Abstract
A system for high-level expression of heparinase I, heparinase II, heparinase III, chondroitinase AC, and chondroitinase B in Flavobacterium heparinum is described. hepA, along with its regulatory region, as well as hepB, hepC, cslA, and cslB, cloned downstream of the hepA regulatory region, was integrated in the chromosome to yield stable transconjugant strains. The level of heparinase I and II expression from the transconjugant strains was approximately fivefold higher, while heparinase III expression was 10-fold higher than in wild-type F. heparinum grown in heparin-only medium. The chondroitinase AC and B transconjugant strains, grown in heparin-only medium, yielded 20- and 13-fold increases, respectively, in chondroitinase AC and B expression, compared to wild-type F. heparinum grown in chondroitin sulfate A-only medium. The hepA upstream region was also studied using cslA as a reporter gene, and the transcriptional start site was determined to be 26 bp upstream of the start codon in the chondroitinase AC transconjugant strain. The transcriptional start sites were determined for hepA in both the wild-type F. heparinum and heparinase I transconjugant strains and were shown to be the same as in the chondroitinase AC transconjugant strain. The five GAG lyases were purified from these transconjugant strains and shown to be identical to their wild-type counterparts.
Flavobacterium heparinum (Cytophaga heparina [3], Sphingobacterium heparinum [36], and Pedobacter heparinus [33]), is a nonpathogenic soil bacterium that was isolated by Payza and Korn (23). The bacterium was described as a strictly aerobic, gram-negative, nonsporing rod that produces a yellow pigment when grown on agar plates (33). It synthesizes five enzymes, three heparinases, and two chondroitinases that degrade heparin and acidic mucoheteropolysaccharides with sulfate groups from various animal tissues and uses them as sole sources of carbon, nitrogen, and energy (6, 11, 17).
Heparinases from F. heparinum have been studied extensively. Three heparinases, heparinase I (HepI), HepII, and HepIII, have been purified to homogeneity and characterized (19, 39). HepI is described as a 43-kDa enzyme that degrades mainly heparin, HepII is a 85-kDa enzyme that depolymerizes both heparin and heparan sulfate, and HepIII is a 71-kDa enzyme that degrades mainly heparan sulfate (19). The heparinase genes, hepA (coding for HepI) (27), hepB (coding for HepII), and hepC (coding for HepIII) (34), were cloned and sequenced. Molecular analysis of the three heparinases revealed no significant homology either at the DNA or protein levels, nor were they closely linked on the F. heparinum chromosome (34). The heparinase genes were expressed recombinantly in Escherichia coli with intact biological function (27, 34). Structural and functional studies employing chemical modifications and site-directed mutagenesis were also conducted for both HepI and HepII and revealed that, in both cases, a histidine residue played a critical role in their catalytic function (10, 28). In addition, two putative calcium binding sites were identified in HepI that were shown to be essential to HepI's catalytic function (18, 29). However, due to the absence of a genetic system for the introduction of DNA into F. heparinum, these studies were performed with E. coli.
Two chondroitinases from F. heparinum, chondroitinases AC (ChnA) and B (ChnB), were purified and characterized. It was shown that ChnA is a 75-kDa enzyme degrading both chondroitin sulfate A and C and that ChnB is a 55-kDa enzyme degrading only chondroitin sulfate B (11). The cslA and cslB genes coding for ChnA and ChnB, respectively, were cloned, sequenced, and expressed in E. coli with biological function (37). Molecular analysis indicated that the cslA and cslB genes shared no significant homology either at the DNA or peptide level but were separated by approximately 5 kbp on the F. heparinum chromosome and were translated in the same orientation (37). In addition, both enzymes were crystallized and their structure was resolved (7, 8, 13, 16). These studies suggested that the chondroitinases were very different with respect to their structures and catalytic mechanisms.
These glycosoaminoglycan-degrading enzymes from F. heparinum display another particularity. They are posttranslationally modified by glycosylation. It was shown that HepI, HepII, and ChnB carried one carbohydrate moiety, while ChnA possessed two [M. Laliberte, B. Eggimann, J. J. F. Zimmermann, L. Huang, and H. Van Halbeek, 10th Symp. Protein Soc., Protein Sci. 5(Suppl. 1):435s, 1996]. The glycosylation site(s) was identified for each enzyme and contained the consensus sequence Asp-Ser or Asp-Thr, which resembles the sequence described for Flavobacterium meningosepticum (24). Structural analysis of the carbohydrate moiety from HepI using nuclear magnetic resonance and mass spectroscopy showed it to be an O-linked branched heptasaccharide with a molecular mass of 1,161 Da (12).
GAG lyases from F. heparinum are presently being developed for therapeutic applications. HepI has been used clinically to neutralize the anticoagulant properties of heparin (1). HepI and HepIII have been shown to regulate various cellular processes in vitro, such as adhesion, differentiation, migration, and proliferation (14, 15, 30). ChnA and ChnB were shown to inhibit fibroblast proliferation and tumor cell invasion, proliferation, and angiogenesis (4). The limited availability of these enzymes has been the main hurdle to conducting in-depth in vitro and preclinical studies to fully explore their potential as therapeutic agents. It has been shown that the heparinases and chondroitinases could be produced in E. coli recombinantly; however, many problems associated with protein expression, protein degradation, and refolding from inclusion bodies made the process inefficient. The development of a gene expression system in F. heparinum would thus offer the best alternative.
