Abstract
Centromere DNA element II (CDEII) of budding yeast centromeres is an AT-rich sequence essential for centromere (CEN) function. Sequence analysis of Saccharomyces cerevisiae CDEIIs revealed that A5–7/T5–7 tracts are statistically overrepresented at the expense of AA/TT and alternating AT. To test the hypothesis that this nonrandom sequence organization is functionally important, a CEN library in which the CDEII sequences were randomized was generated. The library was screened for functional and nonfunctional members following centromere replacement in vivo. Functional CENs contained CDEIIs with the highly biased An/Tn run distribution of native centromeres, while nonfunctional CDEIIs resembled those picked from the library at random. Run content, defined as the fraction of residues present in runs of four or more nucleotides, of the functional and nonfunctional CDEII populations differed significantly (P < 0.001). Computer searches of the genome for regions with an A + T content comparable to CDEIIs revealed that such loci are not unique to centromeres, but for 14 of the 16 chromosomes the AT-rich locus with the highest An≥4 + Tn≥4 run content was the centromere. Thus, the distinctive and nonrandom sequence organization of CDEII is important for centromere function and possesses informational content that could contribute to the determination of centromere identity.
CENTROMERES are essential for the proper segregation of chromosomes at mitosis and meiosis. Mammalian centromeres as well as those of higher plants are composed of megabases of highly repetitive AT-rich satellite DNA (Sullivan 2001; Hosouchi et al. 2002), while at the other extreme, the so-called point centromeres of budding yeasts are only 125 bp or so in length and contain specific DNA sequence motifs that determine centromere identity (Hegemann and Fleig 1993). Despite the lack of conservation at the DNA sequence level, all centromeres share a universal chromatin structure. Specifically, centromere DNA is packaged into specialized nucleosomes in which histone H3 is replaced by the centromere-specific H3 variant, CenH3 (CENP-A in humans, Cse4 in yeast) (Choo 2001). Drosophila and Arabidopsis CenH3's are adaptively evolving in regions of the protein thought to affect DNA-binding specificity, suggesting that CenH3 molecules have coevolved with the rapidly evolving satellites with which they interact (Malik and Henikoff 2001). The basis of that DNA-binding selectivity is not understood, but it is unlikely to be dependent on a specific DNA sequence (Henikoff and Dalal 2005).
Saccharomyces cerevisiae centromeres are recognizable by their conserved DNA elements (CDEs) (Hieter et al. 1985). CDEI, at the left-hand end of the centromere (CEN) DNA, is the degenerate octanucleotide RTCACRTG. Although CDEI is 100% conserved, neither it nor the factor that binds it (Cbf1/Cep1) is essential (Baker and Masison 1990; Mellor et al. 1990). CDEIII, located at the right-hand end of the centromere, is a 24-bp sequence with partial twofold symmetry. CDEIII is the binding site for CBF3, a complex of four essential proteins, Ndc10, Cep3, Ctf13, and Skp1 (Lechner and Carbon 1991; Stemmann and Lechner 1996). CDEIII is absolutely essential for CEN activity. Point mutations of the central CCG of CDEIII do not bind CBF3 (Lechner and Carbon 1991), fail in kinetochore assembly (Meluh and Koshland 1997), and abolish mitotic centromere function (McGrew et al. 1986). Separating CDEI and CDEIII is 79–88 bp of highly AT-rich DNA, designated CDEII. The function of CDEII is not known, although it has been proposed that it binds one or more essential kinetochore proteins (see discussion). The presence of this AT-rich element is arguably the only commonality between S. cerevisiae CENs and the AT-rich satellite DNA-laden centromeres of higher organisms.
CDEII is essential for S. cerevisiae centromere function. Reducing the length of CDEII or increasing its G + C content compromises CEN activity (Cumberledge and Carbon 1987; Gaudet and Fitzgerald-Hayes 1987), and an isolated CDEIII sequence integrated into the chromosome retains little or no CEN function (Carbon and Clarke 1984). Previous mutational studies of CDEII were limited by the then existing in vitro mutagenesis technologies. As a result, most constructed mutations altered both length and G + C content simultaneously, and few well-controlled studies in which the specific sequence of CDEII was analyzed have been carried out—not that it would be obvious what sequence changes to make, since no actual consensus sequence for CDEII has been proposed. In their original description of CDEII DNA (not so named at the time), Fitzgerald-Hayes et al. (1982) noted that “[CDEII] is arranged primarily into stretches of A residues followed by several T residues as opposed to runs of alternating A:T base pairs.” Substituting 49 bp of CDEII with mitochondrial DNA of equivalent A + T content but lacking the A and T tracts resulted in only a fourfold increase in mitotic chromosome loss, leading to the conclusion that the specific arrangement of A's and T's was not important (Cumberledge and Carbon 1987). Murphy et al. (1991), using totally synthetic CEN DNAs, concluded that the ability of CDEII DNA to form a static bend was important; “bent” and “unbent” CEN DNAs, differing at only six CDEII nucleotides, displayed a 60-fold difference in mitotic chromosome loss rates.
In this study, we tested the hypothesis that CDEII sequences contain a nonrandom sequence “code” that is important for centromere function. We performed a statistical analysis of the endogenous CDEII sequences to look for nonrandom patterns in the arrangement of CDEII nucleotides, we employed a genetic strategy to search for correlation between CDEII sequence content and centromere function, and we used computer programs to scan the genome for CDEII-like sequences. The results showed that centromere function positively correlates with the homopolymer run content of CDEII and that AT-rich sequences having both the high A + T content and homopolymer run bias of CDEII sequences are found predominantly at centromeres. The results suggest that a similar type of sequence “coding” could, in part, explain centromere identity in higher organisms.
MATERIALS AND METHODS
Media and cell growth:
Rich and defined media for S. cerevisiae growth were described previously (Baker and Masison 1990). Color indicator medium contained 6 mg/liter adenine (one-third usual concentration). Cells were grown at 30°. DNA transformations were performed by the lithium acetate method (Schiestl and Gietz 1989).
Construction of the randomized CDEII library:
A single-stranded 110-mer of sequence 5′-CCCCCACGTG[X]82CATATGATCTGCGTAGCC-3′, where X = 3.5% G, 3.5% C, 46.5% A, and 46.5% T, was synthesized by Oligos Etc. (Wilsonville, OR). The region of sequence degeneracy was flanked by PmlI and NdeI restriction sites, respectively (underlined). The oligonucleotide was made double stranded by annealing the 18-mer 5′-GGCTACGCAGATCATATG-3′ and extending with Klenow polymerase. The product was ligated into pPCR-Script vector (Stratagene, La Jolla, CA) and transformed into XL10-Gold Escherichia coli to obtain ∼9000 insert-containing transformants. Plasmid DNA was prepared from the pooled transformants and cleaved with NdeI and PmlI to release the CDEII insert, which was gel purified and ligated into NdeI/PmlI-cut pRB387 vector to regenerate a complete CEN sequence. Plasmid pRB387 is pUC8 containing a 243-bp fragment of CEN3 (chromosome III coordinates 114,560–114,318) fused to the 346-bp HindIII-BamHI fragment of pBR322. This insert is essentially the HindIII-SalI fragment of dl314 (McGrew et al. 1986) with the HindIII site changed to SphI. The CEN3 insert in pRB387 was also modified by deleting the CEN3 CDEII sequence and replacing it with “stuffer” DNA carrying PmlI and NdeI restriction sites at the CDEI and CDEIII boundaries, respectively (see Figure 2). The resulting library of regenerated CEN sequences (named pRB507) was approximately threefold redundant relative to the original CDEII library. Sixteen individual clones from the pRB507 library were sequenced to verify the structure of the CEN sequence. As expected, all differed at CDEII; the average A + T content of the degenerate regions was 94.1%, very close to the 93.0% programmed. There was a T bias in the library sequences, probably the result of higher than expected coupling efficiency of T (vs. A) during synthesis. The sequences contained 55.4% T and 38.6% A. The sequences were analyzed for randomness in dinucleotide frequencies (see below) and no statistically significant deviation was found.
