site recognized by the three restriction enzymes. The molecular size is shown in kb.
Agdsx regulatory elements for the splicing of the female-specific exon
In Dmdsx, the inclusion or the exclusion of the female-specific exon depends on the presence of a weak 3’ splice site in the preceding intron. This weak 3’ splice site contains a polypyrimidine tract that is interrupted by purine residues (only 6 residues out of 12 are pyrimidines), making it inefficiently recognized by the splicing machinery. It is thought that TRA and TRA2 activate this weak 3’ splice site by binding to nearby dsxREs and PRE elements present in the 3’ UTR of the female-specific exon, to bring about the female mode of splicing (Inoue et al., 1992; Lynch and Maniatis, 1995). The absence of TRA in males leads to a default male-specific splicing in which the weak 3’ splice site is not recognized and the next downstream acceptor site is chosen by the splicing machinery, causing the splicing out of the female-specific exon. Genomic sequence and transcript analysis indicate that in A. gambiae, splicing into sex-specific transcripts does not depend on the choice of an alternative 3’ splice site. In Agdsx, two introns flank the female-specific region (exon 5, Fig. 3) to form a cassette exon, which is either retained (in females) or excised (in males). Importantly, the 3’ acceptor site preceding exon 5 does not appear to be weak, as its polypyrimidine stretch (8/12 pyrimidine residues) does not deviate significantly from the consensus number for splice acceptor sites (8.69), which is based on a tabulation of 215 A. gambiae intron sequences (Table 1). Furthermore, although a group of six 13 nt long sequences with 9-11 nt identity with the dsxRE from D. melanogaster and two potential PREs are found in the 3’ UTR of exon 5, they are located immediately upstream of the 5’ donor site of the following intron (intron 5), much further downstream from the 3’ acceptor splice site of intron 4 than in D. melanogaster (Table 2). These observations suggest that a different type of regulation may be occurring in A. gambiae, with the female splicing pattern brought about by the activation of the 5’ donor site of intron 5 through the action of TRA and TRA2.
Table 1.
Splice acceptor sequences of Agdsx introns. The intron/exon boundaries at the 3′ of intron 2, 3, 4 and 5 were amplified from genomic A. gambiae DNA, cloned and sequenced to analyze the 3′ splice acceptor sites. The number of pyrimidines in the 12 bp preceding the 3′ acceptor site (NYAG) is indicated. The consensus number of pyrimidines (8.69) is derived from the analysis of 215 A. gambiae splice acceptor sites. Intron 4 represents the intron preceding the female-specific exon 5 and its polypyrimidine stretch does not deviate significantly from the consensus, suggesting it may not be a weak acceptor site.
| Intron | Acceptor Sequences | Number of Pyr | |
|---|---|---|---|
| Agdsx | 2 | TTGCTCTCCTTT TCAG | 11 |
| 3 | TTCCGCCCCGTT TCAG | 10 | |
| 4 | TTTATGTTTAAC ACAG | 8 | |
| 5 | TGTAACCCCCAA AAAG | 7 | |
| Consensus | 8.69 ± 2.07 |
Table 2.
Regulatory elements in the 3′UTR of the female-specific exon in Agdsx. A) Sequence of six putative AgdsxRE elements with high homology to the splice enhancer dsxRE elements from Dmdsx, identified near the end of the female-specific exon 5. The upper case letters match the consensus dsxRE from Dmdsx. The distance of these elements from the 3′ splice acceptor site of intron 4 (bp to 3′) and the 5′ splice donor site of intron 5 (bp to 5′) are indicated, as well as the distance of the dsxRE region from the weak 3′ acceptor site of intron 3 in Dmdsx (Fig. 3). The dsxRE elements in Agdsx are found much further downstream from the 3′ splice acceptor site than in Drosophila, immediately upstream of the 5′ donor site of intron 5, a situation seen in the sex-specific splicing of the D. melanogaster fruitless gene (Dmfru). The D. melanogaster fruRE elements are shown as well as their distance from the alternative 3′ splice acceptor and the female-specific 5′ donor site of the following intron (33). B) Sequence of two potential purine-rich PREs identified in the region of the dsxRE in Agdsx, and their comparison with the Dmdsx PRE.