Until recently, it was not possible to introduce DNA into F. heparinum (35). We have constructed a DNA transfer system for this microorganism by assembling (i) an E. coli mobile plasmid for conjugative DNA transfer, (ii) a DNA fragment from the F. heparinum chromosome to facilitate homologous recombination, and (iii) an F. heparinum function-selective marker that was constructed by placing the trimethoprim resistance gene under the control of the hepA regulatory region. This plasmid was successfully introduced into F. heparinum by conjugation where it integrated into the chromosome by homologous recombination (35). In order to transform this plasmid system into a high-level gene expression system, a strong promoter remained to be identified.
Little information is available on the regulation of gene expression in F. heparinum. The only thorough studies have been those conducted on the expression of these glycosaminoglycan-degrading enzymes. It was shown that heparin- and heparan sulfate-degrading activities are present at very low levels in F. heparinum grown in glucose minimal medium. The activities increased with the addition of the inducer, heparin, and peaked when cells were grown in heparin-only medium (9). The analysis of these heparinases, synthesized from F. heparinum cells grown on heparin-only medium, showed that HepI was the most abundant, with an estimated expression level of 0.3% of total cellular protein (19). The hepA promoter seemed to offer the characteristics required in a gene expression system.
In this paper, we describe a high-level recombinant protein expression system for F. heparinum. The hepA upstream region was used to regulate gene expression. We demonstrated that high levels of hepB, hepC, cslA, and cslB expression could be achieved using this system. In addition, a significant increase in hepA expression was observed upon introduction of a second chromosomal copy of the gene. Studies on the regulation of the hepA upstream region could not reveal the mechanism responsible for such high levels of GAG lyase expression. The transcriptional start site of hepA was determined for wild-type F. heparinum and for the hepA and cslA expression strains. It was found that the hepA gene transcripts, either from the wild-type or hepA and cslA transconjugant strains, started 26 bp from hepA's start codon. The GAG lyases from these recombinant F. heparinum strains were purified and shown to be identical to their native counterparts.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in Luria broth (LB) at 37°C unless stated otherwise. The growth medium for F. heparinum (FH) was described by Zimmermann et al. (40) and Su et al. (35). For the study of GAG lyase gene expression in F. heparinum, FM (40) was supplemented with 1% heparin (MH) or 1% chondroitin sulfate A (MA). For the study on regulation of gene expression, F. heparinum strains were grown in the appropriate medium for 48 to 72 h, diluted to an optical density at 600 nm (OD600) of 0.2 in fresh medium, and grown for an additional 32 h at 23°C.
TABLE 1.
Bacterial strains, λ phages, and plasmidsa
| Strain, phage, or plasmid | Genotype and relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| S17-1 | hsdR17(rK−mK−) recA RP4-2(Tcr::Mu-Kmr::Tn7 Strr) | 31 |
| XL-1 Blue | Stratagene | |
| F. heparinum | ||
| ATCC 13125 | Wild type | ATCCb |
| FIBX1 | pIBXF1 integrated into the F. heparinum chromosome Tprf | 35 |
| FIBX3 | pIBXF3 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX4 | pIBXF4 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX5 | pIBXF5 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX6 | pIBXF6 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX7 | pIBXF7 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX8 | pIBXF8 in the F. heparinum chromosome Tprf | This study |
| FIBX9 | pIBXF9 in the F. heparinum chromosome Tprf | This study |
| FIBX10 | pIBXF10 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX11 | pIBXF11 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX12 | pIBXF12 integrated into the F. heparinum chromosome Tprf | This study |
| FIBX13 | pIBXF13 integrated into the F. heparinum chromosome Tprf | This study |
| λ phages | ||
| λIB2 | λ phage containing hepB gene | 34 |
| λIB4 | λ phage containing hepC gene | 34 |
| Plasmids | ||
| pUC21 | Apre | 38 |
| R751 | IncP, Tpre | 20 |
| pIB10 | pACYC184 containing 10-kbp hepA DNA fragment | 34 |
| pIB15 | pBluescript containing 6-kbp cslA gene, Apre | 37 |
| pIB16 | pBluescript containing 5-kbp cslB gene, Apre | 37 |
| pIB17 | 2.2-kb, BamHI-HindIII, hepA DNA fragment in pUC21, Apre | 35 |
| pIB18 | 830-bp hepA upstream sequence in pTZ/PC, Apre | 35 |
| pIB23 | 2.1-kbp, NdeI-XbaI, hepB DNA fragment in pUC21, Apre | This study |
| pIB24 | 1.5-kbp, NdeI-XbaI, hepC DNA fragment in pUC21, Apre | This study |
| pIB25 | 830-bp hepA upstream region cloned into pIB23, Apre | This study |
| pIB26 | 830-bp hepA upstream region cloned into pIB24, Apre | This study |
| pIB27 | 2.1-kb, EcoRI-BamHI, cslA DNA fragment in pUC21, Apre | This study |
| pIB28 | 1.3-kbp 5′ end of cslB DNA fragment in pUC21, Apre | This study |
| pIB29 | 2.3-kb cslB DNA fragment in pUC21, Apre | This study |
| pIB30 | 830-bp hepA upstream region cloned into pIB27, Apre | This study |
| pIB31 | 830-bp hepA upstream region cloned into pIB29, Apre | This study |
| pIB32 | 50-bp hepA upstream region fused to cslA gene, Apre | This study |
| pIB33 | 250-bp hepA upstream region fused to cslA gene, Apre | This study |
| pIB34 | 461-bp hepA upstream region fused to cslA gene, Apre | This study |
| pIB35 | 810-bp dhfrII EcoRI-XmnI DNA fragment in pUC21, Apre | This study |
| pIB36 | 800-bp dhfrII in plasmid pIB30, Apre | This study |
| pIB37 | 3.5-kbp hepA upstream region fused to cslA gene in plasmid pIB35, Apre | This study |
| pIBXF1 | 10-kbp HindIII DNA fragment in pIB21, Apre, Tprf | 35 |
| pIBXF2 | pBBCmRI derivative with mob, Apre, and Tprf | 35 |
| pIBXF3 | 3.1-kbp DNA fragment containing hepBHU coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF4 | 2.9-kbp DNA fragment containing hepCHU coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF5 | 2.3-kbp DNA fragment containing hepA cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF6 | 2.9-kbp DNA fragment containing cslAHU coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF7 | 3.1-kbp DNA fragment containing cslBHU coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF8 | 2.9-kbp DNA fragment containing cslAHU coding region cloned into plasmid pIBXF2, Apre, Tprf | This study |
| pIBXF9 | 3.1-kbp DNA fragment containing cslBHU coding region cloned into plasmid pIBXF2, Apre, Tprf | This study |
| pIBXF10 | 2.2-kbp DNA fragment containing 50-bp hepA upstream region and cslA coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF11 | 2.4-kbp DNA fragment containing 250-bp hepA upstream region and cslA coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF12 | 2.6-kbp DNA fragment containing 460-bp hepA upstream region and cslA coding region cloned into plasmid pIBXF1, Apre, Tprf | This study |
| pIBXF13 | 6.5-kbp DNA fragment containing 3.5-kb hepA upstream region, cslA coding region, and 800-bp dhfrII from R751 cloned into plasmid pIBXF1, Apre, Tprf | This study |
“re” superscript indicates expression in E. coli; “rf” indicates expression in F. heparinum.
