Abstract
Phase variation of the outer membrane protein Ag43 in E. coli requires deoxyadenosine methylase (Dam) and OxyR. Previously, it was shown that OxyR is required for repression of the Ag43-encoding gene, agn43, and that Dam-dependent methylation of three GATC target sequences in the regulatory region abrogates OxyR binding. Here we report further characterization of agn43 transcription and its regulation. Transcription was initiated from a σ70-dependent promoter at the G residue of the upstream GATC sequence. Template DNA and RNA polymerase were sufficient to obtain transcription in vitro, but DNA methylation enhanced the level of transcription. Analyses of transcription in vivo of agn′-lacZ with mutated Dam target sequences support this conclusion. Since methylation also abrogates OxyR binding, this indicates that methylation plays a dual role in facilitating agn43 transcription. In vitro transcription from an unmethylated template was repressed by OxyR(C199S), which resembles the reduced form of OxyR. Consistent with this and the role of Dam in OxyR binding, OxyR(C199S) protected from DNase I digestion the agn43 regulatory region from −16 to +42, which includes the three GATC sequences. Deletion analyses of the regulatory region showed that a 101-nucleotide region of the agn43 regulatory region containing the promoter and this OxyR binding region was sufficient for Dam- and OxyR-dependent phase variation
Phase variation is a regulatory mechanism that allows for heritable but reversible gene expression, which occurs in both gram-positive and gram-negative bacterial species. It often, but not exclusively, regulates the expression or modification of adhesins and other outer membrane proteins. Thus, a cell can switch between a state expressing the phase-variable protein (ON) and a state in which the protein is not expressed (OFF). The frequency of this switch varies and is typical of the regulatory mechanism involved. A variety of mechanisms for phase variation have been identified, and most of them involve a change in the DNA sequence. However, in Escherichia coli and Salmonella spp. phase variation can occur without a DNA sequence change (19, 25). This mechanism, instead, requires DNA modification by deoxyadenosine methylase (Dam).
One of the genes regulated by Dam-dependent phase variation in E. coli is agn43, which codes for the outer membrane protein Ag43. Expression of Ag43 induces autoaggregation of the cells, which led to its original designation as the “fluffing” (flu) locus, and is required for mature biofilm formation under certain growth conditions (11, 12, 15, 17, 18, 22). Ag43 expression is coordinately regulated with fimbrial expression, so that if fimbriae are expressed Ag43 expression is repressed (36). A family of highly homologous agn43 genes have been identified in genomes of various E. coli isolates and in Shigella flexneri (1, 34).
Phase variation of agn43 is regulated at the transcriptional level and requires Dam and the oxidative stress response protein OxyR (15, 18). Dam is the maintenance methylase in E. coli and methylates the adenine residue of GATC sequences. There is no cognate restriction enzyme for Dam and no known demethylating activity. Thus, most if not all of the 18,000 GATC sequences in the E. coli genome are methylated. An E. coli dam mutant is viable, but several important cellular processes, like mismatch repair and proper timing of initiation of DNA replication, require Dam expression (27). The second regulator of Ag43 phase variation is OxyR, which is a DNA binding protein that acts as a global regulator to mediate protection against oxidative stress. Specifically, in the presence of hydrogen peroxide, OxyR becomes oxidized, which results in a change in its structure, which in turn contributes to a change in protein-DNA contacts. Members of the OxyR regulon are, in general, either repressed or activated by specifically one form of OxyR or the other (9, 40, 42, 47).
In the current model for Ag43 phase variation, OxyR represses agn43 transcription by binding to a region upstream of the coding sequence, resulting in the OFF phase. The ON phase is obtained by methylation of the three GATC sequences in the regulatory region, which abrogates the binding of OxyR at that GATC-containing region (19). This model is supported by the observation that in an oxyR mutant as well as in an isolate in which Dam is overexpressed, Ag43 expression is locked ON. In contrast, in a dam mutant expression is locked OFF (15, 18). In addition, OxyR can bind to the agn43 regulatory region, but only when three GATC sequences in the regulatory region are unmethylated. Consistent with this, the methylation state of the agn43 GATC sequences correlates to the expression phase. Specifically, the sequences are unmethylated in phase OFF cells but methylated in oxyR mutant locked-ON cells, as well as phase ON cells of a phase-varying isolate (15). Furthermore, OxyR binding blocks Dam-dependent methylation in vitro (10). At present agn43 is the only identified gene that is under the control of phase variation in a Dam- and OxyR-dependent manner. No accessory regulators have yet been identified (18, 19).
To further understand the regulatory mechanism of Ag43 phase variation, we characterized agn43 transcription and the roles of Dam and OxyR. The results presented here show that Dam- and OxyR-dependent phase variation requires only a promoter and an appropriately positioned OxyR binding site that contains Dam target sequences.
MATERIALS AND METHODS
Growth conditions and strains.
Luria-Bertani (LB) broth, LB agar, and M9 media and agar were prepared as described previously (26, 38). Glycerol was added as a carbon source to M9 medium at 0.2% (vol/vol), unless mentioned otherwise. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside was added as required. Antibiotics were added to the media at the following concentrations: kanamycin, 30 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 34 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 15 μg/ml. To obtain single colonies of isolates containing the oxyR::Ω(Spec) mutation, 75 μl of a 10-mg/ml solution of catalase (Sigma Biochemicals) was spread on the surface of the LB agar.