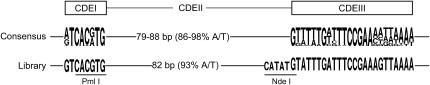
Figure 2.
Design of replacement CENs containing randomized CDEII. The middle line shows the consensus sequence of the 16 endogenous S. cerevisiae centromeres where the height of each letter is proportional to its frequency of occurrence at that position (Schneider and Stephens 1990). The sequence logos were generated by WebLogo (Crooks et al. 2004). Below the consensus sequence is shown the DNA sequence of the replacement centromeres generated by ligation of the randomized CDEII segments into the pRB507 vector (see materials and methods).
DNA of the pRB507 CEN library was prepared and cleaved with SphI and SalI, and the 590-bp SphI/SalI fragment containing the CEN sequence was ligated into the CEN3 replacement vector pJII (Murphy et al. 1991) to obtain the CEN replacement library pRB508. Although the original pRB508 library had only 600 members, it proved to be sufficiently complex for its purpose here, and no attempt was made to enlarge it.
CEN replacement and quantitative analysis of chromosome loss:
Centromere replacements were made in diploid strain R99 (Baker et al. 1998), homozygous for ura3 and ade2-1ochre. R99 was transformed with EcoRI-digested pRB508 DNA, and transformants were selected on uracil dropout agar containing reduced adenine. The CEN3- targeting segment of pRB508 carries insertions of URA3 and SUP11 on opposite sides of the cloning site into which the replacement CEN sequence is inserted; therefore, complete replacement of the resident CEN3 region with the incoming recombinant DNA results in colonies that are Ura+ and pink due to partial suppression of the homozygous ade2 by the single copy of SUP11. Gene conversion at the ura3 locus or recombination events at CEN3 that fail to coconvert the SUP11 marker result in red colonies. Pink transformant colonies were picked, diluted, and plated on nonselective color indicator plates to score missegregation of the marked chromosome III, observed as the appearance of red and white sectors within the predominantly pink colonies. Red sectors arise when the SUP11-URA3-marked chromosome is lost from the diploid, resulting in a 2N − 1 aneuploid and reverting the phenotype to Ade2− (red). White sectors result from gain of the marked chromosome (2N + 1 aneuploidy) and complete suppression of the Ade2− phenotype.
A rapid assay based on red-sectoring frequency was used to measure chromosome loss rates of the CEN replacement strains. Colonies on color indicator agar were viewed under a dissecting microscope by both authors on two consecutive days and assigned a score on a scale of 1 to 9. Reference strains with known chromosome loss rates were used to standardize the scoring (see Figure 3). Scorers were blinded with respect to the other person's scores and results from the previous day. Upon completion of scoring, the four scores were averaged. Subsequently, fluctuation analysis was performed to obtain a statistically robust measurement of chromosome loss rate. For each strain to be analyzed, five colonies were picked and grown overnight to late logarithmic phase in liquid complete minimal medium with reduced adenine. Cultures were diluted and plated on color indicator plates that were then incubated to allow colonies to form. The number of red colonies and total colonies on each plate was determined and used to calculate the chromosome loss rate using the method of median (Lea and Coulson 1949). Standard deviations of loss rates determined by this method were 10–20%. Loss rates of the reference strains (Figure 3) were determined using the more accurate 10-plate fluctuation protocol described by Hegemann et al. (1988). In all fluctuation tests, the rate at which white colonies arose was also determined and found to be about the same as that of red colonies; therefore, the observed chromosome loss events resulted predominantly from mitotic nondisjunction, i.e., 2:0 segregation.
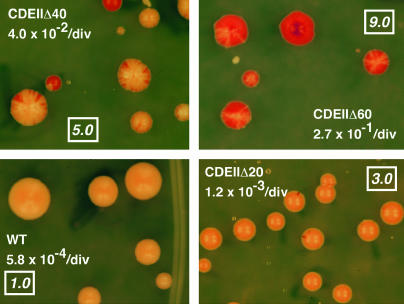
Figure 3.
Visual assay of chromosome loss rate. A wild-type and three CDEII deletion CENs were used to replace CEN3 in diploid strain R99 as described in materials and methods. The CDEII deletions had a common endpoint at the CDEII-CDEIII junction. The sectoring phenotype and the mitotic loss rate of the marked chromosome determined by fluctuation analysis are shown. The boxed numbers are the numerical scores assigned to that phenotype.
Characterization of high- and low-loss centromere populations:
From three independent transformations of the CEN replacement library DNA, a total of 60 pink transformant colonies were picked for analysis. One transformant yielded only red colonies upon restreaking and was discarded. Sectoring scores were determined for the others, and several strains with scores at both extremes of the distribution were subjected to fluctuation analysis to verify the loss rate. Twenty-three strains with sectoring scores ≥3.0 were chosen for the high-loss group. The mean sectoring score for the group was 4.9 ± 0.9 (SD). The marker chromosome loss rate was measured for five members of this group and found to be 0.032 ± 0.005 (mean ± SD) events/division. Fluctuation analysis was performed on all strains having a sectoring score of ≤2.5. Fourteen strains were found to have loss rates <2.4 × 10−3 events/division and were chosen for the low-loss group. The mean loss rate of the low-loss strains was 1.2 ± 0.8 (SD) × 10−3 events/division. Genomic DNA from the high- and low-loss strains was purified using Genomic tip 500/G columns (QIAGEN). The chromosome III replacement CEN loci were amplified by PCR using the primers 5′-TGTGGGTTTAGATGACAAGGG-3′ (URA3) and 5′-CCTAGTCGCGGTTTGTTATACC-3′ (vector-CEN junction). The resulting 930-bp products were purified using Qiaquick cartridges (QIAGEN) and subjected to automated DNA sequencing using the primer 5′-CTGGAGCCACTATCGACTAC-3′ (located in the pBR322 DNA immediately flanking the replacement centromere).
Computational methods:
The DNA sequences of the 16 S. cerevisiae chromosomes were downloaded from the University of California at Santa Cruz (UCSC) Genome Browser database (http://genome.ucsc.edu/) (Karolchik et al. 2003). The data were based on sequence dated October 1, 2003, in the Saccharomyces Genome Database (http://www.yeastgenome.org/). DNA sequence analysis was performed using computer programs written with MATLAB version 4.2c.1 (The Mathworks) and executed on an Apple Macintosh G4 desktop computer running in Classic mode under OS 10.3. For randomization trials, subject sequences were randomly permuted using the MATLAB randperm function and used as input to the respective analysis programs. The process was repeated 100 times to determine a mean and standard deviation for the result. Statistical analysis was performed using Prism 4 software (GraphPad Software).
To search for AT-rich regions in the S. cerevisiae genome, the number of A + T nucleotides within a sliding 85-nucleotide window was determined for each position of all 16 chromosomes. The coordinates of windows meeting a set threshold of A + T content were saved, and windows meeting the threshold but separated by ≤10 nucleotides were catenated along with the intervening “spacer.” The 10-nucleotide spacing requirement was imposed to reduce noise. The assembled sequences were used as input to other programs that analyzed nucleotide, dinucleotide, and run content. In the genome analysis, An or Tn runs of >11 nucleotides were ignored in calculating run content to avoid bias due to long runs not characteristic of centromere CDEII sequences. Such runs are not very common. For example, in the analysis shown in Figure 8, only 5 of the 60 AT-rich loci contained runs longer than 11 nucleotides (runs of 24, 21, 19, 12, and 12 nucleotides, all on different chromosomes).