ATCC, American Type Culture Collection.
Conjugation.
The conjugation procedure was used as described previously (35).
Molecular biology techniques.
Isolation of chromosomal DNA, cloning, and DNA manipulation techniques were performed as described by Sambrook et al. (25). T4 DNA ligase and restriction endonucleases were purchased from New England Biolabs (Mississauga, Canada). DNA fragments destined for ligation were first separated by agarose gel electrophoresis, and the DNA was extracted from the agarose with a Geneclean I kit (Bio 101, La Jolla, Calif.). DNA was introduced into E. coli by electroporation using a Gene Pulser electroporator (Bio-Rad, Mississauga, Canada) as recommended by the manufacturer. E. coli transformants were first analyzed by colony cracking (25), and plasmid DNA was isolated for restriction analysis using the RPM kit (Bio 101). DNA was prepared for Southern blotting (32) by digestion with the appropriate restriction endonucleases and separation by electrophoresis on a 0.8% agarose gel followed by transfer to a nylon membrane (Hybond-N; Amersham Pharmacia Biotech, Oakville, Canada). The probes were labeled with [α-32P]dATP using the RediprimeII random primer labeling system from Amersham Pharmacia Biotech; hybridizations and washes were performed as previously described (35). Oligonucleotide primer synthesis, preparation of genomic F. heparinum DNA or plasmid DNA for use as template, and analysis of PCR-generated products were all performed as previously described (34). PCR amplification was performed according to Mullis et al. (21) with the modifications described previously (34). Pfu DNA polymerase (Stratagene) was used to generate the hepB, hepC, cslA, and cslB gene products from the appropriate templates. The annealing temperature was 50°C. DNA sequences were determined by the dideoxy chain termination method of Sanger et al. (26).
Purification of GAG lyases.
Purification of the heparinases and chondroitinases from F. heparinum was described previously (11, 34).
Enzyme assay.
Heparin-, heparan sulfate-, chondroitin sulfate A-, chondroitin sulfate C-, and dermatan sulfate-degrading activity assays were performed as previously described (34, 37).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis.
The procedure used for these analyses was described previously (34).
Molecular weight determination.
The molecular weights were determined by electrospray mass spectroscopy on a Sciex API III Plus instrument (Perkin-Elmer Sciex, Norwalk, Conn.). The protein samples were desalted by reverse-phase high-performance liquid chromatography (water/acetonitrile solvent) and were dried by evaporation. After reconstitution of the samples in a 50:50 (vol/vol) acetonitrile/water solution containing 5% acetic acid, they were infused into the instrument at a flow rate between 2 and 5 μl/min. Profile data were accumulated over 30 to 80 scans, and protein molecular weight determinations were performed with the HyperMass algorithm (Perkin-Elmer Sciex). The instrument was calibrated with a standard mixture of polypropylene glycol, and a myoglobin standard was used as a system suitability check.
Protein concentration analysis.
Protein concentrations in purified samples were calculated from the UV absorption at 280 nm and their respective extinction coefficients.
Construction of plasmids pIBXF3 and -4.
Two pairs of primers, FGHII-1 (5′GGCATATGAAAAGACAATTATACCT3′) and FGHII-2 (5′CTTCTAGATGCTACAGCCAGTAGAATGG3′) and FGHIII-1 (5′GGCATATGACTACGAAAATTTTTAAAAGG3′) and FGHIII-2 (5′GGTCTAGAGGGATATATCACTTTAATCAGGG3′), were used to amplify a 2.3-kbp PCR fragment from the λIB2 phage and a 2.0-kbp fragment from the λIB4 phage, respectively. Both DNA fragments were then digested with NdeI and XbaI, gel purified, and inserted into the corresponding sites of pUC21, yielding plasmids pIB23 and pIB24, respectively. An 830-bp, gel-purified, SacI-NdeI DNA fragment from pIB18 containing the hepA upstream region was then cloned into the corresponding sites of pIB23 and pIB24, forming plasmids pIB25 and pIB26, respectively. Finally, a gel-purified, 3.1-kbp, SpeI DNA fragment, from pIB25 containing the hepA upstream region and the hepB gene, and a gel-purified, 2.9-kbp, SpeI DNA fragment, from pIB26 containing the hepA upstream region and the hepC gene, were inserted into the XbaI site of plasmid pIBXF1 (35), forming plasmids pIBXF3 and pIBXF4, respectively.