The strains and plasmids used are listed in Table 1. Not included in this table are the dam-13::Tn9 oxyR::Ω(Spec) or dam oxyR::Ω(Spec) double-mutant derivatives that were constructed for this study; they are listed with their genotypes in Tables 2, 3, and 4; nor are the derivatives containing pTP166 listed in this table. Details on plasmids and strains that were made as intermediates in the construction of the single-copy lysogens containing the agn′-lacZ transcriptional fusions are available on request. The numbering of the agn43 regulatory region in the strain description (Table 1) follows that of GenBank accession number AE000291 (3). In the text, reference to specific nucleotide locations is also made relative to the transcription start site (+1), which we identified at nucleotide (nt) 9251.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or Reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD ΔlacU169 rpsL thi-1 | 8 |
| MV198 | MC4100 λMV195 lysogen, [agn(8785-9525)′-lacZ] | 15 |
| MV201 | MV198 oxyR:: Ω(Spec) | 15 |
| MV202 | MV198 dam-13::Tn9 | 15 |
| MV203 | MV198 pTP166 | 15 |
| MV253 | MC4100 pTP166 | This study |
| MV289 | MC4100 [agn(8785-9525)(AATC-III)′-lacZ]; λMV284 (λRS45-pMV124 recombinant) lysogen | This study |
| MV301 | MC4100 λMV139 lysogen, [agn(9212-9525)′-lacZ] | This study |
| MV310 | MC4100 [agn(8785-9525)(AATC-II)′-lacZ]; λMV294 (λRS45-pMV130 recombinant) lysogen | This study |
| MV336 | MC4100 [agn(8785-9525)(GATG-I)′-lacZ]; λMV330 (λRS45-pMV137 recombinant) lysogen | This study |
| MV348 | MC4100 λMV139 lysogen [agn(9296-9525)′-lacZ] | This study |
| MV353 | MC4100 [agn(8785-9525)(GATG-I; AATC-II, AATC-III)′-lacZ]; λMV142 (λRS45-pMV142 recombinant) lysogen | This study |
| MV365 | MC4100 λMV141 lysogen, [agn(9248-9525)′-lacZ] | This study |
| MV383 | MV203 katF13::Tn10 | This study |
| MV415 | RZ201 λMV198 lysogen (F− Δ(lac-proAB)x111ara thi rpsL CSH26 recA Strr) | 21, this study |
| MV416 | RZ6520 λMV198 lysogen (F− Δ(lac-proAB)x111ara thi rpsL CSH26 recA StrrrpoD800 zgh-3075:Tn10) | 21, this study |
| MV425 | MV415 pTP166 | |
| MV426 | MV416 pTP166 | This study |
| MV441 | MC4100 λMV145 lysogen (agnK1′-lacZ); contains E. coli K1 agn43 DNA homologous to nt 8785-9525 from E. coli K-12 | This study |
| MV442 | MV441 dam-13::Tn9 | This study |
| MV444 | MV441 oxyR:: Ω(Spec) | This study |
| MV496 | pMV151 in XL-1 Blue | This study |
| MV497 | pMV151 in GM2929 (dam-13::Tn9 dcm-6 hsdR2 recF143 McrA− McrB−) | This study; Marinus (unpublished) |
| MV500 | MC4100 λMV149 lysogen, [agn(8785-9312)′-lacZ] | This study |
| MV556 | MC4100 λMV167 lysogen, [agn(9212-9312)′-lacZ] | This study |
| UM122 | HfrH thi katF13::Tn10 | 30 |
| Plasmids | ||
| pTP166 | pBR322 with dam under Tac promoter control | 28 |
| pRLG593 | pKM2 flanked by terminator sites (pRLG770) containing an EcoRI-HindIII fragment with the lacUV5 promoter region | 35 |
| pMV124 | pRS550 with agn(8785-9525) insert containing a point mutation at nt 9283 resulting in agn(AATC-III) | 39, this study |
| pMV130 | pRS550 with agn(8785-9525) insert containing a point mutation at nt 9270 resulting in agn(AATC-II) | 39, this study |
| pMV137 | pRS550 with agn(8785-9525) insert containing a point mutation at nt 9254 resulting in agn(GATG-I) | 39, this study |
| pMV142 | pRS550 with agn(8785-9525) insert containing point mutations at nt 9254, 9270, and 9283 resulting in agn(GATG-I; AATC-II, AATC-III) | 39, this study |
| pMV151 | pRLG770 with an EcoRI-HindIII fragment containing agn43(8785-9389). | 35, this study |
TABLE 2.
Quantitation of LacZ expression of isolates with agn′-lacZ fusions containing deletions of the regulatory region as shown in Fig. 4
| Strain | Colony phenotypea | Relevant genotypeb | Mean ± SDc β-galactosidase activity (MU) | % Lac+d |
|---|---|---|---|---|
| MV348 | agn(9296-9525)′-lacZ | 4 | NAd | |
| MV356 | MV348 dam-13::Tn9 | 6 | NA | |
| MV357 | MV348 oxyR::Ω(Spec) | 3 | NA | |
| MV358 | MV348 pTP166 | 2 | NA | |
| MV365 | agn(9248-9525)′-lacZ | 6 | NA | |
| MV367 | MV365 dam-13::Tn9 | 4 | NA | |
| MV366 | MV365 oxyR::Ω(Spec) | 6 | NA | |
| MV368 | MV365 pTP166 | 19 ± 1 | NA | |
| MV301 | Lac+ | agn(9212-9525)′-lacZ | 656 ± 22 | 74 |
| Lac− | 25 ± 1 | 7 | ||
| MV311 | MV301 dam-13::Tn9 | 10 ± 1 | NA | |
| MV317 | MV301 oxyR::Ω(Spec) | 1,039 ± 62 | NA | |
| MV312 | MV301 pTpP166 | 795 ± 79 | NA | |
| MV500 | Lac+ | agn(8785-9312)′-lacZ | 86 ± 3 | 92 |
| Lac− | 15 ± 1 | 18 | ||
| MV501 | MV500 dam-13::Tn9 | 7 ± 3 | NA | |
| MV502 | MV500 oxyR::Ω(Spec) | 81 ± 8 | NA | |
| MV505 | MV500 pTP166 | 138 ± 6 | NA | |
| MV556 | Lac+ | agn(9212-9312)′-lacZ | 43 ± 3 | 95 |
| Lac− | 7 | 13 | ||
| MV557 | MV556 pTP166 | 68 ± 4 | NA | |
| MV201 | MV198 oxyR::Ω(Spec) | 1,312 ± 100 | NA | |
| MV203 | MV198 pTP166 | 906 ± 88 | NA |
Cultures of phase-varying isolates were started with a Lac+ or Lac− colony as indicated.
Isolates containing pTP166 have an increased level of Dam activity.
SD was less than 1 MU if no value for the SD is given.
The percentage of Lac+ cells at the time of the assay. NA, not applicable; colonies have uniform Lac phenotype.
Genetic techniques and methodology.
Standard genetic manipulations and techniques were performed as described previously (26, 29). Sequencing was performed by the Genetics Core Facility at the University of Pennsylvania. Chromosomal mutations were introduced into isolates using phage P1L4-mediated transduction as described elsewhere (29). The dam-13::Tn9 and oxyR::Ω(Spec) alleles were introduced into isolates by P1L4-mediated transduction of the resistance markers from GM2929 (M. Marinus, unpublished data) and N9716 (B. Gillette, unpublished data), respectively.
All transcriptional analyses were done with isolates with a single-copy transcriptional fusion of the regulatory region of agn43 to lacZ on the chromosome. The agn′-lacZ fusions were constructed by cloning relevant PCR-derived BamHI-EcoRI fragments into pRS550 (39). Forward and reverse primers had additional sequences at the 5′ end containing BamHI and EcoRI sites, respectively, to facilitate cloning into pRS550. Lysogens of recombinant λ containing the agn′-lacZ fusions were isolated as described previously (39).