Figure 8.
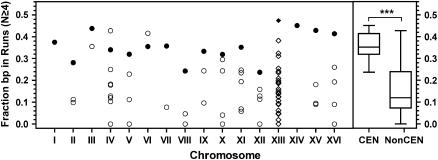
Run content of AT-rich chromosomal loci. Each circle represents a locus having a local A + T content of ≥90.6%. Chromosome XIII loci (diamonds) have an A + T content of ≥85.9% (see text). Solid symbols designate the centromere loci. Box plots of the run content of centromere and noncentromere loci (as in Figure 6) are shown on the right. ***, P < 0.001 (two-tailed t-test).
RESULTS
Analysis of endogenous CDEII sequences:
Table 1 shows the 16 S. cerevisiae CDEII sequences. They vary between 79 and 88 bp in length and between 86 and 98% A + T, averaging 85.7 bp and 93.2% A + T. A statistical analysis of the observed tendency of A or T nucleotides to occur together was made by counting the occurrence of AA and TT dinucleotides in the CDEII sequences and comparing the results with the mean of counts obtained when the sequences were individually randomized (100 trials). The actual occurrence of AA and TT dinucleotides in the 16 CDEII sequences as a group exceeded the random expectation by several multiples of the standard deviation (Figure 1). The probability of such a sequence arrangement occurring at random would be 10−8–10−11 (Z = 5.6–6.7). AT and TA dinucleotides were underrepresented to about the same extent, as must be the case, since the total number of A + T nucleotides is constrained. The occurrence of none of the other 12 possible dinucleotides differed from that expected at random (data not shown). Thus, the arrangement of A's and T's in CDEII sequences is highly nonrandom and appears to favor homopolymer runs at the expense of alternating A and T.
TABLE 1.
S. cerevisiae CDEII sequences
| CEN | Sequence | Length (bp) | AT (%) | Runsa |
|---|---|---|---|---|
| I | ACATAATAATAAATAATTTTAAAAATATAAAATATTTTTAATAGTTTTTAAATATTTTACAGTTTATTTTTTAAATTTATTTATAT– | 86 | 95.3 | 0.38 |
| II | ACTTATTTATTTAATTATTATTAAGTAAAAAAGATTTTCTATTTAAATTTATTAATTAATTTTTTTTCTTAAATAATTATTTTAT— | 85 | 94.1 | 0.26 |
| III | ATGATATTTGATTTTATTATATTTTTAAAAAAAGTAAAAAATAAAAAGTAGTTTATTTTTAAAAAATAAAATTTAAAATATTAGT— | 85 | 92.9 | 0.53 |
| IV | CTTATAATCAACTTTTTTAAAAATTTAAAATACTTTTTTATTTTTTATTTTTAAACATAAATGAAATAATTTATTTATT——— | 79 | 92.4 | 0.37 |
| V | CTTTTTAAAAAATATAAATTTAATTTCATTTTCTATTTCAATATTTATTAAATAAAAAATTTGAAAAATATATAAAAATTGTAGCA– | 86 | 90.7 | 0.36 |
| VI | CTATAAAAATAATTATAATTTAAATTTTTTAATATAAATATATAAATTAAAAATAGAAAGTAAAAAAAGAAATTAAAGAAAAAATA– | 86 | 94.2 | 0.34 |
| VII | TTATATTTACTATATAAAAATTCAATAAATAAAAAGTTAGAAGATAAAAATTATATTATACATATTTTTATTTTTATTATAATTTTT- | 87 | 93.1 | 0.35 |
| VIII | ACTAATAATTCTTTTAATTTTAATTAATTTAATAAAATTAAAATAATATATATACTAAATTGTTTATTAAAAATGATTAAACATTG– | 86 | 91.9 | 0.24 |
| IX | AAAATTTTTATATTTTTAATTAAATTTTTATAATATTATAAATTATTATAATATTGATATTTAAAATTAAAAACAAATTATTAAT— | 85 | 97.6 | 0.33 |
| X | TTAAATAATTAATTTACTTTAAAATTTATTTTTTAATATAAAATATTTATTCTTTTTATTTAAAAATAAAAAACACAAAAAAACAAT- | 87 | 94.3 | 0.43 |
| XI | ATAAAAACATATTTAAAATTTTAAAAAAATTAATTTTCAAAATAAATTTATTATATTTTTTTAATTACATAATCATAAAAATAAAT– | 86 | 95.3 | 0.47 |
| XII | TAATAAATATTATTAAAAAGTTTATTAAAATAAAATAATAATTTAAATTACTATTTTTAAATAAGTTTTATTTTTTAATAACACTATT | 88 | 94.3 | 0.33 |
| XIII | ACTACCTAACAAAATATTTATTTTTCTTTTTTAATATTTGAAAATACTAAAATATTTTTGTTGTTTTTTGAAAAAAGGATTTTTAAT- | 87 | 86.2 | 0.52 |
| XIV | CAGCTTTTTAAAAATATTTTAAAACATTTTAAAAAATATACATTTTTTTATTATTTTTTTATATATTAATGTTAAAATTTATTTAT– | 86 | 93.0 | 0.53 |
| XV | AACTTATTTTGCATTTAAAAAAAAGTAAAAACTATTTGCTAAAATATATTTTTTTAATTTTTAAAAATAATGTTTTAATTATTTAAT- | 87 | 90.8 | 0.49 |
| XVI | ATATATTTTTTATTTTTAATTTTTTTTAATTATAAAAATAATTTTTTTCTTTAAATTAAACAAAAATAAAAAATTGTTTTTTGTT— | 85 | 95.3 | 0.56 |
| Mean | 85.7 | 93.2 | 0.41 | |
| SD | 2.0 | 2.6 | 0.10 |
Fraction of base pairs in homopolymer runs of N ≥ 4.
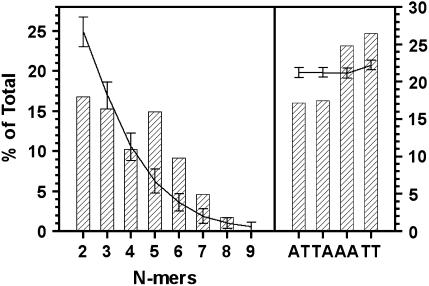
Figure 1.
An/Tn run and dinucleotide content of S. cerevisiae CDEII sequences. CDEII is defined as shown in Figure 2. Results are shown only for AT, TA, AA, and TT dinucleotides (right). All results are expressed as the percentage of total nucleotides. Curves show the means of randomization trials with error bars showing the standard deviation of 100 trials.
To determine if homopolymer runs of a particular length were statistically favored, the CDEII randomization trials were repeated, counting the occurrence of A2–16 and T2–16. The results are shown in Figure 1. Runs of length 2 and 3 were underrepresented, runs of length 5–7 were overrepresented, and runs of length 4 occurred at about the frequency expected for random arrangement of CDEII nucleotides. Runs of ≥8 nucleotides were found so infrequently that statistical analysis was not meaningful. [Three runs of 8 nucleotides occur in the endogenous CDEIIs (T8 in CEN2 and CEN16, A8 in CEN15), while 1.8 ± 1.1 occurrences would be expected. No runs of length >8 occur and, on the average, less than one would be expected.] The greatest overrepresentation was in runs of length 5 and 6, where the observed occurrences exceeded expectation by 5.8 and 5.1 standard deviations, respectively (P < 2 × 10−7). The relative abundance of A5/T5 and A6/T6 runs came mostly at the expense of A2/T2 runs, which were underrepresented by 4.4 standard deviations (P < 10−5). There was no significant difference in the distribution of An runs vs. Tn runs (data not shown), just as there is no significant A or T bias in CDEII sequence content (46.0% A, 47.3% T). These results indicate that CDEII sequences are not simply AT-rich DNAs; rather, their A and T residues are arranged in a highly nonrandom pattern characterized by homopolymer runs of 5–7 nucleotides, implying selection for some specialized function.