Plasmid pIBXF5 construction.
Plasmid pIBXF5 was constructed by inserting a 2.3-kbp SpeI DNA fragment containing the hepA gene from pIB17 into the XbaI site of pIBXF1 (35)
Construction of plasmids pIBXF6 and -7.
Two pairs of primers, AC-1 (5′GGGAATTCCATAAGAAATTATTTGTAACCTG3′) and AC-2 (5′CGCGGATCCCCTAGATTACTACCATCAAAA3′) and B-1 (5′GGAATTCCATATGAAGATGCTGAATAAACTA3′) and B-2 (5′GGAATCAATTCACCGGGATGATC3′), were used to amplify a 2.1-kbp DNA fragment from plasmid pIB15 and a 1.3-kbp DNA fragment from plasmid pIB16, respectively. The amplicons were digested with EcoRI and BamHI, gel purified, and inserted into the corresponding sites of pUC21, yielding plasmids pIB27 and pIB28, respectively. Plasmid pIB29 was constructed by inserting a gel-purified, 1.5-kbp, MscI fragment from plasmid pIB16 into the MscI and EcoRV sites of plasmid pIB28, resulting in an intact cslB. An 830-bp, gel-purified EcoRI-NdeI DNA fragment from pIB18 was cloned into the corresponding sites of pIB27 and pIB29 to form plasmids pIB30 and pIB31, respectively. Finally, the gel-purified, 2.9- and 3.1-kbp, SpeI DNA fragments, containing the hepA upstream region fused to cslA or cslB from plasmids pIB30 and pIB31, respectively, were cloned into the XbaI site of plasmid pIBXF1 to form plasmids pIBXF6 and pIBXF7, respectively.
Construction of plasmids pIBXF8 and -9.
Plasmids pIBXF8 and pIBXF9 were constructed by inserting the gel-purified, 2.9- and 3.1-kbp, SpeI DNA fragments, containing the hepA upstream region with cslA and cslB from plasmids pIB30 and pIB31, respectively, into plasmid pIBXF2 (35).
Construction of plasmids pIBXF10, -11, -12, and -13.
Primers H-2 (5′GGCATATGTCTTTAGTTTTTATTGG3′) and AC-2 were used to generate a 2.2-kbp amplicon, corresponding to a 50-bp hepA upstream region and cslA from plasmid pIB30. The gel-purified, 2.2-kbp amplicon, digested with EcoRI and BamHI, was cloned into the corresponding sites in plasmid pUC21, forming plasmid pIB32. The primers, H-1 (5′CGGAATTCAAGCTAAAAACAGGCACCAT3′) and HU2 (5′GGCATATGTCTTTAGTTTTTATTGG3′), were used to generate a 250-bp amplicon. The gel-purified, EcoRI-NdeI-digested, 250-bp amplicon was inserted into the corresponding sites in plasmid pIB27, forming plasmid pIB33. Plasmid pIB34 was constructed by deleting a 269-bp, MscI-SmaI DNA fragment from plasmid pIB30. Plasmid pIB34 contains a 461-bp hepA upstream region and the cslA coding region. Plasmid pIB37 was constructed in two parts, first, by introducing an 800-bp, gel-purified, SpeI DNA fragment from plasmid pIB35 (which was constructed by inserting an 800-bp EcoRI-XmnI fragment from plasmid R751 into the EcoRI-EcoRV sites of pUC21) into plasmid pIB30 to form plasmid pIB36 and then by introducing a gel-purified, 2.7-kbp, BglII DNA fragment from plasmid pIB10 into the BglII sites of plasmid pIB36, thereby deleting a 50-bp BglII DNA fragment from this plasmid to form plasmid pIB37. The chromosomal organization of the hepA upstream region in plasmid pIB37 was confirmed as being similar to plasmid pIB10. Finally, a gel-purified, 6.5-kbp, SpeI DNA fragment containing the 3.5-kbp hepA upstream region, the 2.2-kbp cslA, and the 800-bp dhfrII from plasmid pIB37, a gel-purified, 2.6-kbp, SpeI DNA fragment containing the 460-bp hepA upstream region and cslA from plasmid pIB34, a gel-purified, 2.4-kbp, SpeI DNA fragment containing a 250-bp hepA upstream region and cslA from plasmid pIB32, and a gel-purified, 2.2-kbp, SpeI DNA fragment containing a 50-bp hepA upstream region and clsA from plasmid pIB33 were inserted into the XbaI site of plasmid pIBXF1 to form plasmids pIBXF10, pIBXF11, pIBXF12, and pIBXF13, respectively.
Strain construction.
All F. heparinum transconjugant strains were constructed by introducing the corresponding plasmid by conjugation as described previously (35).
RNA isolation and transcriptional analysis.
Total RNA was isolated from F. heparinum strains by disrupting the bacterial cell wall using the GramCracker reagent and the RNAqueous kit (Ambion, Austin, Tex.). The RNA samples were further treated to remove DNA contaminants using the DNA free kit (Ambion).