Point mutations in the GATC sequences of the agn43 regulatory region were introduced by sequential PCR (2). Separate mutagenic primers were designed for each mutation. Specifically, by using the GATC designation as depicted in Fig. 6, mutagenesis of nt 9254 changed GATC-I to GATG and mutagenesis of nt 9270 and nt 9283 changed GATC-II and GATC-III, respectively, to AATC. Single mutations were introduced into pMV116, which is pUC19 containing the same BamHI-EcoRI fragment with the agn43 (nt 8785 to 9525) region that is present as an agn′-lacZ fusion in MV198. The triple mutant was made by using the GATC-III mutant DNA as a template to introduce the GATC-II mutation. The GATC-I mutation was combined with these through cloning, making use of the unique BsaBI site. The presence of the desired mutations was confirmed by sequencing. Single-copy transcriptional fusions of the mutagenized BamHI-EcoRI fragments to lacZ were made as described above.
FIG. 6.
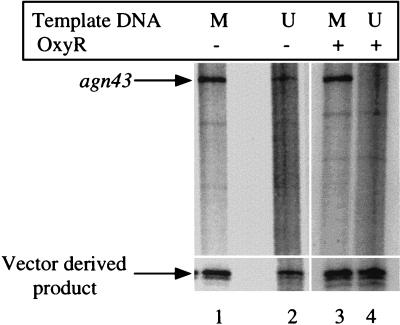
In vitro transcription in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 8 nM purified OxyR(C199S) with the following DNA templates: methylated agn43 (lanes 1 and 3) and unmethylated agn43 (lanes 2 and 4). The lower part of the figure shows the transcription obtained from the vector-derived promoter. The size of the agn43 transcription product is 220 nt.
The regulatory region of agn43 from various pathogenic and nonpathogenic E. coli isolates was amplified with primers hybridizing to the agn43 and yeeP coding sequences. We cloned and sequenced the product obtained from the E. coli K1 isolate E44, which is a spontaneous rifampin-resistant mutant derived from a neonatal meningitis isolate, E. coli K1 RS218 (45). This sequence was used to construct the agnK1′-lacZ transcriptional fusion present in MV441 and its derivatives as described above. The sequence of the agnK1 regulatory region which we obtained was identical to that in the sequenced genome of E. coli K1 RS218 (http://www.genome.wisc.edu/).
Assay of β-galactosidase activity and calculation of the switch frequency.
β-Galactosidase activities of cultures grown on LB with an optical density at 600 nm (OD600) between 0.7 and 1.0 were determined as described previously (29). The assay was performed on at least two independent cultures of each isolate, and each sample was measured in triplicate. For isolates that showed phase variation, cultures were inoculated from single colonies that were known to be in the ON or OFF phase based on the Lac phenotype. Results shown for phase-varying isolates are those of a single, representative assay. The percentage of Lac+ cells and switch frequencies were determined as previously described (6).
DNase I footprint analysis.
The DNase I footprinting assay was performed with an end-labeled 260-bp PCR-derived fragment of the agn43 regulatory region, running from nt 9129 to 9389. Assays were performed in 20 μl of final volume in OxyR binding buffer (25 mM Tris-HCl [pH 7.5], 50 mM KCl, 5 mM MgCl2, 5% glycerol, 50 μg of bovine serum albumin per ml, 1 mM dithiothreitol [DTT], 0.5 mg of herring sperm DNA per ml), with DNA at 60,000 cpm/reaction. OxyR(C199S) was added and incubated for 30 min at room temperature, after which 0.01 U of DNase I (Promega) was added. After 1 min the reaction was stopped by adding stop solution (50% glycerol, 60 mM EDTA). The DNA was recovered through precipitation with 120 μl of precipitation solution (0.75 M ammonium acetate, 90% ethanol, 36 μg of tRNA/ml). Samples were resuspended in formamide loading buffer (1 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol FF, 98% formamide) and analyzed on a 6% polyacrylamide gel in parallel with either a G/A sequencing ladder or a sequencing reaction. Gels were analyzed by using a PhosphoImager and IMAGEQUANT (Molecular Dynamics).
The DNase I protection assay was performed with OxyR(C199S) protein that was purified as described previously, by using a HiTrap heparin column (24), from MV471[oxyR:: Ω(Spec)], which contains plasmid pMV108. In pMV108 the oxyR(C199S) gene is under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)- and arabinose-inducible promoter (10).
Primer extension.
Total RNA was isolated from cultures grown in LB at 37°C when they were in mid-log phase (OD600 of 0.5 to 0.8) using Trizol Reagent (Gibco BRL). To determine the role of rpoD, MV425 and MV426 were grown in LB broth at 30°C until an OD600 of 0.5 to 0.8 was reached, at which time half of the culture was shifted to 42°C. After 1 h, total RNA was isolated by using Trizol Reagent (Gibco BRL) and used for primer extension analysis. Primer extension was performed as described previously, using 10 μg of total RNA and end-labeled, gel-purified primer oMV179 (5′-CTGTACGTATCTGTGCATGTTACTG-3′) (50,000 cpm), which starts at nt 9334 of AE000291 (2, 3). A sequencing ladder was obtained by using the fmol DNA Cycle Sequencing System (Promega). Samples were analyzed as described above using a 5% polyacrylamide gel.
In vitro transcription.
Transcription was determined from pRLG593 containing the lacUV5 promoter and its derivative pMV151 (35). A 604-bp fragment (nt 8785 to 9389) of agn43 DNA was PCR amplified with primers containing an EcoRI and a HindIII restriction site at the 5′ ends of the forward and reverse primers, respectively. The product was cloned into pRLG770, resulting in plasmid pMV151. This agn43 region consists of 466 bp upstream of the +1 (nt 9251) start site and 138 bp of the leader region. Multiple-round in vitro transcription reactions were performed at 37°C for 20 min as described earlier, with minor variations (14). Reaction volumes (25 μl) contained 2 nM supercoiled DNA template, 50 mM Tris-HCl, 200 mM KCl, 10% glycerol, 1 mM DTT, 50 μg of bovine serum albumin per ml, 0.1 mM EDTA, 5 mM MgCl2, 40 μM ATP, 5 μM GTP, 5 μM CTP (Roche Molecular Biochemicals), 5 μM [32P]UTP (Amersham), 1 U of RNase inhibitor (Promega) per μl, and 2 nM E. coli RNA polymerase (Roche). Transcription products were purified by extracting once with phenol-chloroform-isoamyl alcohol and precipitated with 1 ml of 100% ethanol, 0.35 M ammonium acetate, and 0.02 μg of tRNA per ml. Samples were analyzed on a 5% polyacrylamide gel as described above.
In vitro transcription was performed with both methylated and unmethylated pMV151 DNA and methylated pRLG593 as templates. Plasmid pMV151 was isolated from MV497(dam-13::Tn9) to obtain unmethylated DNA and from MV496 for methylated template. To ensure complete methylation of pMV151 isolated from MV496, the plasmid was further methylated in vitro using Dam according to instructions (New England Biolabs). The effect of OxyR on in vitro transcription was studied by including 8 nM of purified OxyR(C199S) in the reaction mixture described above.