| A) | |||||
|---|---|---|---|---|---|
| Gene | dsxRE Sequence | Identity | bp to 3′ | bp to 5′ | |
| Dmdsx | UC(U/A)(U/A)CAAUCAACA | 295-566 | - | ||
| Agdsx | UCgcCgAUCAACc | 9/13 | 1176 | 544 | |
| cCAUCguUCAACc | 9/13 | 1197 | 523 | ||
| UCAACA-UCAuCg | 10/13 | 1220 | 500 | ||
| UCUcCAAUCAAuc | 10/13 | 1343 | 377 | ||
| aCAUCAAUCAAuA | 11/13 | 1606 | 114 | ||
| aCAUCAAUCAAuc | 10/13 | 1694 | 26 | ||
| Dmfru | UCAUCAAUCAACA | 13/13 | 1352 | 238 | |
| UCUUCAAUCAACA | 13/13 | 1387 | 203 | ||
| aCUUCAAUCAACA | 12/13 | 1540 | 50 | ||
| B) | |||||
| Gene | PRE Sequence | ||||
| Dmdsx | AAAGGACAAAGGACAAAA | ||||
| Agdsx | CGAGAAAAGGGGAGAGCAAA | ||||
| ACAAACGAGAGCAAGGAAAA |
Discussion
In this study, we report the isolation of the sex-specific splicing transcripts of the A. gambiae dsx gene as well as the characterization of its locus structure. Bioinformatic analysis of the A. gambiae genome identified a clone (P4060) with significant homology to Dmdsx in the functional domains (DBD/OD1, OD2) and in the female-specific coding region, which are highly conserved in other homologues. Nevertheless, comparative genomics could not help in identifying the male-specific region in Agdsx due to the lack of sequence conservation of this region amongst organisms. Furthermore, the distinctive exon/intron organization of the Agdsx locus and splicing pattern of the female and male transcripts could not be inferred from the available sequence data. Therefore, it was necessary to perform an in-depth molecular analysis to obtain the sequence of the sex-specific transcripts of Agdsx, and to understand its exon/intron organization.
Agdsx is organized in an exon/intron structure spanning an 85 kb region of chromosome 2R, with similarities to Dmdsx and homologues from other insects. The female transcript consists of a 5’ common segment, a female-specific part and a 3’ common region, while the male transcript comprises only the 5’ and 3’ common segments. The 5’ common region is organized into four exons (1-4), the female-specific segment corresponds to exon 5, while the 3’ common segment consists of exons 6 (which encodes for the male-specific part) and 7. In both females and males, translation initiates at the same start codon in exon 2, while it terminates within the female-specific exon 5 in AgdsxF transcripts and within exon 6 in AgdsxM transcripts. The male-specific region is therefore transcribed as a UTR in females, a situation seen in M. scalaris (Kuhn et al., 2000) and B. mori (Ohbayashi et al., 2001; Suzuki et al., 2001) dsx homologues.
Although this situation differs from D. melanogaster, where female transcription terminates before the appearance of male-specific exons, the composition of the proteins is the same in the two species. The AgDSXM and AgDSXF proteins consist of a common N-terminus and sex-specific C-termini. The N-terminus features a highly conserved zinc finger-like DBD/OD1 as well as the non-sex-specific part of the OD2. While the female-specific region is very highly conserved, the male-specific part shows little sequence conservation amongst all DSXM homologues. The only conserved features are the greater length with respect to the female regions, and the presence of strings of repeated amino acids and certain ‘PEST’ amino acids in some of the DSXM homologues.
Sex-specific transcripts were detected in both Northern blots and RT-PCR analyses. Two female-specific bands of different size were detected in the Northern blot experiments, probably representing transcripts containing alternative polyadenylation sites located downstream of the 3’ end of the female F2 and F3 cDNA clones. The presence of alternative polyadenylation sites has been reported in dsx homologues from other organisms (Kuhn et al., 2000).The size of the female clones isolated from the cDNA library is considerably smaller than expected from the size of the female transcripts detected in the Northern blots. However, RT-PCR experiments showed that female and male transcripts share similar 3’ regions that extend beyond the 3’ end of the female cDNA clones, suggesting that these did not contain the entire 3’ UTR. Indeed, including the 3’ UTR from clone M1 in the calculation for the length of the female cDNA transcripts results in a good match with the size of the bands detected in Northern blot analyses in female adults. (Fig. 2G). On the other hand, the cDNA clone M1 is likely to represent the full male transcript as its size is consistent with the band detected in the Northern analysis.