Primer extension analysis was performed using an Automated Laser Fluorescence (A.L.F.) DNA sequencer (Pharmacia) with Cy5-labeled antisense primers as previously described (2, 22). Antisense primer H-5 (5′TCTCGCTCTGCTTAGCACTGTC3′), complementary to nucleotides 110 to 132 of the hepA open reading frame, was used in conjugation with total RNA from wild-type F. heparinum or strain FIBX5 grown with glucose or heparin as the carbon source. Antisense primer AC-4 (5′CGCAAAGGCTTTTTAAGGTCCAG3′), complementary to nucleotides 109 to 131 of the cslA open reading frame, was used in conjugation with total RNA from strain FIBX6, grown with heparin. For the reaction, the Retroscript kit (Ambion) was used as recommended by the manufacturer with the following modification: dATP, dCTP, dTTP, and deaza-dGTP were at 1 mM each. The reaction mixtures were treated with 20 μg of RNase/ml for 30 min at 37°C, extracted with phenol-chloroform, and precipitated with ethanol. Four microliters of water and stop buffer from the AutoRead Sequencing Kit (Pharmacia) were added, and the entire sample was loaded, along with sequencing reactions, onto an 8% sequencing gel. Sequencing reactions with pIB17 and pIB30, performed with primers H-5 and AC-4, respectively, were used as the reference for the 5′ end determination. The transcriptional start sites were determined by comparing the retention times of the primer extension products with those of the products of the sequencing reactions.
RESULTS
GAG lyase gene expression in F. heparinum.
It has been reported that HepI is one of the dominating fractions of F. heparinum cell extracts grown in MH medium (19). It is thus conceivable that hepB, hepC, cslA, and cslB expression in F. heparinum could be increased by placing these genes under the control of hepA's regulatory elements. As described in Materials and Methods, hepB, hepC, cslA and cslB were fused to the 830-bp hepA upstream region (as hepBHU, hepCHU, cslAHU, and cslBHU, respectively) and the expression cassettes were cloned into the unique XbaI site of plasmid pIBXF1, a conjugative-integrative plasmid for DNA transfer into F. heparinum (35), to create plasmids pIBXF3, pIBXF4, pIBXF6, and pIBXF7, respectively. These plasmids were then introduced into F. heparinum by conjugation, and several stable Tpr transconjugants were obtained. The transconjugants obtained from pIBXF3, pIBXF4, pIBXF6, and pIBXF7 donor strains were named FIBX3, FIBX4, FIBX6, and FIBX7, respectively. These strains were confirmed to be derivatives of F. heparinum by their ability to grow on MH medium, and plasmid insertion was demonstrated by PCR and Southern hybridization (data not shown) using the methods described earlier (35).
Heparin-, heparan sulfate- and chondroitin sulfate-degrading activities were measured in strains FIBX3, FIBX4, FIBX6, and FIBX7 grown in MH medium (Table 2). A significant increase in heparan sulfate-degrading activity, 844 mIU ml−1 OD600−1, was observed for the hepC transconjugant strain FIBX4, while wild-type F. heparinum and FIBX1 had levels of 107 and 93 mIU ml−1 OD600−1, respectively. A moderate increase in heparan sulfate-degrading activity was also observed for the hepB transconjugant strain FIBX3. The most remarkable increase in level of activity over that of wild-type F. heparinum was observed for FIBX6. In MH medium, chondroitin sulfate A- and C-degrading activities reached 2,376 and 1,510 mIU ml−1 OD600−1, respectively, an approximate 20-fold increase when compared to wild-type F. heparinum grown in MA medium. Activity levels in FIBX6 were further increased in MA medium, reaching chondroitin sulfate A- and C-degrading activities of 4,057 and 2,620 mIU ml−1 OD600−1, respectively. The level of dermatan sulfate-degrading activity in heparin-induced FIBX7 reached 927 mIU ml−1 OD600−1, a 13-fold increase over that of wild-type F. heparinum grown in MA medium. Unlike the result for strain FIBX6, there was no additional increase of dermatan sulfate-degrading activity when FIBX7 was grown in MA medium.
TABLE 2.
Heparin-, heparan sulfate-, and chondroitin sulfate-degrading activities in native and transconjugant F. heparinum strainsa
| Growth medium | Activity (mIU ml−1 OD600−1)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hc
|
HS | CSA
|
CSC
|
DS
|
|||||
| MHd | MA | MH | MH | MA | MH | MA | MH | MA | |
| Native | 158 ± 12 | 102 ± 10 | 107 ± 9 | 15 ± 1 | 99 ± 9 | 13 ± 1 | 72 ± 9 | 30 ± 2 | 64 ± 5 |
| FIBX1 | 126 ± 8 | 100 ± 4 | 93 ± 7 | 14 ± 2 | 115 ± 4 | 12 ± 2 | 82 ± 5 | 27 ± 2 | 96 ± 14 |
| FIBX3 | 144 ± 31 | NDe | 174 ± 24 | ND | ND | ND | ND | ND | ND |
| FIBX4 | 179 ± 13 | ND | 844 ± 37 | ND | ND | ND | ND | ND | ND |
| FIBX5 | 775 ± 89 | ND | 63 ± 4 | ND | ND | ND | ND | ND | ND |
| FIBX6 | 119 ± 19 | 110 ± 7 | ND | 2,376 ± 135 | 4,057 ± 254 | 1,510 ± 87 | 2,620 ± 399 | 59 ± 4 | 126 ± 9 |
| FIBX7 | 143 ± 12 | 104 ± 10 | ND | 23 ± 3 | 111 ± 4 | 14 ± 3 | 72 ± 2 | 927 ± 134 | 958 ± 97 |
Values are the average of three separate experiments ± the standard error.