RESULTS
Identification and characterization of the phase-varying agn43 promoter.
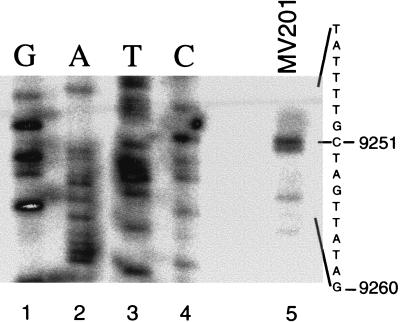
Isolates with a chromosomal agn′-lacZ fusion show phase variation of LacZ expression but are locked ON in the absence of oxyR or in the presence of plasmid pTP166, which contains the dam gene. Expression is locked OFF in a dam mutant (15). The most upstream 3′ end of the products from RNA isolated from MV201[oxyR::Ω(Spec)] corresponds to nt 9251, which is the G residue of GATC-I (Fig. 1). This product was also present in MV203(pTP166) and in ON cultures of the phase-varying isolate MV198 (results not shown). This is not specific for the reporter fusion since it was also present when the full agn43 gene at 43′ of the chromosome is present but not the reporter fusion (MV253). The products were absent from the dam mutant MV202 and from cultures of MV198 consisting predominantly of cells in the OFF phase (results not shown). A sequence highly homologous to the consensus σ70 promoter sequence is located at the −10 and −35 position relative to 9251, which is consistent with transcription initiation originating at 9251 (see below and Fig. 6).
FIG. 1.
Primer extension identifies the agn43 transcription start site at nt 9251. Lanes 1 to 4 show sequencing reactions as indicated, and in lane 5 primer extension products on mRNA isolated from MV201[oxyR::Ω(Spec)] are shown. The corresponding sequence of the template strand of agn43 and relevant nucleotide number (AE000291) are indicated.
In the samples that contained product originating at 9251, additional primer extension products with 3′ ends at residues 9338 and 9437 were obtained with two primers that anneal further downstream in the 222-nt untranslated region (UTR) (data not shown). No recognizable promoter sequence upstream of those sites was present, and these products probably represent premature termination of the primer extension reactions. Indeed, nt 9437 is located at the base of a putative hairpin structure.
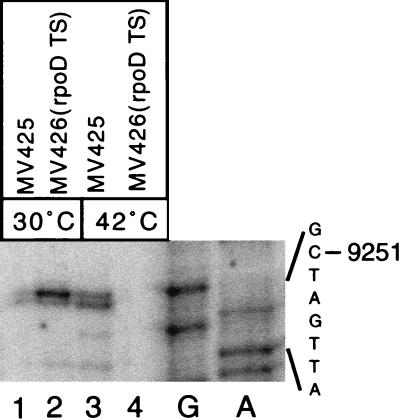
To determine if σ70 is required, we analyzed transcription in MV415 (agn′-lacZ) and its derivative with a temperature-sensitive σ70 allele, MV416 [agn′-lacZ, rpoD800(Ts)]. Based on the Lac phenotype of the colonies, transcription occurred at 30°C in both isolates but only in MV415 at 37°C, which is close to the nonpermissive temperature for RpoD(Ts) (data not shown). This was confirmed by primer extension on RNA isolated before and after a shift to the nonpermissive temperature from the locked ON derivatives MV425 (agn′-lacZ, pTP166) and MV426 [agn′-lacZ rpoD(Ts), pTP166] (Fig. 2). A product originating at 9251 was detected in each of the two isolates grown at 30°C but only in MV425 at 42°C (Fig. 2, lanes 1, 2, and 4).
FIG. 2.
Transcription of the agn′-lacZ fusion is dependent on σ70. Primer extension was performed on total RNA isolated from MV425 (lanes 1 and 3) and MV426[rpoD8000(Ts)] (lanes 2 and 4) cultures grown at 30°C (lanes 1and 2) and after 1 h at 42°C (lanes 3 and 4). The sequence of the template strand is also shown.
The stationary-phase sigma factor encoded by katF does not appear to contribute to agn43 transcription. Specifically, the levels of expression of agn′-lacZ in late logarithmic growth phase were similar in the locked-on isolate MV203(pTP166) and in its isogenic katF derivative, MV383, at 1,049 ± 63 Miller units (MU) (MV203) and 1,049 ± 52 MU (MV383) (mean ± standard deviation [SD]). Taken together, these data show that transcription originating from the phase-varying promoter originates at nt 9251 and is σ70 dependent.
The binding site of OxyR(C199S) at agn43 is consistent with its role in Dam-dependent repression.
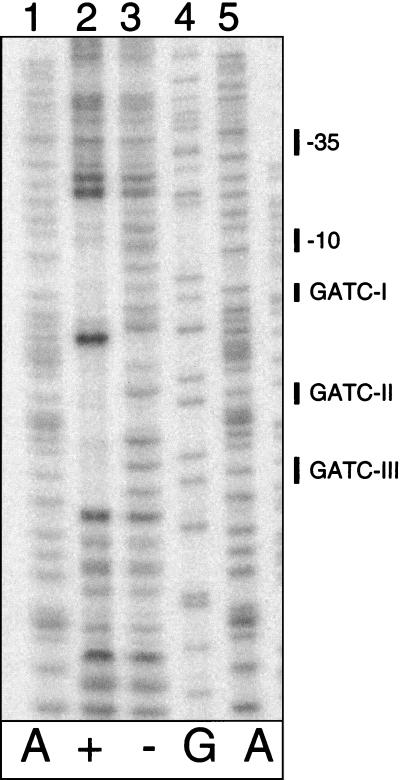
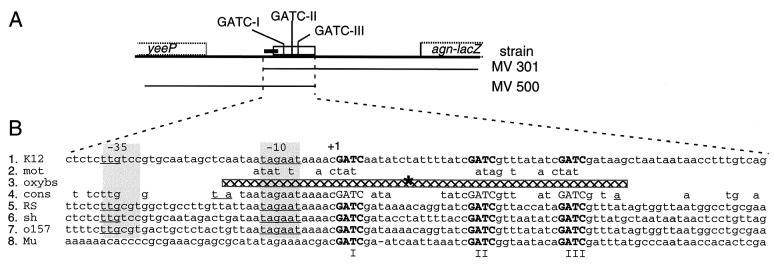
OxyR(C199S) resembles the reduced form of OxyR in DNA binding and regulatory properties, binds unmethylated agn43 DNA in vitro, and is sufficient to repress agn43 transcription in vivo (15, 42). No binding is obtained with methylated DNA (15) (J. Correnti, results not shown). The binding site of OxyR(C199S) at agn43 was determined by DNase I footprint analysis using unmethylated agn43 DNA and purified OxyR(C199S) (Fig. 3). The region of DNA that was protected from DNase I cleavage by OxyR(C199S) runs from −16 to +42 relative to the transcription initiation site, which includes the −10 region of the phase-varying promoter and the three GATC sequences. This is consistent with the fact that OxyR binding is Dam dependent and with the requirement of OxyR for repression (15, 18). The hypersensitive site in the middle of the region protected from DNase I is typical for binding of the reduced form (42). In addition, there is a high degree of conservation (15 of 20 nt) between the binding motif for reduced OxyR and the sequence at agn43 (Fig. 7B, rows 1 to 3) (42).