Genetic screen for functional CDEIIs:
A genetic screen was devised to test the hypothesis that the distinctive sequence organization of CDEII is critical for centromere function. A library of CEN DNAs in which individual sequences differed only at CDEII was generated. The CDEII regions all had the same length (87 bp) and A + T content (93%) as the endogenous CDEIIs, but the actual sequences were random. The library was constructed in a vector that allowed for in vivo replacement of the endogenous CEN3 by the library CEN linked to a color marker that provided the ability to rapidly assay mitotic function of the marked CEN. We reasoned that if only A + T content were important for CDEII function, then all library CENs would have more or less wild-type function. If, on the other hand, homopolymer run content was critical, then poorly functioning CENs screened from the library would contain random sequence CDEIIs, while CENs having wild-type or near wild-type function would contain CDEII sequences resembling those of the endogenous centromeres.
Figure 2 shows the structure of the library centromeres. The randomized CDEII elements were derived from a synthetic, single-stranded 110-bp oligonucleotide for which the central 82 nucleotide positions, flanked by PmlI and NdeI restriction sites, were synthesized using mixed precursors such that each site was predicted to contain 93% A + T and 7% G + C. After primer extension to generate double-stranded DNA and cleavage by PmlI and NdeI, the CDEII segments were ligated into an acceptor vector containing flanking CDEI and CDEIII elements (see materials and methods). The consensus CDEI sequence was the same as that of CEN14. The CDEIII sequence was derived from CEN3. The NdeI site introduces a C residue into the CDEII sequence 5 bp from the start of CDEIII, but two endogenous centromeres (CEN8 and CEN12) have a C at this position. The integration vector was the original CEN3 replacement vector of Clarke and Carbon (1983) as modified by Murphy et al. (1991). The CEN sequence is flanked by URA3 and SUP11. Heterozygous CEN3 replacement strains are readily obtained by transforming a ura3 ade2-1 diploid host, selecting Ura+ transformants on color indicator plates, and picking pink colonies. The pink colony color results from partial suppression of the ade2-1 red color phenotype by the heterozygous CEN-linked SUP11 marker. When diluted and plated on nonselective indicator plates, the pink Ura+ colonies give rise to pink colonies with red and white sectors owing to missegregation (loss and gain, respectively) of the marked chromosome III carrying the replacement CEN.
A rapid, semiquantitative method was devised to assay the CEN replacement strains obtained after transformation of the randomized CEN library. Sectoring phenotypes were assigned a numerical score of 1–9 corresponding to the frequency of red sectors within the pink colonies. The system was standardized using a series of model CENs constructed in the same vector as the library (Figure 3). The model centromeres were wild-type CEN3 and CEN3 derivatives with increasing lengths of CDEII deleted. The mitotic loss rates of chromosomes carrying these centromeres varied 470-fold, from 5.8 × 10−4 loss events/division for wild-type CEN3 to 0.27 loss events/division for CDEIIΔ60. The sectoring phenotype of the wild-type centromere was assigned a score of 1, the essentially acentric segregation behavior of CDEIIΔ60 was assigned a score of 9, and intermediate levels of sectoring were scaled accordingly. This semiquantitative method proved to be quite reliable in predicting actual loss rate as determined by fluctuation analysis.
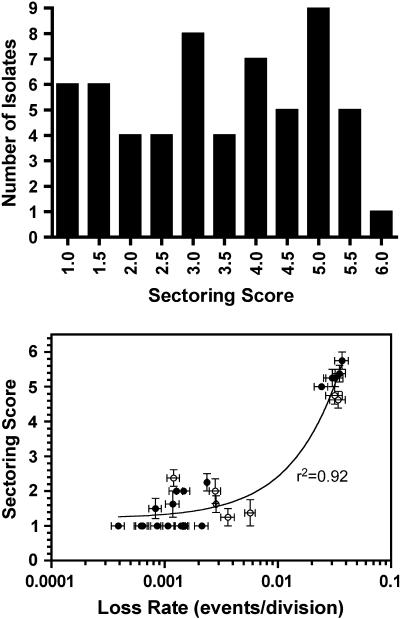
Figure 4 (top) shows the distribution of sectoring scores determined for 59 pink Ura+ library transformants picked at random. The median of the distribution was 3.6, and <10% of transformants contained a replacement centromere exhibiting wild-type function in this assay. Mitotic loss rates of the marker chromosome in several of the library replacement strains were determined by fluctuation analysis. There was high correlation between the sectoring score and the measured loss rates (Figure 4, bottom). In all cases, the observed loss events resulted from nondisjunction (2:0 segregation) of the marked chromosome, rather than from simple loss (1:0 segregation), as would be expected for centromere defects. The broad distribution of nondisjunction rates and low recovery of centromeres with wild-type or near wild-type mitotic function indicated that only a minority of the library CENs are fully functional, despite having CDEIIs of equally high A + T content. Thus, some additional characteristic of CDEII DNA sequence organization is required for mitotic CEN function.
Figure 4.
Histogram of sectoring scores determined for randomly picked transformants carrying replacement centromeres from the randomized CDEII CEN library (top) and correlation between sectoring score and loss rate determined by fluctuation test (bottom). The curve (bottom) represents the least-squares linear regression fit of the data. Open circles indicate strains omitted from further analysis.
Homopolymer run content correlates with CDEII function:
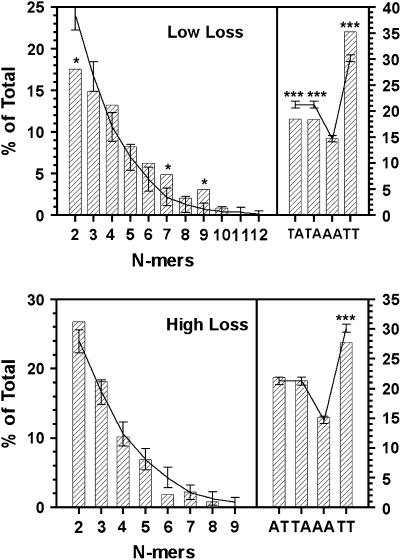
CEN replacement strains falling at the extreme ends of the loss rate continuum were placed into low-loss and high-loss groups (see materials and methods). Fourteen strains were selected for the low-loss group. The loss rate for the group was 1.2 ± 0.6 × 10−3 events/division (mean ± SD), approximately twice the loss rate of this marker chromosome carrying a wild-type centromere. The high-loss group consisted of 23 strains having a loss rate of 0.032 ± 0.005 (mean ± SD) events/division, an increase of ∼60-fold over the wild-type control. The library centromere present in each of the high- and low-loss strains was amplified from genomic DNA and sequenced. The CDEII sequences are shown in Tables 2 and 3. As expected, the two populations had the same A + T content, 93.5 and 93.8% for the high- and low-loss groups, respectively; however, clear differences were revealed when the sequences were analyzed for dinucleotide and homopolymer run content (Figure 5). Like the CDEII sequences of endogenous S. cerevisiae centromeres, AT and TA dinucleotides were underrepresented in the low-loss CDEIIs and TT was overrepresented. The AA dinucleotide content of both populations was less than that of TT due to the overall excess of T in the synthesized sequences (see materials and methods). The homopolymer run profiles of the two populations also differed. In low-loss sequences, runs of two and three nucleotides were underrepresented, while runs of length 4–9 were overrepresented. The difference between expected and observed values was statistically significant (P < 0.05) only for run lengths of 2, 7, and 9, perhaps due to sample size limitation, but in the aggregate, the run occurrence was markedly nonrandom and resembled the profile observed for the endogenous CDEIIs, i.e., underrepresentation of N = 2 and N = 3 runs and overrepresentation of N ≥ 4. In contrast, the run profile of the high-loss sequences was similar to that expected for a random arrangement of nucleotides, with the exception of di- and trinucleotide runs that were somewhat overrepresented and 6-mer runs that were underrepresented. None of the differences was statistically significant.