One international unit of GAG-degrading activity is defined as the amount of enzyme that will produce 1 μmol of product per min at 30°C at pH 8.
Substrates for GAG lyases' activity assay: H, heparin; HS, heparan sulfate; CSA, chondroitin sulfate A; CSC, chondroitin sulfate C; and DS, dermatan sulfate.
Media used in this study. MH, FM supplemented with 1% heparin; and MA, FM supplemented with 1% chondroitin sulfate A.
ND, not determined.
The corresponding GAG lyase activities from these transconjugant strains were surprisingly high. It was expected that the transconjugant strains would yield levels of activities comparable to the level of heparin-degrading activity of wild-type F. heparinum. When the activity values were normalized for variable factors, such as molecular weight and specific activity of each enzyme, the levels of activity in strains FIBX4, -6, and -7 were expected to reach 150 mIU ml−1 OD600−1 of heparan sulfate-degrading activity, 390 mIU ml−1 OD600−1 of chondroitin sulfate A-degrading activity, and 150 mIU ml−1 OD600−1 of chondroitin sulfate B-degrading activity, respectively. In fact, activity levels reached seven times the expected levels, while the activity of the other GAG lyases remained comparable to that of the wild-type strain.
The addition of a single copy of hepCHU, cslAHU, and cslBHU, integrated into the F. heparinum chromosome, resulted in a significant increase in these GAG lyase-degrading activities. It was thus conceivable that the integration of a second hepA gene cassette would confer a significant increase in heparin-degrading activity, if the activity increase was mediated by the integrative plasmid system. A hepA transconjugant strain, FIBX5, was thus constructed as described in Materials and Methods. Heparin- and heparan sulfate-degrading activities of FIBX5 are displayed in Table 2. An approximate fivefold increase in heparin-degrading activity over that found in wild-type F. heparinum (775 versus 157 mIU ml−1 OD600−1) was observed, or a sixfold increase over activity found in strain FIBX1 was observed (775 versus 126 mIU ml−1 OD600−1) levels. No increase in heparan sulfate-degrading activity was observed.
Analysis of GAG lyase expression in F. heparinum.
It was shown that the five transconjugant strains FIBX3, FIBX4, FIBX5, FIBX6, and FIBX7 displayed significant increases in heparin- or heparan sulfate-degrading activities or high levels of chondroitin sulfate-degrading activities. To demonstrate that these increased activities were the result of a modulation in GAG lyase synthesis in the transconjugant F. heparinum strains, experiments were performed to evaluate protein expression levels in wild-type F. heparinum and in each of the transconjugant strains. As shown in Fig. 1A, B, and C and 2A, a marked increase in the intensity of the protein bands in the soluble cell extracts of strains of FIBX5, FIBX3, and FIBX4, corresponding to purified HepI, HepII and HepIII, respectively, and in the soluble cell extracts of strains of FIBX6 and FIBX7, corresponding to ChnA and ChnB, respectively, was seen.
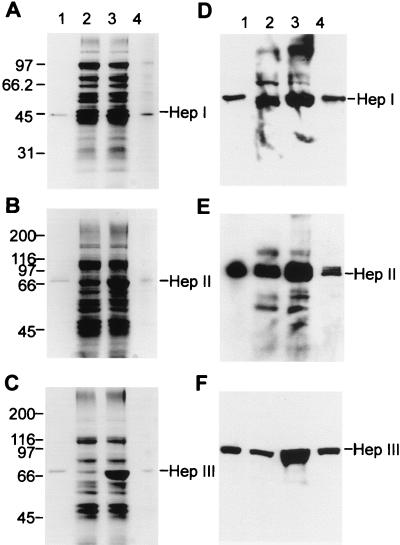
FIG. 1.
HepI, -II, and -III expression in native and transconjugant F. heparinum strains grown in MH medium. Coomassie blue R-250-stained gels were used. Numbers of kilodaltons are given on left of gels. (A) SDS-10% PAGE gel. Lanes: 1, 0.5 μg of purified native HepI; 2, 0.125 OD600 units of soluble extract from native F. heparinum; 3, same amount of extract from FIBX5; and 4, 0.025 OD600 units of soluble extract from FIBX5. (B) SDS-7.5% PAGE gel. Lanes: 1, 0.5 μg of purified native HepII; 2, 0.125 OD600 units of soluble extract from native F. heparinum; 3, same amount of extract from FIBX3; and 4, 0.0125 OD600 units of soluble extract from FIBX3. (C) SDS-7.5% PAGE gel. Lanes: 1, 0.5 μg of purified native HepIII; 2, 0.125 OD600 units of soluble extract from native F. heparinum; 3, same amount of extract from FIBX4; and 4, 0.0125 OD600 units of soluble extract from FIBX4. Western blot analysis was performed. (D) Results were transferred from an SDS-10% PAGE gel. Lane 1, 0.2 μg of purified native HepI; lane 2, 0.05 OD600 units of soluble extract from native F. heparinum; lane 3, same amount of extract from FIBX5; and lane 4, 0.01 OD600 units of soluble extract from FIBX5. (E) Results were transferred from an SDS-7.5% PAGE gel. Lane 1, 0.2 μg of purified native HepII; lane 2, 0.05 OD600 units of soluble extract from native F. heparinum; lane 3, same amount of extract from FIBX3; and lane 4, 0.01 OD600 units of soluble extract from FIBX3. (F) Results were transferred from an SDS-7.5% PAGE gel. Lane 1, 0.2 μg of purified native HepIII; lane 2, 0.05 OD600 units of soluble extract from native F. heparinum; lane 3, same amount of extract from FIBX4; and lane 4, 0.01 OD600 units of soluble extract from FIBX4.