FIG. 3.
DNase I footprint of the bottom strand of unmethylated agn43 regulatory region in the presence (lane 2) or absence (lane 3) of OxyR(C199S) (10 nM). Sequencing ladders are shown in lanes 1, 4, and 5. The positions of the three GATC sequences and the −10 and −35 regions of the promoter are indicated.
FIG. 7.
Fifty conserved nucleotides contain the three elements required for Dam- and OxyR-dependent expression. In panel A, an overview of the regulatory region is given. Symbols are as described for Fig. 4. In panel B, the DNA sequence of the required 101-nt region is shown. The −10 and −35 regions of the promoter are shaded, and the GATC Dam target sequences are bold and capitalized. Row 1 (K12), shared sequence between MV301 and MV500; row 2 (mot), binding motif for the reduced form of OxyR, from Kullik et al. (24); row 3 (oxybs), DNase I footprint with OxyR(C199S) (Fig. 2) indicated by a hatched bar (∗, DNase I hypersensitive site); row 4 (cons), nucleotides conserved between rows 1, 5, 6, and 7 (underlined nucleotides are conserved only between rows 1 and 5); rows 5 to 8, sequences from E. coli RS218 (RS), S. flexneri (AF200692) (1) (sh), E. coli O157: H7 (O157) (AE005174) (31), and phage Mu (Mu) (V01463) (20), respectively (see text).
A 101-nt region is sufficient for Dam- and OxyR-dependent regulation.
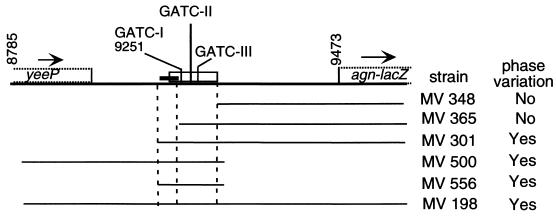
Our analyses suggested that phase variation is a result of only OxyR-dependent repression and Dam-dependent derepression and activation of the identified promoter. To support this conclusion, a minimal sequence that still contains the information required for phase variation was identified by deletion analyses (Fig. 4; Table 2). The 101-nt region that is present in the agn′-lacZ fusion of MV556 contains only the promoter region of the nt 9251 transcription initiation site and the OxyR binding region that contains the Dam target sequences (see Fig. 7). LacZ expression in MV556 is still under the control of phase variation (Fig. 4; Table 2). However, the level of expression in MV556 and the locked ON derivative MV557 was less than 15% of that obtained with the full regulatory region (MV301, MV198, and their locked ON derivatives). Further deletion analyses showed that the identified promoter but not the region upstream of this promoter is required for transcription (Table 2, MV301, MV348, MV365, and derivatives) (Fig. 4). This supports the conclusion that the identified promoter is the only one in the agn43 regulatory region. In contrast, analyses of MV500 and its derivatives showed that the 151-nt region of 5′-UTR downstream of the OxyR binding site is required for full LacZ expression (Fig. 4; Table 2). Thus, except for the OxyR binding region, the leader region is dispensable for phase variation but contributes to a high level of expression. In summary, the 101-nt region that contains the promoter and the OxyR binding site which contains the three GATC sequences is sufficient for Dam- and OxyR-dependent phase variation
FIG. 4.
A 101-nt region is sufficient for Dam- and OxyR-dependent phase variation of agn′-lacZ. Shown is an overview of the sequence included in the various deletion strains indicated by MV numbers; whether phase variation of Lac expression occurred in the isolate containing the fusion is also indicated. The relative positions of the open reading frames yeeP and the agn′-lacZ fusion and the direction of transcription (arrows) are shown, as well as the three GATC sequences in this region. The open rectangle indicates the OxyR binding region, and the closed rectangle indicates the promoter region. A partial DNA sequence of this region is presented in Fig. 7, row 1.
Methylation of the agn43 GATC sequences is required for full agn43 transcription.
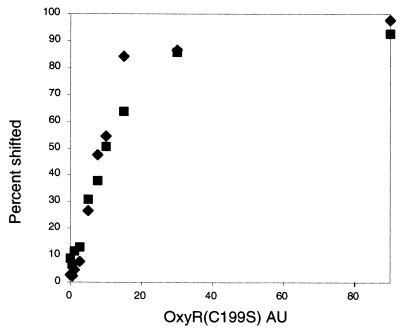
To determine whether all three GATC sequences are required for Dam-dependent phase variation , we determined the effects of point mutations in each of the GATC sequences. Specifically, the GATC-I sequence was changed to GATG, and GATC-II and GATC-III were changed to AATC sequences. The GATC-I and GATC-II mutations were introduced outside the predicted OxyR-DNA contact sites to minimize direct effects of the sequence change on OxyR binding (Fig. 7). Indeed, in an electrophoretic mobility shift assay (EMSA) with purified OxyR(C199S), no difference in binding was observed between unmethylated wild-type DNA and mutant DNA as a template (Fig. 5).
FIG. 5.
Binding of OxyR(C199S) to GATC-I, -II, and -III mutant DNA (▪) is comparable to binding to unmethylated wild-type DNA (⧫). Quantified data of an EMSA are shown.
In MV353, in which all three GATC sequences of agn′-lacZ are mutagenized, expression of LacZ was OFF and remained OFF even in the presence of the Dam-encoding plasmid pTP166 (Table 3). This was expected, since methylation cannot take place, and therefore OxyR binding cannot be abrogated. To determine whether a specific GATC sequence can confer this effect, we also analyzed the effect of each mutation separately. In an otherwise wild-type background, a single GATC-II or GATC-III mutation resulted in a locked OFF phenotype, whereas a GATC-I mutation conferred a phase-varying phenotype, but with altered switch frequencies that resulted in a preference for the OFF phase (Table 3). Thus, even a single GATC mutation causes a change in, or abrogation of, phase variation. However, introduction of the dam-encoding plasmid pTP166 in each of these mutant isolates still resulted in a locked ON agn′-lacZ phenotype (Table 3).
TABLE 3.