TABLE 2.
Low-loss CDEII sequences
| ID | Sequence | Length (bp) | AT (%) | Runsa |
|---|---|---|---|---|
| L1 | TAAAATTTTATACATAAAATTAAATTAATTTAATATTTTGTAATTAATTTTAATTTTTAAAATATTATTTTTTTAATTTTAT | 82 | 97.6 | 0.49 |
| L2 | TTTAAATAAATTTAAAATATTTTTAAAAAAATATTTTGATGTATTTTTTTTTGACTAATTAATTTTTATTGTTTGATTAAAA | 82 | 92.7 | 0.46 |
| L3 | TATTTATATAAATAATGTTGTTAAAATTTTTTTATTTTCCATTGTATTTTTATTTTTTTATAATTTTTAATATATTTCTTTT | 82 | 92.7 | 0.44 |
| L4 | ATATAGAAAAAAATTTTTTATTTAATTTAAATTTTATATTTTTAATATTTTTTATATATAAGTATTTTTTTACTTAATAATT | 82 | 96.3 | 0.43 |
| L5 | TTAAAATTCAGTGTTTAAAATATTTTTATTTTTTTTTATTTAGAAAATAGTTAAAATATTTATTTTATTTAAGTTAGAATAT | 82 | 91.5 | 0.41 |
| L6 | TATTTTTTAAAATAGATTAGAAGTTTTTAAATATTTTAAATGATTATGCATAAAAATTATTTAATTATTATTTATATTTTTT | 82 | 92.7 | 0.37 |
| L7 | TAATAATATTTTATGTTTAATTATATATATATATTATATTTTATAAATTTTTTTTATATATTTTTTTTTATTATGTAAATAA | 82 | 97.6 | 0.30 |
| L8 | TTGTTATTTTTTAGATGATTATTTTTTACATTTATGTTTATATTTTATTAAATTTTACTTCGTGTAATTGATCTAAATTTTT | 82 | 86.6 | 0.30 |
| L9 | GTATAGCTTTTATATAAATTTAATTTTTTAAAATTTTTTATATTTATTTTTTAATTGTTTATCTTTACTATTATAATATTTT | 82 | 92.7 | 0.37 |
| L10 | AAAATAATATTATTATTTATTTATTTTAAATTTATAATATATTAAATTTTTGTATAGTTTTTATTTTTTTTAATAATTTTTT | 82 | 97.6 | 0.39 |
| L11 | TATTTTAATAAATTGATTAATTTAATATAATTAAATTTTTATTTATTTAAAATAATATTTTTTGTTTAAAAGAATTTAAAAA | 82 | 96.3 | 0.34 |
| L12 | ATAATTTAAATTAAATTGTAAAATAATATTTTTTTTAGTATATTTTTTATTATCATTTTAATTTAATATATTTTTAATTTTT | 82 | 96.3 | 0.39 |
| L13 | TTTAAATAAATTTAAAATATTTTTAAAAAAATATTTTGATGTATTTTTTTTTGACTAATTAATTTTTATTGTTTGATTAAAA | 82 | 92.7 | 0.46 |
| L14 | TTATATTATTCATTATTTAATTAATTATTTTTTTATTCTTTTTGGATCGTATTTTTTTTTTATTAAATGAGATTTATAATTA | 82 | 90.2 | 0.27 |
| Mean | 93.8 | 0.39 | ||
| SD | 2.6 | 0.10 |
Fraction of base pairs in homopolymer runs of N ≥ 4.
TABLE 3.
High-loss CDEII sequences
| ID | Sequence | Length (bp) | AT (%) | Runsa |
|---|---|---|---|---|
| H1 | AAAATATATTAATTTAATTTATATTATTAAAATTACATATTTTATATTTTTATTTATTAATTTTAATAGTATATTAAATGAC | 82 | 95.1 | 0.26 |
| H2 | ATTAATTAATAGTTAAATTAAATTTTATTAATTATATATATTTTTATTTTTATTTTATTTAAGAGTTTTATAATAATTTTTA | 82 | 96.3 | 0.33 |
| H3 | AATTAATAGTTATTTTTATATAAAAGATATTTATATGAAATAGAGTTTTATAATTAATATATTAAATATAAGTATTTTTAAA | 82 | 92.7 | 0.22 |
| H4 | TTCATAAATTTTATTAGATAATATAAGTTAAAAATATGAAATGAATATTTAATGTAACGAAATTTTTTTATTATTTTAATTA | 82 | 90.2 | 0.24 |
| H5 | TTTCTAATTATTTTTATTAAGTGAATAATACTAAAAAAAATAATTATTATTACTTTATTAATTGATTTTAATAAATTTTTAA | 82 | 92.7 | 0.27 |
| H6 | TTTTTTATTATTTATATTTAATTTTATTTAAAATAAATATTATTTATTTCTTAATTATATTATATAAGTATATGTAATATTT | 82 | 96.3 | 0.17 |
| H7 | ATTTATTTTTATTTTTATTGAATTTTTAATAAAATTGTATTTATTTCAATAGTTAAAATATATTTTGATTATTTAGTTTAAA | 82 | 92.7 | 0.33 |
| H8 | AAAATTTAAATATAATTTATATTTAATTTAATTGATTATTTTAATAAATTAGTAGTAATTTATTAATTATTATTATTTTTA- | 81 | 96.3 | 0.16 |
| H9 | AAATATTTAATATAATTTTTTTATTTATAATTATATTAATTAAAATTAATTATCTATTTTAATAATTATAGTACTTAATAAA | 82 | 96.3 | 0.18 |
| H10 | TTATATAAATTTATATAGAAATAAGAAAATTTGTTTAACTAATATATATTTTATTTTTTATTTATATATTTTCATAGTAGTT | 82 | 91.5 | 0.22 |
| H11 | TTTATTTATTTTTAACTTAAATTTTTAATAAGTCAATTAATTAGATAAAAATAATATTATTATATTTTATTATTTTTTTAAT | 82 | 95.1 | 0.32 |
| H13 | TAAAATAAAAAATGAAATATTAATTGAATTTAAGAATAAATTTATAATTACACATAGAATAAATTTATTAATCAAATTAAAA | 82 | 91.5 | 0.17 |
| H14 | AATTATAATAATAATTTGAAATTATATATTTTAAATTTTTTTATTTATTATATTAATATATAATTTATTGATATATTTTAAT | 82 | 97.6 | 0.18 |
| H15 | TTATATATTTAATTATAATATATTTTTTAATTTAGAATATATGTTTAGGATATAATTAATTTAAATTTACGGTTTATTATTA | 82 | 91.5 | 0.07 |
| H16 | ATAAATTATTATATATATTTGATTTATATAAAATAAAATATCTAATAATTATATTTAATTTAATTAATTATTTGTGTTTTTT | 82 | 95.1 | 0.17 |
| H17 | ATTTAATTTAATAATCATTTTAACATGGATTTAACGTATTCTTTTTCAAAATGATAGAATTTGATTGAATTAATTATATTT- | 81 | 85.2 | 0.16 |
| H18 | TTTATTTTTTTAAAAATATATTTTTATTTGTAATTTTTCTTGATTCTTTTTATTATTAAATCAATATTTCAACTATTTTAAA | 82 | 91.5 | 0.38 |
| H19 | TTAAAATTATTGTGTTTTTGTTATATAATTATTTTATATTTAATTATATAAATGATAATGATTTATATTTTATTTATAATCT | 82 | 92.7 | 0.21 |
| H20 | AATATAATGATTTACCATTATATGTAATTAACAAAAGAGAATATTTATTAATTATTTTTAAAAATTTATATTATTTTATTTA | 82 | 91.5 | 0.22 |
| H22 | AATAAATATTTATAAATAATATATAGTTTTTTTTATATTTGTATTATTTTAATTTTATGTTTTTTTATGAATTTAAATTCAT | 82 | 93.9 | 0.28 |
| H23 | TTTTATATAAATATTATATTTAATTTAACTCATATTTATTTAATTATATTATTTTTGTTAAATTAATATTAGTTATTAATTT | 82 | 95.1 | 0.11 |
| H24 | ATAATTAAATAATTATTTCTTGTATTTTATAATGTTATTATTCAATGATATTATAATAATTTTATTTTGATTTCTTTATATT | 82 | 91.5 | 0.15 |
| H25 | TAATAATTAAATATTTTGATAATTAATATAAATTATAATTATTTTAATATTTTTATTAAATTAAAATTAATTATCTTTTTT- | 81 | 97.5 | 0.28 |
| Mean | 93.5 | 0.22 | ||
| SD | 2.9 | 0.08 |
Fraction of base pairs in homopolymer runs of N ≥ 4.