Western blot analyses confirmed the identity of each GAG lyase, and densitometric analysis allowed an estimation of HepI, HepII, and HepIII protein expression levels in strains FIBX5, -3, and -4, respectively. As shown in Fig. 1D, E, and F, fivefold and tenfold dilutions of the soluble extracts of strains FIBX5 and FIBX3, respectively, displayed HepI and HepIII bands of intensity similar to those of wild-type F. heparinum. A 10-fold dilution of the FIBX4-soluble extract showed a band with approximately half the intensity of that displayed by the wild-type strain. The expression of ChnA and ChnB in strains FIBX6 and FIBX7, respectively, was also confirmed by Western blot analysis (Fig. 2B and C). The intense bands seen in the SDS-PAGE gel (Fig. 2A) were shown to react with the antibodies specific for ChnA and ChnB. It was also shown that both ChnA and ChnB were expressed in very low levels when wild-type F. heparinum was grown in MA medium and that there was no detectable expression when this strain was grown in MH medium.
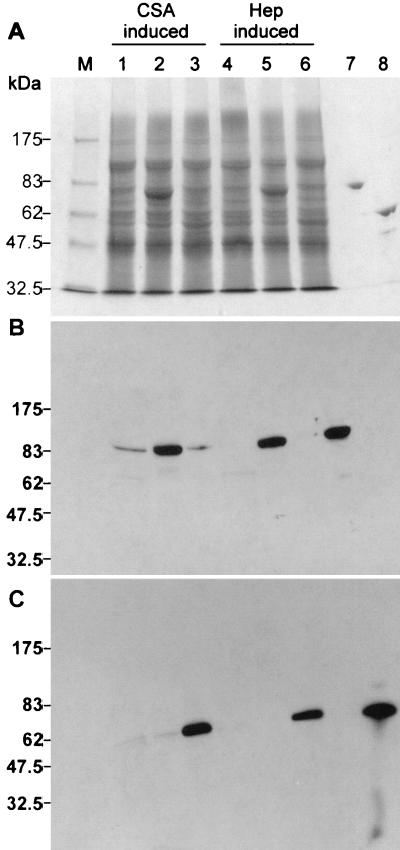
FIG. 2.
ChnA and -B expression in the native and transconjugant F. heparinum strains. Amounts of fractions (0.15 OD600 and 0.05 OD600 units of soluble fractions from wild-type and transconjugate strains, respectively) were loaded onto SDS-7.5% PAGE reducing gels. (A) Coomassie blue R-250-stained gel. (B and C) Western blots of triplicates of gel A transferred to nitrocellulose membrane filters and developed with polyclonal antibodies against ChnA (B) or ChnB (C). Lanes: M, molecular weight markers; 1 and 4, wild-type F. heparinum; 2 and 5, strain FIBX6 (cslAHU); 3 and 6, strain FIBX7 (cslBHU); 7, purified native ChnA (0.5 μg) from wild-type F. heparinum; and 8, purified native ChnB (0.5 μg) from wild-type F. heparinum. lanes 1, 2, and 3 represent strains grown in MA medium, while lanes 4, 5, and 6 represent strains grown in MH medium.
Chondroitinase gene expression in pIBXF2 plasmid.
Plasmid pIBXF1-mediated chromosome integration of the hepA upstream region fused with a GAG lyase gene resulted in a significant increase in expression levels. Whether such an increase in expression levels would be observed with an episomal plasmid vector was not known. To investigate this, cslAHU and cslBHU were cloned into plasmid pIBXF2 (35), a plasmid shown to replicate in F. heparinum, to yield plasmids pIBXF8 and pIBXF9, respectively. Both were introduced in F. heparinum by conjugation to create strains FIBX8 and FIBX9, respectively. The existence of both plasmids in these strains was confirmed by using the plasmid DNA preparation from these strains to transform E. coli and was further confirmed by analysis of the plasmid DNA from E. coli (data not shown). The chondroitin sulfate-degrading activities from FIBX8 and FIBX9 were measured and shown to be similar to the activity profiles of strains FIBX6 and FIBX7 (data not shown).
HepA regulation studies.
It was shown that cslAHU resulted in a high level of cslA gene expression. It was also shown that cslA expression was higher when FIBX6 was grown in MA medium than in MH medium, which was not the case for cslB expression in strain FIBX7. To study this further, wild-type F. heparinum and strain FIBX6 were grown in various media and heparin- and chondroitin sulfate A-degrading activities were measured. As seen in Table 3, heparin-degrading activity was observed in both wild-type F. heparinum and FIBX6 in the presence of heparin. However, the presence of glucose resulted in a significant decrease in activity and was further decreased to nondetectable levels in the absence of heparin (growth media 3 and 5). A similar regulation pattern was observed for chondroitin sulfate A-degrading activity, except for a more significant increase in MA medium for FIBX6 (Table 3). The data indicated that cslA expression in strain FIBX6 grown in heparin-containing media was regulated in a fashion similar to heparin-degrading activity in wild-type F. heparinum. The higher chondroitin sulfate A-degrading activity seen in strain FIBX6 grown in chondroitin sulfate A medium could not be explained.
TABLE 3.
