Abstract
Expression of the Escherichia coli napFDAGHBC operon (also known as aeg46.5), which encodes the periplasmic molybdoenzyme for nitrate reduction, is increased in response to anaerobiosis and further stimulated by the addition of nitrate or to a lesser extent by nitrite to the cell culture medium. These changes are mediated by the transcription factors Fnr and NarP, respectively. Utilizing a napF-lacZ operon fusion, we demonstrate that napF gene expression is impaired in strain defective for the molybdate-responsive ModE transcription factor. This control abrogates nitrate- or nitrite-dependent induction during anaerobiosis. Gel shift and DNase I footprinting analyses establish that ModE binds to the napF promoter with an apparent Kd of about 35 nM at a position centered at −133.5 relative to the start of napF transcription. Although the ModE binding site sequence is similar to other E. coli ModE binding sites, the location is atypical, because it is not centered near the start of transcription. Introduction of point mutations in the ModE recognition site severely reduced or abolished ModE binding in vitro and conferred a modE phenotype (i.e., loss of molybdate-responsive gene expression) in vivo. In contrast, deletion of the upstream ModE region site rendered napF expression independent of modE. These findings indicate the involvement of an additional transcription factor to help coordinate nitrate- and molybdate-dependent napF expression by the Fnr, NarP, NarL, and ModE proteins. The upstream ModE regulatory site functions to override nitrate control of napF gene expression when the essential enzyme component, molybdate, is limiting in the cell environment.
Molybdenum is an essential component of the molybdopterin cofactor in nearly all species, including bacteria, plants, and animals, where it is located at the active center of a certain oxidoreductases including nitrate reductase, dimethyl sulfoxide (DMSO) reductase, trimethylamine-N-oxide (TMAO) reductase, and biotin sulfoxide reductase. Escherichia coli has evolved a regulatory scheme to coordinate molybdenum uptake, in the form of molybdate, which is utilized for cofactor synthesis and assembly into the mature molybdoenzymes. The key regulatory element in this scheme is the ModE protein, a molybdate-responsive transcription factor the structure of which was recently determined (14). ModE was first identified in E. coli as the negative regulator of the high-affinity molybdate uptake system, encoded by the modABCD operon (11, 23, 33). Utilizing a combination of in vivo and in vitro approaches, ModE was shown to bind the modA promoter in a molybdate-dependent fashion (2, 11, 23). ModE also binds and regulates expression of the moaADCDE and dmsABC operons, which encode enzymes involved in the first steps of molybdate assimilation into molybdopterin and the DMSO reductase, respectively (1, 20, 21, 23). Recently ModE was shown to play a minor role in regulating the hyc and nar operons in E. coli (29). Finally, ModE orthologues have been identified in a wide number of bacteria, including Azotobacter vinelandii, Rhodobacter capsulatus, Ralstonia eutropha (Alcaligenes eutrophus), and Thiosphaera pantotropha (Paracoccus denitrificans) (reviewed in reference 13). ModE has been shown to regulate various molybdate-associated operons in several of these cases (18, 24, 32).
In this study, we examine the role of ModE in regulating the expression of the napFDAGHBC operon of E. coli. Sequence and biochemical analysis indicates this operon encodes a molybdenum-containing periplasmic nitrate reductase (10). Expression of the E. coli napFDAGHBC operon is positively regulated in response to anaerobiosis by Fnr and by the presence of nitrate and/or nitrite by NarP (5, 7, 26, 34). Here we show that in the absence of ModE, expression of the napF operon in response to both nitrate and nitrite is dramatically reduced. A ModE site was identified well upstream of the Fnr and NarP binding sites at the napF promoter, and we demonstrate that ModE binds this site in a molybdate-responsive manner to thereby coordinate enzyme synthesis with molybdate availability. We further demonstrate that by deleting the region containing the ModE binding site or by replacing modE+ with a molybdate-independent modE allele (modE*), induction of napF-lacZ expression in response to nitrate is rendered molybdate independent.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and culture conditions .
The strains, phages, and plasmids used are listed in Table 1. P1 transductions were performed as described previously (17). For β-galactosidase assays, cells were grown either aerobically or anaerobically at 37°C with glucose (20 mM) in sodium phosphate-buffered minimal medium (pH 7) (6). Where indicated, sodium molybdate, sodium nitrate, and sodium nitrite were added at 100 μM, 40 mM, and 2.5 mM, respectively (6).
TABLE 1.