Lac phenotypes of colonies and switch frequencies of agn′-lacZ-containing isolates with GATC mutations in an oxyR+ dam+ background and in the presence of the Dam-encoding plasmid pTP166
| agn43 sequence | Strain | Lac phenotypea (switch frequency) | Strain with pTP166 | Lac phenotypea |
|---|---|---|---|---|
| Wild type | MV198 | ON⇒OFF (2.5 × 10−3) | MV203 | ON |
| OFF⇒ON (8.7 × 10−4) | ||||
| GATC-I mutant | MV336 | ON⇒OFF (4.0 × 10−2) | MV342 | ON |
| OFF⇒ON (4.5 × 10−4) | ||||
| GATC-II mutant | MV310 | OFF | MV315 | ON |
| GATC-III mutant | MV289 | OFF | MV295 | ON |
| GATC-I, -II, -III mutant | MV353 | OFF | MV375 | OFF |
Cultures from ON colonies had LacZ activity ranging from 1,100 to 2,000 MU; those from OFF colonies had <20 MU of LacZ activity.
Previous results indicated that, besides abrogating OxyR binding, Dam-dependent methylation plays an additional role in activating agn43 transcription, since transcription of agn′-lacZ in a dam oxyR double mutant is about 10% of that in an oxyR background (15). This could be a result of methylation of the agn43 Dam target sequences or an indirect result of a requirement of methylation elsewhere in the genome. To address this, transcription from mutant DNAs was analyzed in oxyR mutant derivatives and compared to that in oxyR dam double mutants (Table 4). The results show that when the agn43 GATC sequences are mutagenized, a dam mutation no longer caused a decrease in transcription (Table 4, MV369 and MV391). Thus, Dam facilitates agn43 transcription specifically and only through methylation of the agn43 sequences. This effect is not a result of one specific GATC sequence, since in isolates containing a single point mutation, methylation still enhances transcription (Table 4).
TABLE 4.
Levels of LacZ expression of oxyR::Ω(Spec) and dam-13::Tn9 oxyR::Ω(Spec) isolates containing an agn′-lacZ fusion with point mutations in the GATC sequences
| Strain | agn′-lacZ mutation(s) | Specific host strain mutation(s) | Mean ± SD β-galactosidase activity (MU) | Ratio of dam oxyR to oxyR |
|---|---|---|---|---|
| MV369 | GATC-I, -II, -III | oxyR | 298 ± 26 | 0.98 |
| MV391 | GATC-I, -II, -III | dam oxyR | 291 ± 22 | |
| MV343 | GATC-I | oxyR | 1311 ± 60 | 0.23 |
| MV354 | GATC-I | dam oxyR | 308 ± 34 | |
| MV316 | GATC-II | oxyR | 605 ± 27 | 0.21 |
| MV351 | GATC-II | dam oxyR | 125 ± 5 | |
| MV308 | GATC-III | oxyR | 1210 ± 13 | 0.21 |
| MV350 | GATC-III | dam oxyR | 258 ± 14 | |
| MV201 | Wild type | oxyR | 1027 ± 102 | 0.08 |
| MV230 | Wild type | dam oxyR | 81 ± 1 |
The three point mutations unexpectedly resulted in an increase of transcription, independent of any effects of methylation or OxyR binding, as is evident from a comparison of MV369(oxyR) (298 MU) and MV230(dam oxyR) (81 MU) (Table 4). This effect cannot be attributed to one specific mutation but appears to be cumulative, as is evident from the increased level of transcription compared to wild type in a dam oxyR background of each fusion carrying a single mutation (Table 4). The molecular mechanisms underlying these OxyR-independent effects of the sequence and methylation of the 5′-UTR on transcription are not known at present.
In vitro transcription analysis of agn43.
The data described above suggest that no accessory factors are required to obtain Dam-dependent transcription and OxyR-dependent repression of the agn43 promoter. This was tested by performing an in vitro transcription assay with methylated and unmethylated DNA, in the presence and absence of OxyR(C199S) (Fig. 5, lanes 1 and 2). In the data shown, the level of transcription originating from the agn43 promoter from the unmethylated template, normalized to a vector-specific band, was 50% lower than from the methylated template. In additional experiments the level obtained with an unmethylated template was consistently between 15 and 50% of the level obtained with methylated DNA. This indicates that methylation of the agn43 GATC sequences directly facilitates transcription, which is consistent with our analyses of transcription in vivo (Table 4) (15).
In vitro transcription of both methylated and unmethylated templates was also performed in the presence of OxyR(C199S). OxyR(C199S) was able to completely repress transcription of unmethylated DNA (Fig. 6, lane 4). Addition of OxyR(C199S) also caused a slight decrease in the level of transcription from methylated template. This could be attributed to a low level of binding of OxyR to methylated template in the transcription buffer, which lacks nonspecific competitor DNA, as was observed by EMSA with 8 nM OxyR(C199S) (data not shown). Taken together, these in vitro analyses show that no accessory factors are required for transcription, that Dam-dependent methylation facilitates transcription, and that OxyR(C199S) is sufficient for repression of the phase-variable agn43 promoter.
Conservation of specific nucleotides in the 101-nt region of the Ag43 family of genes.
A comparison of the sequence of the agn43 regulatory region of E. coli K-12 to that of E. coli RS218, which is a neonatal meningitis isolate (45), shows that in the 101-nt required region only 54 nt are conserved (Fig. 7, rows 1, 4, and 5). Transcription was analyzed from an agn43K1′-lacZ fusion in a MC4100 background (MV441). Colonies of MV441 had variable Lac phenotypes, but no distinct Lac− colonies were obtained, which may be a result of altered switch frequencies between the ON and OFF phase. Consistent with this hypothesis, cultures that were inoculated with a colony showing a lower level of LacZ expression yielded LacZ activity levels (1,059 ± 25 MU) very similar to those of cultures inoculated with a distinct Lac+ phenotype colony (1,379 ± 42 MU). Nevertheless, this transcription was Dam- and OxyR-dependent, because in the oxyR mutant MV444, transcription was at 1,394 ± 193 MU, whereas in the dam mutant MV442 expression was repressed (31 ± 5 MU).
Similar regulatory regions were found in the sequenced genomes of E. coli O157:H7 and S. flexneri (Fig. 7, rows 6 and 7). In both cases, the regulatory region was upstream of a gene of the agn43 family. In E. coli O157:H7 this is located in the isolate-specific pathogenicity island no. 43, whereas in S. flexneri the gene was designated sap and is located on the she pathogenicity island (1, 31). Of the 54 conserved nucleotides described above, 51 are also conserved in the S. flexneri 2a and the E. coli O157:H7 sequences. These 51 nt include the −35 and −10 regions of the promoter, the three GATC sequences, and nucleotides that are predicted to be essential for binding of OxyR.