Figure 5.
An/Tn run and dinucleotide content of low- and high-loss CDEIIs. The NdeI site common to all sequences and technically part of CDEII (see Figure 2) was not included in the analysis. Results are shown only for AT, TA, AA, and TT dinucleotides (right). All results are expressed as the percentage of total nucleotides. Curves show the means of randomization trials with error bars showing the standard deviation of 100 trials. *, P < 0.05; ***, P < 0.001.
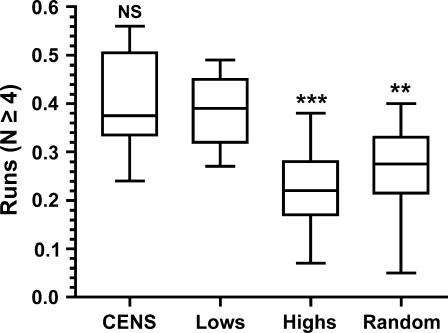
As a simple means to describe the homopolymer run content of individual sequences, the total number of nucleotides present in Tn or An runs of length N ≥ 4 were counted and expressed as a fraction of total nucleotides. Figure 6 shows plots of the results for CDEIIs of the high and low-loss populations as well as for 16 library CENs picked at random and for the 16 endogenous S. cerevisiae CDEIIs. The run content of the low-loss CDEIIs does not differ significantly from that of the endogenous CENs; however, the difference in run content between the low- and high-loss CDEIIs is significant at the P < 0.001 level. The endogenous centromere CDEIIs also differ from the high-loss CDEIIs (P < 0.001, not shown). There was no absolute threshold of run content that determined high- or low-loss phenotype. While no high-loss CDEII had a run content >0.38 and no low-loss CDEII had a run content <0.27, there was overlap within this range (Tables 2 and 3). The statistical significance of the difference derives from comparing populations, not individuals. Since the assignment of sequences into the two test groups was on the basis of chromosome loss phenotype only, we interpret the significant difference observed in run content to mean that high homopolymer run content in CDEII is important for mitotic centromere function.
Figure 6.
Box plots of homopolymer run content of CDEII sequences. Horizontal lines indicate the median, with the box showing the 25th and 75th percentile bounds; error bars show the extremes. The results of Bonferroni's multiple comparison test comparing the “Lows” group with all other groups are shown above the plots. CENS, endogenous CDEIIs; Lows, low-loss CDEII population; Highs, high-loss CDEII population; Random, 16 random CDEII clones; NS, not significant; **, P < 0.01; ***, P < 0.001.
Genomic searches for CDEII-like sequences:
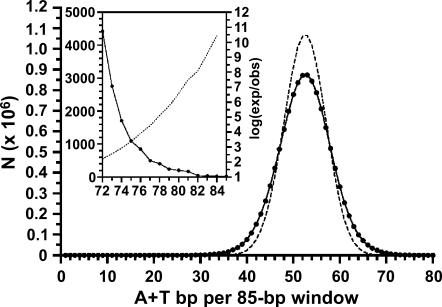
The distinctive arrangement of CDEII nucleotides into runs of A and T and the importance of this nonrandom sequence organization for centromere function prompted us to ask if similar blocks of AT-rich DNA were present in the yeast genome at locations other than centromeres. Figure 7 shows the distribution of A + T content in the S. cerevisiae genome. For each chromosome, the number of A + T nucleotides was counted in a sliding 85-bp window moving the length of the chromosome. The 85-bp window length was chosen because it is close to the average length of the endogenous CDEII sequences. Since every nucleotide must be either A/T or G/C, the random probability of finding any given number of A or T nucleotides within the window is given by the corresponding term of the binomial P = (a + b)85, where a and b are the frequencies of A + T and G + C nucleotides in the genome, 0.617027 and 0.382973, respectively. As can be seen in Figure 7, the actual distribution is symmetrical but flattened, meaning that there is a relative excess of windows at the extremes, but the number of high A + T windows is offset by a similar number of low A + T windows. The distribution of A + T content in individual chromosomes does not differ significantly from that of the genome as a whole (data not shown). The genome average was 52.50 ± 5.49 (mean ± SD) A + T nucleotides/85-nucleotide window, or 61.76 ± 6.46% A + T.
Figure 7.
Genomic distribution of A + T content. Solid symbols indicate the number of 85-bp windows in the yeast genome containing the number of A + T residues given on the x-axis. Dotted line shows the result expected at random. The inset shows the right-hand extreme of the distribution. The dotted line of the inset shows the likelihood of occurrence, plotted as the log10 of the ratio of expected to observed occurrences (values are negative).
Fifteen of the 16 S. cerevisiae CDEIIs have an A + T content >90.6% (Table 1). This corresponds to ≥77 A + T nucleotides in an 85-bp window. A total of 1677 such windows were found in the genome. Thus, while infrequent, sequences having the high A + T content characteristic of CDEIIs are not rare. Closer examination revealed that the windows of high A + T content were clustered and limited to one or a few distinct sites per chromosome (Table 4). For this analysis, windows meeting the threshold but separated by ≤10 bp were combined with the intervening windows of lower A + T content, explaining why the average A + T content of a given locus was slightly less than 90%. The average length of the AT-rich loci defined in this manner was 112 bp, larger than the average CDEII. On chromosomes I and XIV, the only locations at which the local A + T content reaches the 90% threshold is at and adjacent to the centromere.
TABLE 4.