E. coli K-12 strains, plasmids, and bacteriophages
| Strain, plasmid, or phage | Origin | Relevant genotype or phenotype | Source or reference |
|---|---|---|---|
| Strains | |||
| MC4100 | F Δ(argF-lac)U169 | Laboratory stock | |
| PM6 | MC4100 | modC modE::Kanr | 20 |
| PM8 | MC4100 | modE::kanr | 20 |
| Plasmids | |||
| pR415 | pBR322 | lacZ+lacY+lacA+ Ampr | 30 |
| pPM6 | pACYC184 | modE+ Cmr | 20 |
| pPM9 | pACYC184 | modE∗; encodes a molybdate independent modE allele, Cmr | 20 |
| pHW2 | pRS415 | napF-lacZ operon fusion, Ampr | Laboratory stock |
| pPM58 and -59 | pRS415 | napF-lacZ; operon fusions with upstream deletions in napF promoter region, Ampr | This study |
| pPM69 | pPM58 | napF-lacZ; pPM58 with upstream region cloned back in correct orientation, Ampr | This study |
| pPM70 | pPM58 | napF-lacZ; pPM58 with upstream region cloned back in reverse orientation, Ampr | This study |
| pPM71 | pPM69 | napF-lacZ; pPM69 105-bp insert from the cat gene cloned in the EcoRI site, Ampr | This study |
| pPM54 and -55 | pRS415 | napF-lacZ; operon fusions with mutations in the ModE binding motif, Ampr | This study |
| Phages | |||
| λRS45 | lacZ′ lacY+lacA+ | 30 | |
| λHW2 | λRS45 | Φ(napF-lacZ) (operon fusion) | Laboratory stock |
| λPM58 and -59 | λRS45 | Φ (napF-lacZ) (operon fusions with upstream deletions in napF promoter region) | This study |
| λPM69 | λPM58 | Φ(napF-lacZ) (λPM58 with upstream region cloned back in correct orientation) | This study |
| λPM70 | λPM58 | Φ(napF-lacZ) (λPM58 with upstream region cloned back in reverse orientation) | This study |
| λPM71 | λPM69 | Φ(napF-lacZ) (λPM69 with 105-bp insert from the cat gene cloned in the EcoRI site) | This study |
| λPM54 and 55 | λRS45 | Φ(napF-lacZ) (operon fusions with mutations in the ModE binding site) | This study |
Recombinant DNA techniques.
Transformation of E. coli, plasmid isolation, and DNA manipulations were performed as described previously (19). DNA sequencing with the Sequitherm Excel kit (Epicentre Technologies) and PCR amplifications were performed according to the manufacturer's instructions. One strand of all PCR products was sequenced entirely to verify accurate amplification (data not shown).
Plasmid constructions and site-directed mutagenesis.
Segments of the napF promoter were PCR amplified from E. coli MC4100 to introduce flanking EcoRI and BamHI restriction sites. The resulting fragments were cloned into the corresponding sites in plasmid pRS415 to generate the following operon fusions: pHW2, pPM58, and pPM59. Mutations in the ModE binding site were introduced into the promoter fragment cloned in pHW2 by splicing by overlap extension (15). All napF-lacZ fusions were transferred to λRS45 to generate the corresponding prophages, which were then integrated into the chromosome of the indicated strains in single copy as previously described (23, 31).
Gel shift assays and DNase I footprint analysis.
ModE was purified as described previously (23). Gel shift assays and DNase I footprint analysis was performed as described previously (23). DNA fragments were PCR amplified and labeled by end filling with Klenow fragment. Maxam-Gilbert G reactions were run as size markers for the DNase I gel analysis (19).
β-Galactosidase assays.
β-Galactosidase levels were determined by hydrolysis of 2-nitrophenyl-β-d-galactopyranoside (ONPG), and units of activity are expressed as nanomoles of ONPG hydrolyzed per minute per milligram of protein (6). The values presented are the average of three independent experiments that deviated less than 10% from the mean.
RESULTS
ModE is required for normal napF-lacZ expression under all growth conditions.
To determine if ModE plays a role in the regulated expression from the napF promoter, we measured expression from a napF-lacZ operon fusion, λHW2 (Materials and Methods) (Fig. 1) in both wild-type (MC4100) and modE(PM8) backgrounds. Consistent with previous studies (5, 7, 26), napF-lacZ expression in the wild-type strain was increased fivefold in response to anaerobiosis and by an additional fivefold or sevenfold through the addition of either nitrate or nitrite, respectively (Table 2). In contrast, napF-lacZ expression was impaired in a modE strain under all conditions examined. Aerobic and anaerobic expression levels were lowered approximately two- and threefold, respectively. The anaerobic induction of napF-lacZ expression when either nitrate or nitrite was added was lowered by 9- and by 11-fold, respectively. Provision of modE+ in trans by introduction of plasmid pPM6 restored napF-lacZ regulation to levels seen in MC4100 (Table 2).
FIG. 1.