Apart from the highly divergent sequence of the male-specific region, the major difference between Agdsx and Dmdsx resides in the splicing mechanism of the female-specific exon. In D. melanogaster, the inclusion or excision of the female-specific exon depends on the presence of a weak 3’ acceptor site in the preceding intron. Activation of this splice site in females is brought about by TRA and TRA2, which form a multiprotein complex with RNA-binding protein 1 and bind to cis-regulatory elements (dsxREs and PRE) present in the 3’ UTR of the female-specific exon immediately downstream of the 3’ splice acceptor site (Hoshijima et al., 1991; Inoue et al., 1992; Lynch and Maniatis, 1995). In males, the absence of TRA leads to a default male-specific splicing in which the weak 3’ splice site is not recognized and the next downstream acceptor site is chosen by the splicing machinery, resulting in the splicing of the female-specific exon. On the other hand, in A. gambiae, the retention of exon 5 in females seems to depend on the activation of the 5’ donor site of the downstream intron 5. This scenario would resemble the splicing of fruitless (fru) in D. melanogaster, where the TRA/TRA2 enhancer complex activates a female-specific 5’ splice site (Lam et al., 2003). Similarly to Agdsx, fru contains repeat elements (fruREs) nearly identical to the DmdsxREs but located immediately upstream of this alternative 5’ splice donor site, approximately 1.3 kb downstream of the 3’ acceptor site of the preceding intron (Table 2A). The remarkable similarity between Agdsx and Dmfru suggests that splicing of Agdsx follows the same mechanism, with the female mode of splicing occurring though the activation of the 5’ splice site of intron 5 following the binding of TRA and TRA2 to the AgdsxREs. On the other hand, the female transcript may represent the default splicing mode, with the 5’ donor site of intron 5 repressed in males. The region preceding intron 5 shows the presence of putative silencer-binding sites such as guanosine-rich motifs (GGGG and UAGG), which are involved in the regulation of a cassette exon in the glutamate NMDA (N-methyl-D-aspartate) R1 receptor (GRIN1) (Grabowski, 2004). Alternatively, a decoy 3’ acceptor site could engage the 5’ donor site of intron 5 in a non-productive interaction, conferring in turn a competitive advantage to the skipping of exon 5, as found in the caspase-2 pre-mRNA (Cote et al., 2001).
Conservation of the exon/intron structure and of the functional regions (DBD/OD1, OD2) found in all known dsx homologues suggests a role for Agdsx in dimorphic differentiation in A. gambiae. The identification of female and male specific transcripts of Agdsx represents an important step towards the understanding of the sex differentiation process in A. gambiae and will facilitate the development of genetic tools to induce male sterility or manipulate sex ratios in mosquitoes, for instance by constitutively expressing the female-specific form of dsx in the male gonads or by inducing the sex-specific splicing of a dominant lethal.
Acknowledgements
We thank Peter Billingsley for providing the cDNA library and John Lucchesi for reading of the manuscript. We are grateful to Giuseppe Saccone, Catello Polito and Elisa Petris for helpful suggestions. FC was supported by the Wellcome Trust, CS was supported by the Wellcome Trust and CS and QL were supported by Universities UK, with The Overseas Research Students Award Scheme (ORS award).
Abbreviations
- SIT
sterile insect technique
- PRE
purine-rich enhancer
- DBD
DNA-binding domain
- OD1
oligomerization domain 1
- OD2
oligomerization domain 2
- RT-PCR
reverse-transcriptase polymerase chain reaction
- UTR
untranslated region
- ORF
open reading frame
References
- Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 1988;2:477–89. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- Benedict MQ, Robinson AS. The first releases of transgenic mosquitoes: an argument for the sterile insect technique. Trends Parasitol. 2003;19:349–55. doi: 10.1016/s1471-4922(03)00144-2. [DOI] [PubMed] [Google Scholar]
- Boggs RT, Gregor P, Idriss S, Belote JM, McKeown M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987;50:739–47. doi: 10.1016/0092-8674(87)90332-1. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, Crisanti A. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405:959–62. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- Chandler D, McGuffin ME, Piskur J, Yao J, Baker BS, Mattox W. Evolutionary conservation of regulatory strategies for the sex determination factor transformer-2. Mol Cell Biol. 1997;17:2908–19. doi: 10.1128/mcb.17.5.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline TW. The Drosophila sex determination signal: how do flies count to two? Trends Genet. 1993;9:385–90. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- Cote J, Dupuis S, Wu JY. Polypyrimidine track-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J Biol Chem. 2001;276:8535–43. doi: 10.1074/jbc.M008924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci U S A. 1996;93:9004–9. doi: 10.1073/pnas.93.17.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. Embo J. 1993;12:527–35. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gare DC, Piertney SB, Billingsley PF. Anopheles gambiae collagen IV genes: cloning, phylogeny and midgut expression associated with blood feeding and Plasmodium infection. Int J Parasitol. 2003;33:681–90. doi: 10.1016/s0020-7519(03)00055-9. [DOI] [PubMed] [Google Scholar]