DISCUSSION
In E. coli Dam is required for phase variation of the members of the pap-like family of fimbrial adhesins as well as of the outer membrane protein Ag43 (4, 5). Dam-dependent methylation also affects transcription of some non-phase-variable genes, for example, by directly affecting binding of RNA polymerase (27). Nevertheless, not all genes that contain a Dam target sequence within the binding site of its regulatory protein are methylation dependent (27, 43). To gain a more complete understanding of the role of Dam in transcription and phase variation, we analyzed agn43 transcription in further detail.
The agn43 promoter, which is upstream of the Dam target sequences, has a very good homology (9/12) to the canonical E. coli promoter sequence and has optimal spacing between the −10 and −35 sequence. Consistent with this, in vitro transcription occurred with only RNA polymerase and template DNA (Fig. 6). Furthermore, in vitro transcription from only the unmethylated template, which is a binding substrate for OxyR, was repressed in the presence of OxyR(C199S), consistent with the OxyR(C199S) binding region that overlaps the −10 region of the promoter and contains the Dam target sequences (Fig. 3 and 7). Thus, transcription and OxyR-dependent repression of agn43 do not require an accessory factor in vitro. This is in contrast to the only other known Dam- and OxyR-dependent promoter, which is the E. coli phage Mu mom promoter (16, 20, 32, 41). The mom promoter is located downstream of the OxyR binding site and GATC sequences and has poor homology to the canonical promoter sequence, and its transcription requires MuC. OxyR represses mom transcription by inhibiting MuC-dependent recruitment of RNA polymerase (41). Furthermore, mom expression is not known to phase vary. Thus, mom and agn43 regulation have Dam-dependent binding of OxyR in common, but their mechanisms of activation and repression differ.
Binding of OxyR to its recognition sequence is proposed to occur as a dimer of dimers (9, 42). A motif has been proposed for the binding of each of the four monomers (42, 48) (Fig. 7, row 2). The DNA contact sites of the two dimers of the reduced form of wild-type OxyR as well as of mutant OxyR(C199S) are separated by one turn of the helix, resulting in a characteristic hypersensitive site in a DNase I footprint (42). The DNase I footprint that we obtained with OxyR(C199S) and unmethylated agn43 DNA is consistent with these observations (Fig. 3 and 7). In addition, based upon this footprint, we aligned the proposed binding motif to the agn43 sequence, and there is a high degree of homology. The motifs also fall within the conserved sequence between the regulatory regions (Fig. 7, rows 1, 2, and 4). The latter suggests that OxyR-dependent regulation has been conserved.
Methylation of the agn43 GATC sequences appears to contribute to transcription of agn43 by two mechanisms. First, methylation of these sequences abrogates OxyR binding and thus alleviates OxyR-dependent repression (Fig. 6) (15). Our data indicate that each GATC sequence contributes to this effect, with a mutation in either GATC-II or GATC-III resulting in complete abrogation of phase variation, and the GATC-I mutation resulting in altered switch frequency, resulting in a preference for the OFF phase (Table 3). Access of Dam to the mutant DNAs is the limiting factor only when Dam is present at wild-type levels, since an elevated level of Dam resulted in a locked ON phenotype of all three isolates with a single GATC mutation (Table 3). Thus, in this DNA, methylation of the two remaining GATC sequences is sufficient to abrogate OxyR-dependent repression. Consistent with this, OxyR does not bind these fully methylated mutant DNAs in vitro (J. Correnti and M. van der Woude, unpublished data). The basis of the different phenotype of the GATC-I mutant is not known but, based on these results, may lie in an affinity difference for specifically hemimethylated mutant DNA.
Methylation of the GATC sequences in the 5′-UTR is also required for full transcription even in the absence of OxyR. This was observed with both in vivo and in vitro transcriptions (15) (Table 4 and Fig. 6). This enhancement of transcription does not appear to be due to a specific GATC sequence (Table 4). The effect may be mediated by a methylation-dependent change in a physical property of the DNA, like bending or melting temperature, which in turn may affect RNA polymerase binding, isomerization, or elongation (13, 33). Surprisingly, the sequence change associated with the GATC mutations also directly affected the level of transcription, independent of OxyR binding or DNA methylation (Table 4, MV391 versus MV230). The sequence of this region, like methylation, may result in changes in the physical property of the DNA. Alternatively, the effect may be mediated by changes in one of the potential stem-loop structures of the mRNA. Furthermore, the untranslated region downstream of +61 also appears to modulate the level of expression, which may involve mRNA stability (Fig. 4; Table 2). Studies are in progress to identify the molecular mechanisms that result in regulation by the length, sequence, and methylation state of the 5′-UTR of agn43.
Most members of the OxyR regulon are regulated as a result of binding of either the reduced or the oxidized form of OxyR (40, 49). In this paper we have analyzed the role of OxyR(C199S), which resembles the reduced form of wild-type OxyR in the cell that is present in the absence of oxidative stress (37). OxyR(C199S) is sufficient to repress agn43 transcription and block Dam-dependent methylation of the agn43 GATC sequences in vivo and in vitro (Fig. 6) (10, 15). However, in vitro, oxidized OxyR also binds unmethylated agn43 DNA (10, 15). Therefore, studies are in progress to analyze in further detail the role of oxidative stress and oxidized OxyR in Ag43 phase variation. This is of additional interest in light of the model for coordinated regulation of Ag43 and fimbriae recently proposed by Schembri and Klemm, a model in which fimbrial synthesis leads to repression of Ag43 expression by causing a localized oxidative stress response (36).
In summary, the data presented here indicate that the OFF phase is a result of OxyR(C199S)-dependent repression of a σ70-dependent promoter upstream of the GATC-I sequence, which is mediated by OxyR binding at unmethylated agn43 DNA. The ON phase is a result of abrogation of OxyR binding and direct enhancement of transcription, both mediated by methylation of the agn43 GATC sequences. Furthermore, the promoter and the OxyR binding site containing Dam target sequences appear to be necessary and sufficient to obtain phase variation. Based on this, a model for regulation of Ag43 phase variation is presented in Fig. 8. This differs from a model by Henderson et al. in that there is no downstream promoter, nor is there an accessory transactivating factor (18, 19).
FIG. 8.
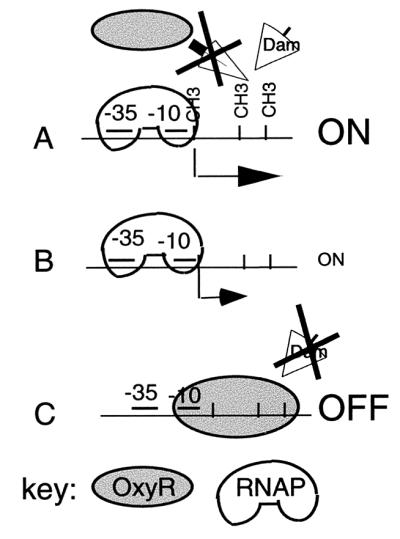
Schematic model for regulation of agn43 transcription. The GATC sequences are indicated by vertical lines, methylation is indicated by CH3, OxyR and RNA polymerase (RNAP) are as indicated, transcription is indicated by a closed arrow, and access of Dam and OxyR is indicated by an open arrow. (A) ON phase obtained with methylated sequence, which results in a maximal level of transcription. (B) ON phase obtained in the absence of OxyR and Dam-dependent methylation, which has a lower level of transcription. (C) OFF phase obtained by OxyR-dependent repression.