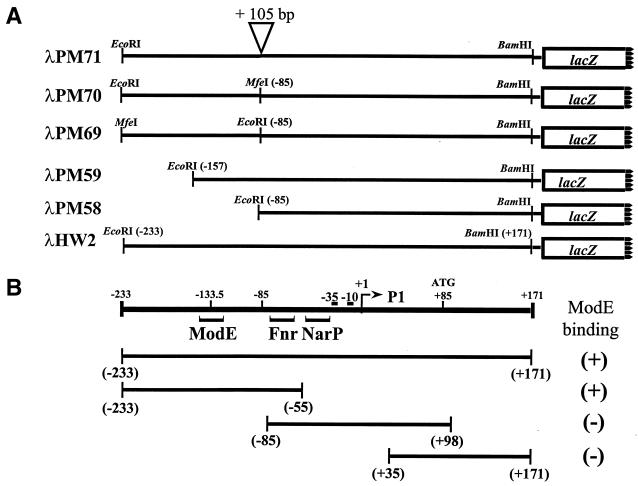
Deletion analysis and mapping of the ModE binding site at the E. coli napF promoter. Shown in panel A are the various DNA fragments, with relevant restriction sites, used in the construction of the napF-lacZ operon fusions detailed in the text. Restriction site locations relative to the start site of transcription are indicated in parentheses. Shown below is a schematic representation of the napF promoter region. The transcription start site is indicated (5), and coordinates relative to this start site are given in base pairs. The locations of Fnr, NarP (7), and ModE binding sites are indicated with brackets. (B) The DNA fragments used to map the ModE binding site are shown. The ability (+) or inability (−) of ModE to bind a particular fragment in a gel shift assay with 128 nM ModE is indicated.
TABLE 2.
Effect of a modE allele on napF-lacZ expression in response to anaerobiosis and addition of nitrate
| Strain | Relevant genotypea | β-Galactosidase activityb
|
|||||
|---|---|---|---|---|---|---|---|
| + O2, NA | −O2
|
||||||
| NA | +NO2− | +NO3− | +Mo | +NO3−, +Mo | |||
| MC4100(pACYC184) | λHW2 | 50 | 280 | 1,900 | 1,250 | 265 | 1,300 |
| PM8(pACYC184) | λHW2 modE | 25 | 85 | 170 | 130 | 85 | 125 |
| PM8(pPM6) | λHW2 modE (modE+) | 55 | 320 | 2,000 | 1,320 | 300 | 1,350 |
| PM6(pACYC184) | λHW2 modE modC | 25 | 90 | NDc | 130 | 90 | 135 |
| PM6(pPM6) | λHW2 modE modC (modE+) | 30 | 425 | ND | 140 | 305 | 1,350 |
| PM6(pPM9) | λHW2 modE modC (modE∗) | 55 | 2,340 | ND | 1,440 | 330 | 1,400 |
λHW2 is a prophage, inserted in the chromosome of the indicated strains in single copy, carrying a napF-lacZ operon fusion. Genes, present on multicopy plasmids are shown in parentheses.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic and anaerobic conditions as described in the text. Sodium nitrate (NO3−), sodium nitrite (NO2−), and sodium molybdate (Mo) were added where indicated at 40 mM, 2.5 mM, and 100 μM, respectively; NA, no addition of nitrate, nitrite, or molybdate.
ND, not determined.
To establish if the ModE control was molybdate responsive, we repeated the assays described above with the isogenic strain PM6, which is both modE and modC. The modC mutation blocks molybdate transport via the high-affinity modABC uptake system and can be phenotypically suppressed by supplementing the medium with large amounts (ca. 100 μM) of molybdate (27). The introduction of the modC mutation into the modE background had no further effect on napF-lacZ expression. However, when modE+ was provided in trans (i.e., on plasmid pPM6), a wild-type pattern of napF regulation was seen, but only if the medium was supplemented with molybdate (Table 2). Interestingly, under anaerobic growth conditions, napF-lacZ expression in strain PM6 containing modE+on plasmid pPM6 was slightly higher than in the wild-type strain grown under the same conditions. The addition of nitrate caused a modest reduction in napF gene expression (Table 2). When these assays were repeated with a strain containing a molybdate-independent modE allele, modE* (expressed from plasmid pPM9), we observed an even greater increase in anaerobic expression. Again, addition of molybdate, and to a lesser degree nitrate, reduced this effect; this hyper-induction phenomenon is addressed below in the Discussion.
ModE binds the napF promoter well upstream of the start site of transcription.