- Grabowski PJ. A molecular code for splicing silencing: configurations of guanosine-rich motifs. Biochem Soc Trans. 2004;32:924–7. doi: 10.1042/BST0320924. [DOI] [PubMed] [Google Scholar]
- Hediger M, Burghardt G, Siegenthaler C, Buser N, Hilfiker-Kleiner D, Dubendorfer A, Bopp D. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol. 2004;214:29–42. doi: 10.1007/s00427-003-0372-2. [DOI] [PubMed] [Google Scholar]
- Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991;252:833–6. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci U S A. 1992;89:8092–6. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Sievert V, Traut W. The sex-determining gene doublesex in the fly Megaselia scalaris: conserved structure and sex-specific splicing. Genome. 2000;43:1011–20. doi: 10.1139/g00-078. [DOI] [PubMed] [Google Scholar]
- Lam BJ, Bakshi A, Ekinci FY, Webb J, Graveley BR, Hertel KJ. Enhancer-dependent 5’-splice site control of fruitless pre-mRNA splicing. J Biol Chem. 2003;278:22740–7. doi: 10.1074/jbc.M301036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–93. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- Meise M, Hilfiker-Kleiner D, Dubendorfer A, Brunner C, Nothiger R, Bopp D. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development. 1998;125:1487–94. doi: 10.1242/dev.125.8.1487. [DOI] [PubMed] [Google Scholar]
- Muller-Holtkamp F. The Sex-lethal gene homologue in Chrysomya rufifacies is highly conserved in sequence and exon-intron organization. J Mol Evol. 1995;41:467–77. doi: 10.1007/BF00160318. [DOI] [PubMed] [Google Scholar]
- Nayler O, Cap C, Stamm S. Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics. 1998;53:191–202. doi: 10.1006/geno.1998.5471. [DOI] [PubMed] [Google Scholar]
- O'Neil MT, Belote JM. Interspecific comparison of the transformer gene of Drosophila reveals an unusually high degree of evolutionary divergence. Genetics. 1992;131:113–28. doi: 10.1093/genetics/131.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi F, Suzuki MG, Mita K, Okano K, Shimada T. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol. 2001;128:145–58. doi: 10.1016/s1096-4959(00)00304-3. [DOI] [PubMed] [Google Scholar]
- Pane A, Salvemini M, Delli Bovi P, Polito C, Saccone G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development. 2002;129:3715–25. doi: 10.1242/dev.129.15.3715. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999a;215:208–20. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, Kusz K, Jaruzelska J, Reinberg Y, Flejter WL, Bardwell VJ, et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum Mol Genet. 1999b;8:989–96. doi: 10.1093/hmg/8.6.989. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–5. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- Saccone G, Peluso I, Artiaco D, Giordano E, Bopp D, Polito LC. The Ceratitis capitata homologue of the Drosophila sex-determining gene sex-lethal is structurally conserved, but not sex-specifically regulated. Development. 1998;125:1495–500. doi: 10.1242/dev.125.8.1495. [DOI] [PubMed] [Google Scholar]
- Saccone G, Peluso I, Testa G, Di Paola Pane A. In Enhancement of the Sterile Insect Technique through Genetic Transformation using Nuclear Techniques. IAEA/FAO; Vienna: 1996. Drosophila sex-lethal and doublesex homologues genes in Ceratitis capitata: searching for sex-specific genes to develop a medfly transgenic sexing strain. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Laboratory; NY: 1989. [Google Scholar]
- Segade F, Hurle B, Claudio E, Ramos S, Lazo PS. Molecular cloning of a mouse homologue for the Drosophila splicing regulator Tra2. FEBS Lett. 1996;387:152–6. doi: 10.1016/0014-5793(96)00496-6. [DOI] [PubMed] [Google Scholar]
- Shearman DC, Frommer M. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol Biol. 1998;7:355–66. doi: 10.1046/j.1365-2583.1998.740355.x. [DOI] [PubMed] [Google Scholar]
- Sievert V, Kuhn S, Paululat A, Traut W. Sequence conservation and expression of the sex-lethal homologue in the fly Megaselia scalaris. Genome. 2000;43:382–90. doi: 10.1139/g99-132. [DOI] [PubMed] [Google Scholar]
- Suzuki MG, Ohbayashi F, Mita K, Shimada T. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem Mol Biol. 2001;31:1201–11. doi: 10.1016/s0965-1748(01)00067-4. [DOI] [PubMed] [Google Scholar]
- Wilkins AS. Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays. 1995;17:71–7. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]