Chromosomal distribution of 85-bp windows having ≥77 A + T residues
| Avg. (%) AT
|
||||||
|---|---|---|---|---|---|---|
| Chromosome | Length (bp) | N | Loci | Avg. length (bp) | Alla | Maximumb |
| I | 230208 | 28 | 1 | 112 | 88.4 | 95.3 |
| II | 813136 | 74 | 4 | 103 | 89.2 | 93.5 |
| III | 316613 | 35 | 2 | 106 | 89.6 | 91.8 |
| IV | 1531914 | 224 | 9 | 110 | 89.5 | 93.1 |
| V | 576869 | 77 | 4 | 107 | 89.5 | 92.4 |
| VI | 270148 | 26 | 2 | 98 | 88.9 | 92.9 |
| VII | 1090944 | 99 | 2 | 134 | 89.7 | 95.3 |
| VIII | 562639 | 53 | 3 | 102 | 88.9 | 92.5 |
| IX | 439885 | 124 | 3 | 128 | 88.6 | 94.1 |
| X | 745446 | 83 | 5 | 104 | 89.2 | 91.8 |
| XI | 666445 | 326 | 6 | 141 | 90.4 | 92.5 |
| XII | 1078173 | 141 | 5 | 113 | 89.9 | 92.7 |
| XIII | 924430 | 160 | 4 | 124 | 89.9 | 94.7 |
| XIV | 784328 | 18 | 1 | 102 | 87.3 | 95.3 |
| XV | 1091285 | 33 | 3 | 94 | 89.4 | 91.2 |
| XVI | 948060 | 176 | 5 | 122 | 89.5 | 93.4 |
| Total: | 12070523 | 1677 | 59 | 112 | 89.2 | 93.3 |
All loci.
Windows of highest A + T content at each locus.
We next asked if chromosomal sites of high A + T content were distinguishable on the basis of sequence organization, specifically, homopolymer run content. The run content of all AT-rich loci described in Table 4 was determined by counting An and Tn runs of length 4 ≤ n ≤ 11. The upper limit of 11 nucleotides was set to avoid biasing the results by the occurrence of a single, long run (see discussion). The results for all except chromosome XIII sequences are shown in Figure 8. For 13 of the chromosomes, the exceptions being IV and VI, the AT-rich locus with the highest content of homopolymer runs was the centromere. The run content of noncentromere loci differed significantly from that of the centromere loci (P < 0.001), although as seen with the library CENs, there was overlap between the populations and no absolute cut-off was observed (Figure 8, right). Chromosome XIII was an interesting exception. At 86.2%, the CDEII of CEN13 has the lowest A + T content of the endogenous centromeres, and it fell below the threshold used to produce the results shown in Table 4. When chromosome XIII was rescanned using a lowered threshold of 85.9% A + T, 27 loci were found, and the locus with the highest run content was CEN13 (Figure 8).
DISCUSSION
Two lines of evidence lead us to conclude that the distinctive arrangement of CDEII nucleotides into runs of An≥4 and T≥4 is important for centromere function in S. cerevisiae. First, CDEII sequences selected from the randomized library solely on the basis of the ability to form functional centromeres contained a significantly higher run content than CDEIIs having compromised centromere function, despite the fact that the overall A + T content of both populations was the same. Second, stretches of DNA having the extremely high A + T content of CDEIIs are found in the genome at locations other than centromeres, but the run content of centromere CDEIIs is significantly higher than that of noncentromeric AT-rich loci. For 14 of the 16 chromosomes, the AT-rich sequence having the highest run content was found at the centromere.
A similar conclusion was reached by Espelin et al. (2003), on the basis of limited data. In a study focusing on CDEIII-independent binding of Ndc10 to CDEII, these investigators used a computer program to scan a region of chromosome III for CDEII-like sequences, defined as having an A + T content >80% and the number of An or Tn stretches “significantly above the genome average.” They found and tested three such sequences and all selectively bound Ndc10. One of the Ndc10-binding sequences was assayed for its ability to function as a CDEII element in the context of a centromere and found to have partial activity, leading Espelin et al. (2003) to conclude tentatively that CDEII function correlated with homopolymer run content, a conclusion rigorously confirmed here. Thus, CDEII is not simply AT-rich spacer DNA separating CDEI and CDEIII. Although CDEII cannot be defined by a specific nucleotide sequence, the requirement for a distinguishable sequence organization, i.e., homopolymer runs, implies that CDEII contributes positively to centromere function, perhaps serving as the preferred binding site for one or more kinetochore proteins.
The most likely candidates for CDEII-binding proteins are Ndc10, Mif2, and the S. cerevisiae CenH3 protein Cse4 (Espelin et al. 2003). Mif2 is an essential protein having homology to the mammalian kinetochore protein CENP-C (Brown 1995). Since Mif2 also has homology to the “AT-hook” motif found in other proteins known to bind AT-rich DNA, it has been suggested that Mif2 binds CDEII (Brown 1995). Indeed, in chromatin immunoprecipitation (ChIP) experiments, Mif2 is found associated with CEN DNA (Meluh and Koshland 1997); however, there is no direct evidence supporting a direct CDEII interaction in vivo, and recombinant Mif2 fails to bind CEN DNA in vitro (Espelin et al. 2003). Cse4 binds centromere DNA as an integral component of one or more centromere-specific nucleosomes. ChIP data seem to show that a single Cse4 nucleosome forms on S. cerevisiae CEN DNA, approximately centered on the CDEI-CDEII-CDEIII sequences (Meluh et al. 1998). If so, CDEII would occupy over one full superhelical turn of the nucleosomal DNA, and CDEII sequence characteristics could strongly influence nucleosome binding. Observed genetic interactions between cis-acting CEN mutations and cse4 alleles support such a model (Keith and Fitzgerald-Hayes 2000). On the other hand, Espelin et al. (2003) interpret the ChIP data differently and suggest that Cse4 nucleosomes are absent from, but instead flank both sides of, the CDEI-CDEII-CDEIII core. In their model, CDEII is bound by dimers of Ndc10, separate from the Ndc10 that binds CDEIII as a part of the CBF3 complex. Given the observed binding selectivity of Ndc10 for the “CDEII-like” sequences defined by Espelin et al. (2003), it would be interesting to test the high- and low-loss CDEIIs from the randomized CDEII library to determine if CDEII function correlates with Ndc10-binding affinity.
While homopolymer run content is highly predictive of CDEII function, no absolute threshold of run content separates the high- and low-loss CDEII populations. Likewise, there is overlap between the run content distribution of the endogenous CDEIIs with that of noncentromeric sequences having similar A + T content. Attempts to devise more refined mathematical models to describe CDEII sequence organization failed to yield results that were more discriminating than simply calculating N ≥ 4 run content. Fourier analysis was used to search for di- and trinucleotide periodicities (Satchwell et al. 1986). Although the endogenous CDEIIs show a statistically significant 5.9-bp periodicity in the occurrence of AA/TT and AAA/TTT, there was no similar signal found in the low-loss library sequences. Given the possibility that CDEII may serve as a nucleosome-positioning sequence for Cse4 nucleosomes, the library sequences were analyzed with a computer program that predicts free energy of nucleosome formation (Anselmi et al. 2000). There was no significant difference in the mean free energy values of the high- and low-loss populations; however, the prediction algorithm has been tested only for canonical nucleosomes and may not be applicable in this case. In calculating run content, only runs of four or more nucleotides were counted. This minimum was chosen because it is the shortest run length overrepresented in the run profile of the selected low-loss CDEII population. Setting the minimum run length at 3 or 5 did not affect the outcome of the analysis. In all cases, the CDEII run contents of the high- and the low-loss subpopulations of the randomized library differed at the P < 0.001 level of significance.
The extreme bias in both A + T content and run content of CDEII sequences relative to genome averages begs the question of whether the two parameters are necessarily dependent. To partially answer this question, chromosomes I and III were scanned for the occurrence of sequences having a An≥4 + Tn≥4 run content of ≥0.41 in a window of 85 nucleotides, independent of total A + T content. The 0.41 threshold corresponds to the average run content of endogenous CDEIIs (Table 1). Thirty-nine such regions were found (data not shown), but they differed from centromere CDEIIs in two characteristics. In many cases, the high run content was due to a single run of ≥10 nucleotides. Additional experiments are needed to determine if runs of this length are compatible with CDEII function; however, no An/Tn run >8 nucleotides in length is found in endogenous CDEII sequences. Second, with the exception of the centromere on each of the two chromosomes analyzed, sequences identified solely on the basis of An≥4/Tn≥4 content had an average A + T content of 74.3%, not greatly higher than the genome average of 61.8% and significantly less than that of CDEIIs. Thus, sequences highly biased with respect to A + T content or An≥4/Tn≥4 content exist independently in the genome, but very few sequences other than CDEIIs have both characteristics.