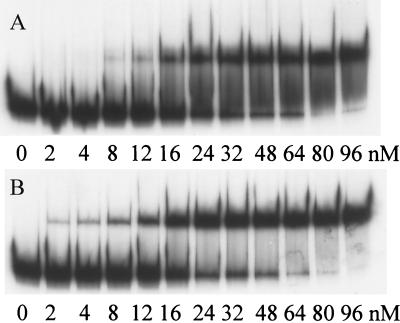
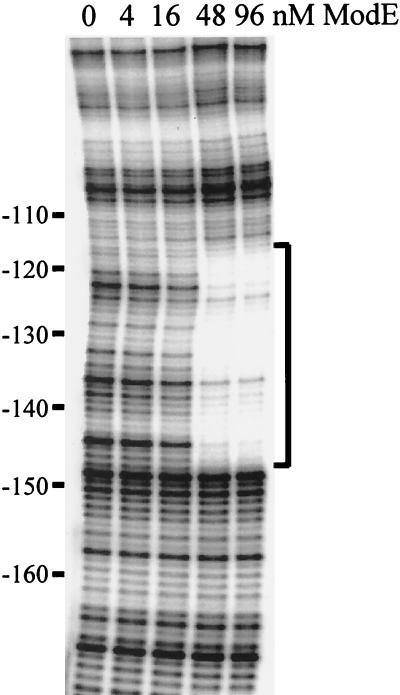
Inspection of the napF promoter region revealed a close match to the proposed E. coli ModE consensus recognition sequence reported by Anderson, McNicholas, and McNicholas (2, 20, 23). The putative binding site is centered at position −133.5 with respect to the napF transcript start site (Fig. 1). To establish that ModE binds the napF promoter fragment contained in λHW2 (Fig. 1), we performed gel shift assays with purified protein. ModE bound this fragment with high affinity and displayed an apparent dissociation constant (Kd) of 35 nM (Fig. 2A). When the gel shift was repeated in the presence of molybdate (100 μM) in the reaction buffer (Fig. 2B), a twofold decrease in the apparent Kd was observed (16 μM), consistent with molybdate binding (12). To rule out the possibility that other ModE binding sites exist elsewhere within the napF promoter region, we repeated the gel shift assays with three truncated promoter fragments (Fig. 1B). These studies localized the ModE binding site between positions −233 and −55 and ruled out the presence of additional ModE sites located near the start of napF transcription (data not shown). To precisely identify where ModE binds, DNase I footprinting was performed (Fig. 3). ModE protected a 30-bp region (nucleotides −147 to −118) centered at position −133.5. Thus, this region contains a ModE binding site typical of others on the chromosome (22). Since several nucleotides were not completely protected (i.e., at positions −123, −125, −136, and −138), ModE may reside on one face of the DNA.
FIG. 2.
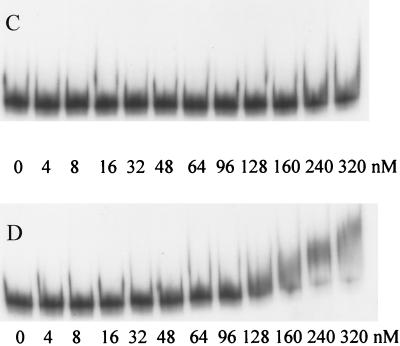
Interaction of ModE with napF promoter DNA. Increasing amounts of purified ModE protein were incubated with a labeled napF promoter fragment from λHW2. (A) Wild-type napF promoter DNA and ModE without molybdate added. (B) Wild-type napF promoter DNA and ModE with 100 μM molybdate added. (C) Mutated napF promoter DNA from λPM54. (D) Mutated napF promoter DNA from λPM55.
FIG. 3.
DNase I footprint analysis of ModE interaction at napF. The pattern of protection when ModE is bound at napF in the presence of 100 μM molybdate is shown. The vertical bracket indicates the region of protection. Coordinates relative to the start site of transcription are given in base pairs.
Deletion of the ModE binding site relieves the need for modE.
It was previously shown that the deletion of napF sequences upstream of position −85, which contains the ModE binding site centered at position −133.5, had no effect on wild-type napF-lacZ expression in response to either anaerobiosis or nitrate addition (7). To confirm this observation, we constructed a similar napF-lacZ fusion; a fragment of DNA spanning nucleotides −233 to −85 (with respect to the transcript start site) was deleted from λHW2 to give λPM58 (Fig. 1A). In the wild-type strain (MC4100) the pattern of napF-lacZ expression from λPM58 was similar to the full-length fusion contained on λHW2 (Table 3). However, in direct contrast to the ModE-dependent expression seen from λHW2, napF-lacZ expression from λPM58 was unaffected in a modE deletion strain. When we reintroduced the upstream DNA segment back into λPM58 to give λPM69, napF-lacZ expression was restored to modE dependency. (Note that in constructing these plasmids, we mutated 3 bp to introduce a unique EcoRI site at the downstream cloning junction [Fig. 1A].) Finally, to establish that DNA sequences 5′ of the ModE binding site were not required for the molybdate response, λPM59 was constructed where the nucleotides from −233 to −157 were deleted. (Nucleotide −147 marks the upstream boundary of the ModE binding site.) The pattern of napF-lacZ expression from λPM59 was identical to that of λHW2 in both wild-type and in modE backgrounds under all conditions tested (data not shown).