This study addresses the regulatory events leading to an ON or OFF phase of Ag43 expression. It is not clear what leads to a switch in expression phase, even though it appears that the outcome of competition between Dam and OxyR for the GATC sequences is the critical event (10, 15). A critical intermediate to obtain the OFF phase would be the competition occurring at the hemimethylated state of DNA, which occurs after DNA replication of fully methylated DNA present in the ON phase. Indeed, OxyR binds wild-type hemimethylated agn43 DNA, but with a decreased affinity compared to unmethylated DNA (10). Other cellular factors and processes that may be required for or affect the switch include the protein SeqA, which binds hemimethylated agn43 DNA, binding of RNA polymerase itself, or the passage of the replication fork (10).
Dam- and OxyR-dependent regulation of agn43 differs strongly from the well-studied paradigm of Dam- and Lrp-dependent phase variation of the pyelonephritis-associated pilus operon (pap). The key feature of pap phase variation is that it requires translocation of the global regulator Lrp between two binding sites in the pap regulatory region, where it acts either as a repressor or as an activator. This translocation is mediated by converse methylation states of two Dam target sequences, one in each Lrp binding site, in conjunction with protein-protein interactions between Lrp and a pap-specific regulatory protein (5, 7, 44, 46). Furthermore, pap transcription is subject to regulation by accessory factors, which allows for input of environmental signals (reviewed in reference 23). A biological relevance for the different Dam-dependent regulatory mechanisms of phase variation remains to be determined.
Acknowledgments
This work was supported in part by grant MCB-0077501 and the Research Experience Undergraduate supplement from the National Science Foundation, by the McCabe Foundation, and by a University of Pennsylvania Cancer Center Pilot Project Award.
We thank Chris Pericone, Heinze Matse, Madhu Kumar, Moon Majumdar, and Bonnie Manaski for technical assistance, Sol Goodgal for critical reading of the manuscript, and Nate Weyand and Rick Gourse for helpful discussions and strains.
REFERENCES
- 1.Al-Hasani, K., K. Rajakumar, D. Bulach, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1. John Wiley and Sons Inc., New York, N.Y.
- 3.Blattner, F. R., G. R. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I., and M. Van der Woude. Regulation and function of phase variation in Escherichia coli, p. 89-113. In M. Wilson (ed.), Bacterial adhesion to host tissues: mechanisms and consequences, in press. Cambridge University Press, Cambridge, England.
- 5.Blomfield, I. C. 2001. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 45:1-49. [DOI] [PubMed] [Google Scholar]
- 6.Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. A. Rolfson, and D. A. Low. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: Evidence for transcriptional regulation. EMBO J. 8:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, B. A., X. Nou, L. S. Kaltenbach, and D. A. Low. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577-588. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. 1976. Transposition and fusion of the lac genes to selected promoters in E. coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 9.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103-113. [DOI] [PubMed] [Google Scholar]
- 10.Correnti, J., V. Munster, T. Chan, and M. van der Woude. 2002. Dam-dependent phase variation of Ag43 in E. coli is altered in a seqA mutant. Mol. Microbiol. 44:521-532. [DOI] [PubMed]
- 11.Danese, P. N., L. A. Pratt, S. Dove, and R. Kolter. 2000. The outer-membrane protein, Ag43, mediates cell-to-cell interactions within E. coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 12.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diekmann, S. 1987. DNA methylation can enhance or induce DNA curvature. EMBO J. 6:4213-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 15.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in E. coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 16.Hattman, S. 1999. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 84:367-388. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, I. R., M. Meehan, and P. Owen. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 149:115-200. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 20.Kahmann, R. 1983. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harbor Symp. Quant. Biol. 47:639-646. [DOI] [PubMed] [Google Scholar]
- 21.Karls, R., V. Schulz, S. B. Jovanovich, S. Flynn, A. Pak, and W. S. Reznikoff. 1989. Pseudorevertants of a lac promoter mutation reveal overlapping nascent promoters. Nucleic Acids Res. 17:3927-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 23.Krabbe, M., N. Weyand, and D. Low. 2000. Environmental control of pilus gene expression., p. 305-321. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 24.Kullik, I., M. B. Toledano, L. A. Tartaglis, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 28.Marinus, M. G., A. Poteete, and J. A. Arraj. 1984. Correlation of DNA adenine methlyase activity with spontaneous mutability in Escherichia coli K-12. Gene 28:123-125. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Mulvey, M. R., P. A. Sorby, B. L. Triggs-Raine, and P. C. Loewen. 1988. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene 73:337-345. [DOI] [PubMed] [Google Scholar]
- 31.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 32.Plastkerk, R. H., H. Vrieling, and P. Van de Putte. 1983. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-encoded transactivator. Nature 301:344-347. [DOI] [PubMed] [Google Scholar]
- 33.Polaczek, P., K. Kwan, and J. L. Campbell. 1998. GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition. Mol. Gen. Genet. 258:488-493. [DOI] [PubMed] [Google Scholar]
- 34.Roche, A., J. McFadden, and P. Owen. 2001. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology 147:161-169. [DOI] [PubMed] [Google Scholar]
- 35.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 36.Schembri, M. A., and P. Klemm. 2001. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 20:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 40.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:88-94. [DOI] [PubMed] [Google Scholar]
- 41.Sun, W., and S. Hattman. 1996. Escherichia coli OxyR protein represses the unmethylated bacteriophage Mu mom operon without blocking binding of the transcriptional activator C. Nucleic Acids Res. 24:4042-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 43.Van der Woude, M. W., W. B. Hale, and D. A. Low. 1998. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J. Bacteriol. 180:5913-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Woude, M. W., L. S. Kaltenbach, and D. A. Low. 1995. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol. Microbiol. 17:303-312. [DOI] [PubMed] [Google Scholar]
- 45.Weiser, J. N., and E. C. Gotschlich. 1991. Outer membrane protein (ompA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weyand, N., and D. Low. 2000. Lrp is sufficient for the establishment of the phase OFF pap DNA methylation pattern and repression of pap transcription in vitro. J. Biol. Chem. 275:3192-3200. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, M., and G. Storz. 2000. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 59:1-6. [DOI] [PubMed] [Google Scholar]
- 48.Zheng, M., X. Wang, B. Doan, K. A. Lewis, T. D. Schneider, and G. Storz. 2001. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J. Bacteriol. 183:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]