In analyzing the local A + T content of S. cerevisiae chromosomes, we noted that the extremely high A + T content of the centromere regions is not limited to CDEII, but extends beyond the centromere boundaries defined by CDEI and CDEIII. That is, the CDEI-CDEII-CDEIII core sequences are embedded in islands of AT-rich DNA characterized by an overall A + T content significantly above the genome average. For example, if a boundary threshold is set at 84% A + T (71 of 85 A or T nucleotides), which occurs in the genome at a frequency of <0.001, the average length of the 16 CEN AT-rich islands is 175 bp, over twice the average length of CDEII. One possibility is that the AT-rich islands are remnants of ancestral centromeres that more resembled the AT-rich repetitive DNA structure commonly seen in eukaryotes rather than the defined-sequence centromeres present in budding yeasts today. The acquisition of binding sites for sequence-specific factors (CDEI and CDEIII), in effect, defined CDEII that then evolved a distinct function requiring the extremely high A + T and homopolymer run content observed now.
Centromere identity in S. cerevisiae appears to be determined by CDEIII. Point mutations in CDEIII abolish centromere function (McGrew et al. 1986), and CBF3 binding is a prerequisite for the association of all other known kinetochore components (Sorger et al. 1994; Ortiz et al. 1999; Cheeseman et al. 2002; Measday et al. 2002). Nonetheless, CDEII is also essential for centromere function. Shortening CDEII or increasing its G + C content compromises mitotic and meiotic centromere function (Cumberledge and Carbon 1987; Gaudet and Fitzgerald-Hayes 1987; Murphy et al. 1991), and isolated CDEIIIs lack mitotic function (Carbon and Clarke 1984) and fail in kinetochore assembly even though CBF3 is bound (Meluh and Koshland 1997; Ortiz et al. 1999). CDEII may act in concert with CDEIII to provide a fail-safe mechanism for establishing CEN identity, ensuring that CBF3 interactions at noncentromere sites of CDEIII homology do not nucleate assembly of functional kinetochores and generate dicentric chromosomes. Such ectopic interactions of CBF3, while less thermodynamically favored, must nonetheless occur. The rudimentary CDEII “code” defined here is highly specific for centromeric CDEII sequences. Requiring such a site to be adjacent to a CBF3 binding site creates the biochemical equivalent of a logical AND gate whose output is kinetochore assembly. The random probability of triggering the process at noncentromeric sites would be vanishingly small.
In contrast to the small, sequence-defined point centromeres of budding yeasts, the centromeres of higher eukaryotes are composed mostly of arrays of repetitive DNA, and no conserved centromere identifier sequence has been found (Karpen and Allshire 1997; Sullivan 2001; Cleveland et al. 2003). Henikoff and Dalal (2005) have suggested that centromere identity is not determined by DNA sequence per se, but by the chromatin structure it organizes, in particular, the incorporation of nucleosomes containing CenH3. If so, then DNA sequence characteristics that affect flexibility, bendability, and major and minor groove dimensions will probably be more important than a specific nucleotide sequence. The CDEII code of S. cerevisiae may be an example of just such a sequence characteristic. Better deciphering of the CDEII code, understanding its consequences for DNA structure, and elucidating the molecular mechanism by which it is recognized may well provide insight into centromere identity and kinetochore formation in higher organisms.
Acknowledgments
We thank Kendra Harris for technical assistance in preparing vectors used for constructing the randomized CDEII library, Anita Scipioni for providing the computer program for predicting sequence-dependent nucleosome thermodynamic stability, and Sam Stoler for helpful comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM61120).
References
- Anselmi, C., G. Bocchinfuso, P. De Santis, M. Savino and A. Scipioni, 2000. A theoretical model for the prediction of sequence-dependent nucleosome thermodynamic stability. Biophys. J. 79: 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R. E., K. Harris and K. Zhang, 1998. Mutations synthetically lethal with cep1 target Saccharomyces cerevisiae kinetochore components. Genetics 149: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R. E., and D. C. Masison, 1990. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10: 2458–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M. T., 1995. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene 160: 111–116. [DOI] [PubMed] [Google Scholar]
- Carbon, J., and L. Clarke, 1984. Structural and functional analysis of a yeast centromere (CEN3). J. Cell Sci. Suppl. 1: 43–58. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I. M., D. G. Drubin and G. Barnes, 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157: 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K. H., 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1: 165–177. [DOI] [PubMed] [Google Scholar]
- Clarke, L., and J. Carbon, 1983. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 305: 23–28. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Y. Mao and K. F. Sullivan, 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421. [DOI] [PubMed] [Google Scholar]
- Crooks, G. E., G. Hon, J. M. Chandonia and S. E. Brenner, 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge, S., and J. Carbon, 1987. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics 117: 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin, C. W., K. T. Simons, S. C. Harrison and P. K. Sorger, 2003. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell 14: 4557–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke and J. Carbon, 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29: 235–244. [DOI] [PubMed] [Google Scholar]
- Gaudet, A., and M. Fitzgerald-Hayes, 1987. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann, J. H., and U. N. Fleig, 1993. The centromere of budding yeast. Bioessays 15: 451–460. [DOI] [PubMed] [Google Scholar]
- Hegemann, J. H., J. H. Shero, G. Cottarel, P. Philippsen and P. Hieter, 1988. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2523–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Y. Dalal, 2005. Centromeric chromatin: What makes it unique? Curr. Opin. Genet. Dev. 15: 177–184. [DOI] [PubMed] [Google Scholar]
- Hieter, P., D. Pridmore, J. H. Hegemann, M. Thomas, R. W. Davis et al., 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42: 913–921. [DOI] [PubMed] [Google Scholar]
- Hosouchi, T., N. Kumekawa, H. Tsuruoka and H. Kotani, 2002. Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 9: 117–121. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., R. Baertsch, M. Diekhans, T. S. Furey, A. Hinrichs et al., 2003. The UCSC genome browser database. Nucleic Acids Res. 31: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G. H., and R. C. Allshire, 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489–496. [DOI] [PubMed] [Google Scholar]
- Keith, K. C., and M. Fitzgerald-Hayes, 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics 156: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the number of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Lechner, J., and J. Carbon, 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64: 717–725. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157: 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew, J., B. Diehl and M. Fitzgerald-Hayes, 1986. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., D. W. Hailey, I. Pot, S. A. Givan, K. M. Hyland et al., 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16: 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor, J., W. Jiang, M. Funk, J. Rathjen, C. A. Barnes et al., 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9: 4017–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., and D. Koshland, 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11: 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland and M. M. Smith, 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. [DOI] [PubMed] [Google Scholar]
- Murphy, M. R., D. M. Fowlkes and M. Fitzgerald-Hayes, 1991. Analysis of centromere function in Saccharomyces cerevisiae using synthetic centromere mutants. Chromosoma 101: 189–197. [DOI] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank and J. Lechner, 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13: 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell, S. C., H. R. Drew and A. A. Travers, 1986. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191: 659–675. [DOI] [PubMed] [Google Scholar]
- Schiestl, R. H., and R. D. Gietz, 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16: 339–346. [DOI] [PubMed] [Google Scholar]
- Schneider, T. D., and R. M. Stephens, 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P. K., F. F. Severin and A. A. Hyman, 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell. Biol. 127: 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann, O., and J. Lechner, 1996. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 15: 3611–3620. [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K. F., 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11: 182–188. [DOI] [PubMed] [Google Scholar]