TABLE 3.
Effects of deletions and insertions in upstream DNA on napF-lacZ expression in response to anaerobiosis and nitrate addition
| Strain | Relevant genotypea | β-Galactosidase activityb
|
||
|---|---|---|---|---|
| + O2, −NO3− | −O2
|
|||
| −NO3− | +NO3− | |||
| MC4100 | λHW2 | 50 | 280 | 1,250 |
| PM8 | λHW2 modE | 25 | 85 | 130 |
| MC4100 | λPM58 | 55 | 300 | 1,505 |
| PM8 | λPM58 modE | 60 | 305 | 1,420 |
| MC4100 | λPM69 | 45 | 260 | 1,150 |
| PM8 | λPM69 modE | 20 | 80 | 115 |
| MC4100 | λPM70 | 40 | 220 | 1,050 |
| PM8 | λPM70 modE | 40 | 210 | 950 |
| MC4100 | λPM71 | 35 | 205 | 870 |
| PM8 | λPM71 modE | 35 | 195 | 845 |
Prophages λPM58, -59, -70, and -71, carrying various alterations in the upstream region of napF and their wild-type progenitor, λHW2, were inserted in the chromosome of the indicated strains in single copy.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic and anaerobic conditions as described in the text. Sodium nitrate (NO3) was added (at 40 mM) where indicated.
A correctly positioned ModE cis-acting site is essential for regulating napF-lacZ expression.
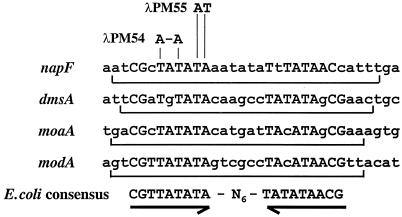
The analysis presented above demonstrated that deletion of upstream DNA sequences containing the ModE binding site alleviated the in vivo requirement for molybdate and ModE for optimal napF-lacZ expression. To confirm that modE has an essential role in regulating napF-lacZ expression, we performed site-directed mutagenesis of conserved residues in the ModE recognition sequence centered at −133.5 relative to the napF promoter (Fig. 1). Two sets of 2-bp substitutions were introduced (Fig. 4), and the resultant promoter fragments were fused to lacZ, generating λPM54 and λPM55 (i.e., the λPM54 and λPM55 fusions each differ from λHW2 by only 2 bp). The effects of the mutations on ModE binding were assayed by in vitro gel shift assays (Fig. 2C and D). The promoter fragment from λPM54 no longer bound ModE (i.e., even when a 10-fold-higher level of ModE was used relative to the amount needed to shift the wild-type fragment). The altered promoter fragment from λPM55 displayed a sevenfold reduction in ModE binding.
FIG. 4.
Alignment of the ModE binding site at the napF, dmsA, modA, and moaA promoters with the proposed ModE consensus sequence. Nucleotides protected from DNaseI digestion are bracketed, and nucleotide matches to the ModE consensus sequence are shown in uppercase (2, 20, 23).
The ModE binding site mutations were also evaluated in vivo by measuring β-galactosidase expression in a wild-type strain lysogenized with either λPM54 or λPM55 (Table 4). When cells were grown aerobically, anaerobically, or anaerobically with nitrate present, the ModE recognition site mutations markedly reduced napF-lacZ expression under each condition. Furthermore, introduction of a modE chromosomal deletion into strains carrying either λPM54 or λPM55 had no effect on gene expression (Table 4). Thus, the cis-acting mutations confer a ModE− phenotype.
TABLE 4.
Effect of introducing mutations in the modE operator site on napF-lacZ expression in response to anaerobiosis and nitrate addition
| Strain | Relevant genotype | Mutations in ModE binding sitea | β-Galactosidase activityb
|
In vitro ModE bindingc | ||
|---|---|---|---|---|---|---|
| + O2, −NO3− | − O2
|
|||||
| −NO3− | +NO3− | |||||
| MC4100 | λHW2 | CGCTATATA-N6-TTTATAACC | 50 | 280 | 1,250 | Wild type |
| PM8 | λHW2 modE | 25 | 85 | 130 | ||
| MC4100 | λPM54 | CGCaAaATA-N6-TTTATAACC | 30 | 90 | 140 | Absent |
| PM8 | λPM54 modE | 25 | 90 | 135 | ||
| MC4100 | λPM55 | CGCTATAat-N6-TTTATAACC | 30 | 100 | 150 | Seven-fold reduction |
| PM8 | λPM55 modE | 30 | 85 | 125 | ||
Prophage λHW2 carries a copy of the wild-type ModE binding site. The mutations in the ModE binding site carried on the prophages λPM54 and -55 are underlined and shown in lowercase. All prophages are inserted in the chromosome of strains MC4100 and PM8 in single copy.
Units are given in nanomoles of ONPG hydrolyzed per minute per milligram of protein. Cells were grown in minimal glucose medium under aerobic and anaerobic conditions as described in the text. Sodium nitrate (NO3−) was added (at 40 mM) where indicated.
In vitro binding was measured via gel shift assays.
Finally, to establish if the relative position of the ModE site was important, a 105-bp insertion was made at position −85 relative to the start of napF transcription to give λPM71 (Fig. 1). Although expression from the λPM71 fusion was slightly reduced when compared to that of the wild-type fusion (λHW2, Table 3), it was independent of ModE (i.e., repositioning the ModE site upstream by 10 helix turns was equivalent to deleting the ModE binding site region). In a similar manner, the orientation of the MfeI-EcoRI fragment in λPM69 was reversed to invert the ModE site and move it from position −135.5 to a new position centered at −78.5 relative to the napF transcript start site (λPM70; Fig. 1). This rearrangement also abolished ModE control (Table 3).
Nitrate induction of napF-lacZ (λPM58) expression is molybdate independent.
As noted above, replacement of the wild-type modE gene with a molybdate-independent allele, modE*, abolished the requirement for molybdate for optimal napF-lacZ expression in response to nitrate addition when the upstream ModE binding site was present (i.e., λHW2; Table 2). To confirm that the molybdate requirement for napF-lacZ expression operates solely through modE, we introduced λPM58 (this fusion has a complete deletion of the ModE binding site) into a modC strain and measured gene expression in response to anaerobiosis and addition of nitrate. The modC mutation had no effect on napF-lacZ expression from λPM58 under any growth condition (data not shown), thus demonstrating that it is modE independent.
IHF plays a minor role in regulating napF-lacZ expression.
A putative integration host factor (IHF) binding site was previously identified in the upstream region of the napF promoter (5). The ModE DNaseI footprinting experiments indicate that this proposed IHF site would overlap the left half of the ModE binding site by 5 bp. (The upstream boundary of the putative IHF site is at position −126, and the downstream boundary of the ModE binding site is at −122.) To determine if IHF plays a role in regulating napF-lacZ expression, we transduced a himA allele into wild-type (MC4100) and modE (PM8) strains that harbor λHW2. Compared to the wild-type strain, napF-lacZ expression in a himA strain was elevated twofold under all growth conditions (data not shown). Increased napF-lacZ gene expression was also seen in a himA modE strain, although the overall expression levels were lower due to the modE mutation (data not shown). Therefore, IHF serves a nonessential role in modulating napF gene expression.
DISCUSSION
The E. coli napFDAGHBC operon encodes a periplasmic nitrate reductase enzyme (10) similar to those encoded by the nap operons of other bacteria, including Rhodobacter capsulatus, Ralstonia eutropha (Alcaligenes eutrophus), and Thiosphaera pantotropha (3, 30). The E. coli enzyme is predicted to contain a molybdopterin moiety that raised the possibility that napFDAGHBC operon expression may be ModE dependent. Utilizing a napF-lacZ operon fusion, we demonstrate that a modE deletion impairs napF-lacZ expression by 10-fold. Sequence analysis of the napF promoter region identified a typical ModE binding site, centered at −133.5 bp with respect to the transcript start site. By utilizing a combination of gel shift and DNase I footprinting assays, we confirmed that ModE binds the napF promoter at this location with high affinity. Whereby the addition of molybdate modulated DNA binding by twofold in vitro (Fig. 2), molybdate addition caused a fivefold change in napF gene expression in vivo (Table 2). It is yet unclear if ModE interactions with molybdate act primarily to modulate DNA binding or, alternatively, to affect ModE interactions with other proteins involved in napF gene expression, including Fnr, NarP, NarL, and RNAP. The molybdate-dependent conformational changes within ModE are consistent with either model (9).
Interestingly, deletion of the ModE binding site at the napF promoter alleviates the requirement for modE without affecting napF-lacZ expression (Table 3). Given the distal 3′ location of the ModE binding site relative to ModE sites at other molybdate-regulated promoters, ModE-dependent napF regulation may somehow involve some type of DNA looping event to bring the bound ModE into contact with the other transcription activators. Since the introduction of a himA allele had only a twofold effect, it is unlikely that IHF plays a major role in this process. One possibility is that another general DNA binding protein occupies part of the ModE site and somehow suppresses napF gene expression when molybdate is limiting. Binding of ModE to the DNA under molybdate-sufficient conditions relives this control. It remains to be determined at the molecular level how ModE exerts its effects at the napF promoter.
Introduction of the molybdate-independent modE* allele, which encodes a molybdate-independent variant of ModE, into a modE modC strain resulted in an unusually large (when compared to the wild-type strain) increase in napF-lacZ expression in response to anaerobiosis. The addition of molybdate and, to a lesser degree, nitrate resulted in a drop in gene expression. One explanation for these findings is that the inhibition of molybdate uptake, caused by the modC mutation, results in inactivation of the cell's complement of functional NarG and NapF nitrate reductase enzymes (i.e., inability to synthesize mature molybdoenzymes). Consequently, the cell is unable to metabolize any trace amounts of nitrate that may be present in the cell growth medium. As noted recently (34), trace amounts of nitrate would result in a large increase in napF-lacZ expression. The requirement for low levels of nitrate would also explain why napF-lacZ expression was lowered when nitrate was added to the medium (34). Thus, the provision of trace molybdate in the medium signals for the synthesis of the periplasmic molybdoenzyme for nitrate reduction under these conditions.
Nitrate induction of napF-lacZ expression in a modE modC double mutant was found to be independent of molybdate when modE+ was replaced by a modE* allele. Similarly, expression from a napF-lacZ fusion, which lacked the ModE binding site (λPM58), was unaffected by the introduction of a modC mutation. This finding is in direct contrast to those of previous studies that reported the cellular response to nitrate addition to be largely abolished in a modC background (6, 16, 25). These data were taken to imply that the Nar regulon senses molybdate as well as nitrate. Our studies strongly suggest that for expression originating from the napF promoter, the only molybdate-requiring component involved in mediating the response to nitrate addition is ModE.
The napF promoter is the fourth promoter at which we have characterized a ModE binding site. Based upon the ModE consensus sequence (Fig. 4), we searched for other putative ModE binding sites by using the PatScan program (8). Matches were checked to see if the site was located within the promoter regions of a gene or operon that encoded either molybdoenzymes or proteins involved in molybdate uptake and/or utilization. In E. coli, we found two additional candidates. One lies immediately upstream of an uncharacterized operon (accession no. g1787870) that encodes proteins that are highly homologous to the products of the dmsABC operon (4). The second match lies within 15 bp upstream of the translational start site of open reading frame 95 (ORF95; accession no. U28377), the product of which is unknown. However, ORF95 is directly upstream of hybO, the first gene of the hybOABCDEFG operon, which encodes the hydrogenase 2 complex (28). In Haemophilus influenzae, we also identified putative ModE binding sites upstream of the moaACDE and modABC operons and upstream of the modA and torC homologues (data not shown). Finally, as already noted (18), we also found matches upstream of the anfA homologue and modABC operons in both Rhodobacter capsulatus and Azotobacter vinelandii. Thus, ModE appears to play a global role in regulating molybdenum homeostasis in a number of bacterial species.
Acknowledgments
We thank Sabine Rech for supplying purified ModE protein, Mandy Mazzotta for help with the β-galactosidase assays and Henian Wang for construction of pHW2.
This work was supported in part by a grant from the National Institutes of Health, AI21678.
REFERENCES
- 1.Anderson, L. A., E. McNairn, T. Leubke, R. N. Pau, and D. H. Boxer. 2000. ModE-dependent molybdate regulation of the molybdenum cofactor operon moa in Escherichia coli. J. Bacteriol. 182:7035-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L. A., T. Palmer, N. C. Price, S. Borneman, D. H. Boxer, and R. N. Pau. 1997. Characterization of the molybdenum-responsive ModE regulatory protein and its binding to the promoter region of the modABCD (molybdenum transport) operon of Escherichia coli. Eur. J. Biochem. 246:119-126. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., D. J. Richardson, A. Reilly, A. C. Willis, and S. J. Ferguson. 1995. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem. J. 309:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997.The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Choe, M., and W. S. Reznikoff. 1993. Identification of the regulatory sequence of anaerobically expressed locus aeg-46.5. J. Bacteriol. 175:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. A., and R. P. Gunsalus. 1989. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J. Bacteriol. 171:3817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darwin, A. J., and V. Stewart. 1995. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J. Mol. Biol. 251:15-29. [DOI] [PubMed] [Google Scholar]
- 8.Dsouza, M., N. Larsen, and R. Overbeek. 1997. Searching for patterns in genomic data. Trends Genet. 13:497-498. [DOI] [PubMed] [Google Scholar]
- 9.Gourley, D. G., A. W. Schuttelkopf, L. A. Anderson, N. C. Price, D. H. Boxer, and W. N. Hunter. 2001. Oxyanion binding alter conformation and quaternary structure of the C-terminal domain of the transcriptional regulator ModE. J. Biol. Chem. 276:20641-20647. [DOI] [PubMed] [Google Scholar]
- 10.Grove, J., S. Tanapongpiat, G. Thomas, L. Griffiths, H. Crooke, and J. Cole. 1996. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol. Microbiol. 19:467-481. [DOI] [PubMed] [Google Scholar]
- 11.Grunden, A. M., R. M. Ray, J. K. Rosentel, F. G. Healy, and K. T. Shanmugam. 1996. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J. Bacteriol. 178:735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunden, A. M., W. T. Self, M. Villain, J. E. Blalock, and K. T. Shanmugam. 1999. An analysis of the binding of repressor protein ModE to modABCD (molybdate transport) operator/promoter DNA of Escherichia coli. J. Biol. Chem. 274:24308-24315. [DOI] [PubMed] [Google Scholar]
- 13.Grunden, A. M., and K. T. Shanmugam. 1997. Molybdate transport and regulation in bacteria. Arch. Microbiol. 168:345-354. [DOI] [PubMed] [Google Scholar]
- 14.Hall, R. D., D. G. Gourley, G. A. Leonard, E. M. H. Duke, L. A. Anderson, D. H. Boxer, and W. N. Hunter. 1999. The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds. EMBO J. 18:1435-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering of hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 16.Iuchi, S., and E. C. C. Lin. 1987. Molybdenum effector of fumarate reductase repression and nitrate reductase induction in Escherichia coli. J. Bacteriol. 169:3720-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons, with emphasis on Tn 10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 18.Kutsche, M., S. Leimkühler, S. Angermüller, and W. Klipp. 1996. Promoters controlling expression of the alternative nitrogenase and the molybdenum uptake system in Rhodobacter capsulatus are activated by NtrC, independent of σ54, and repressed by molybdenum. J. Bacteriol. 178:2010-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.McNicholas, P. M., R. C. Chiang, and R. P. Gunsalus. 1998. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol. Microbiol. 27:197-208. [DOI] [PubMed] [Google Scholar]
- 21.McNicholas, P. M., R. C. Chiang, and R. P. Gunsalus. 1996. The Escherichia coli modE gene: effect of modE mutations on molybdate dependent modA expression. FEMS Microbiol Lett. 145:117-123. [DOI] [PubMed] [Google Scholar]
- 22.McNicholas, P. M., M. M. Mazzotta, S. A. Rech, and R. P. Gunsalus. 1998. Functional dissection of the molybdate-responsive transcription regulator, ModE, from Escherichia coli. J. Bacteriol. 180:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNicholas, P. M., S. A. Rech, and R. P. Gunsalus. 1997. Characterization of the ModE DNA-binding sites in the control regions of modABCD and moaABCDE of Escherichia coli. Mol. Microbiol. 23:515-524. [DOI] [PubMed] [Google Scholar]
- 24.Mouncey, N. J., L. A. Mitchenall, and R. N. Pau. 1996. The modE gene product mediates molybdenum-dependent expression of genes for the high-affinity molybdate transporter and modG in Azotobacter vinelandii. Microbiology 142:1997-2004. [DOI] [PubMed] [Google Scholar]
- 25.Pascal, M.-C., J.-F. Burini, J. Ratouchniak, and M. Chippaux. 1982. Regulation of the nitrate reductase operon: effect of mutations in chlA, B and E genes. Mol. Gen. Genet. 188:103-106. [DOI] [PubMed] [Google Scholar]
- 26.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rech, S., U. Deppenmeier, and R. P. Gunsalus. 1995. Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J. Bacteriol. 177:1023-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sargent, F., S. P. Ballantine, P. A. Rugman, T. Palmer, and D. H. Boxer. 1998. Reassignment of the gene encoding the Escherichia coli hydrogenase 2 small subunit identification of a soluble precursor of the small subunit in a hypB mutant. Eur. J. Biochem. 255:746-754. [DOI] [PubMed] [Google Scholar]
- 29.Self, W. T., A. M. Grunden, A. Hason, and K. T. Shanmugam. 1999. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41-55. [DOI] [PubMed] [Google Scholar]
- 30.Siddiqui, R. A., U. Warnecke-Eberz, A. Hengsberger, B. Schneider, S. Kostka, and B. Friedrich. 1993. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J. Bacteriol. 175:5867-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 32.Solomon, P. S., A. L. Shaw, M. D. Young, S. Leimkuhler, G. R. Hanson, W. Klipp, and A. G. McEwan. 2000. Molybdate-dependent expression of dimethylsulfoxide reductase in Rhodobacter capuslatus. FEMS Microbiol. Lett. 190:203-208. [DOI] [PubMed] [Google Scholar]
- 33.Walkenhorst, H. M., S. K. Hemschemeier, and R. Eichenlaub. 1995. Molecular analysis of the molybdate uptake operon, modABCD, of Escherichia coli and modR, a regulatory gene. Microbiol. Res. 150:347-361. [DOI] [PubMed] [Google Scholar]
- 34.Wang, H., C.-P. Tseng, and R. P. Gunsalus. 1999. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J. Bacteriol. 181:5303-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]