Heparin- and chondroitin sulfate A-degrading activities in native F. heparinum and FIBX6 when grown in various mediaa
| Growth mediume | Activity (mIU ml−1 OD600−1)b
|
|||
|---|---|---|---|---|
| Native F. heparinum
|
FIBX6
|
|||
| Hc | CSAd | H | CSA | |
| 1 | 158 ± 12 | 15 ± 1 | 119 ± 19 | 2,376 ± 135 |
| 2 | 102 ± 10 | 99 ± 10 | 110 ± 7 | 4,057 ± 254 |
| 3 | <5 | 11 ± 2 | <5 | 16 ± 2 |
| 4 | 0 ± 6 | 6 ± 2 | 37 ± 11 | 814 ± 54 |
| 5 | <5 | <5 | <5 | 17 ± 2 |
| 6 | <5 | <5 | <5 | 11 ± 2 |
| 7 | 31 ± 3 | 5 ± 1 | 27 ± 2 | 187 ± 21 |
| 8 | 117 ± 8 | 9 ± 2 | 112 ± 8 | 748 ± 26 |
Values are the average of three separate experiments ± the standard error.
One international unit of heparin- or chondroitin sulfate A- degrading activity is defined as the amount of enzyme that will produce 1 μmol of product per min at 30°C at pH 8.
Substrate heparin.
Substrate chondroitin sulfate A.
Media: 1, FM supplemented with 1% heparin (data taken from Table 2); 2, FM supplemented with 1% chondroitin sulfate A (data taken from Table 2); 3, FM supplemented with 0.4% glucose; 4, FM supplemented with 0.4% glucose and 1% heparin; 5, LB medium; 6, LB medium supplemented with 0.4% glucose; 7, LB medium supplement with 0.4% glucose and 1% heparin; and 8, LB medium supplement with 1% heparin.
Molecular analysis of the hepA upstream region.
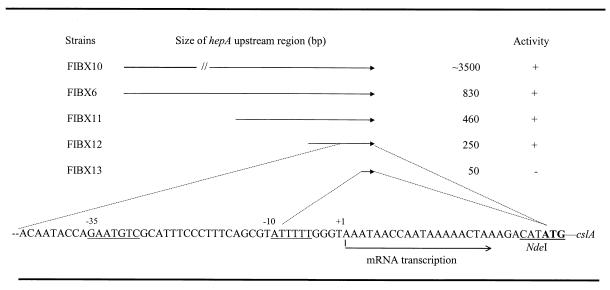
An additional copy of hepA or other GAG lyase genes fused to the hepA upstream region resulted in a significant increase in GAG lyase gene expression. To understand the relationship between the hepA upstream region and the high levels of gene expression, the hepA upstream region was fractionated and fused to the cslA reporter gene. Portions of different sizes of the hepA upstream region from approximately 50 bp to 3.5 kbp were fused to cslA, as shown in Fig. 3, and were introduced in F. heparinum by conjugation. The transconjugant strains were then grown in MH medium, and chondroitin sulfate A-degrading activity was measured. As shown in Fig. 3, similar chondroitin sulfate A-degrading activity was seen in strains FIBX6, FIBX10, FIBX11, and FIBX12 but no activity could be detected in strain FIBX13. The data suggested that the high levels of GAG lyase gene expression observed were not linked to the hepA upstream region and that the hepA promoter region lay approximately 50 bp upstream of the hepA start codon.
FIG. 3.
Analysis of the hepA regulatory region. The activity was measured as chondroitin sulfate A-degrading activity as shown in Table 2. “+” denotes that the activity measured was the same as in Table 2. “−” denotes the absence of any measurable activity. The strains were grown in MH medium as described in Materials and Methods. The sequence of the hepA upstream region was published previously (27), and an NdeI site was incorporated for strain FIBX6. The transcriptional start site is indicated, and the −10 and −35 regions are underlined.
The transcriptional start site of the hepA gene was determined by primer extension analysis with fluorescence-labeled primers corresponding to hepA- or cslA-coding sequences. The experiments were performed with total RNA prepared from cells of native F. heparinum, strain FIBX5, and strain FIBX6 grown in MH medium, which were harvested during the exponential growth phase as described in Materials and Methods. A strong and unique signal, corresponding to 26 bp upstream of the start codon (Fig. 3), was obtained (Fig. 4A) for strain FIBX6 using the primer corresponding to the cslA coding sequence. Putative −10 and −35 boxes were also identified as shown in Fig. 3, which would agree with the lack of cslA activity in strain FIBX13, which carried a hepA promoter region lacking the −35 box. The transcriptional start site of hepA in native F. heparinum and strain FIBX5, using a primer corresponding to the hepA coding sequence, was mapped to the same base (Fig. 4B, graph B1, for strain FIBX5 grown in heparin-only medium and Fig. 4B, graph B3, for native F. heparinum grown in the same medium), but the signal was much weaker for strain FIBX5 and barely detectable for wild-type F. heparinum. In addition, there were two strong signals observed, one just 1 base upstream of translation start site and the other about 35 bp upstream of the transcriptional start site mapped in strain FIBX6. To investigate whether these two signals are related to HepI expression, wild-type F. heparinum was grown in glucose-only medium and total RNA was isolated for mapping. As shown in Fig. 4B, graph B2, these two signals were also present even though HepI was not expressed. Therefore, the location of the hepA transcriptional start site was confirmed at 26 bp upstream of the translational start site. The other two transcriptional signals seen in wild-type F. heparinum and strain FIBX5 were not related to HepI expression, and their presence remains unexplained.
FIG. 4.
Transcriptional start site determination of hepA using an A.L.F. sequencer. Primer extension was carried with 40 μg of total RNA from the FIBX5 and FIBX6 strains and with 120 μg of total RNA from wild-type F. heparinum and Cy5-labeled primers. The sequencing reactions of pIB17 and pIB30 were used as markers. The transcriptional start site, indicated by a line, was determined by comparing the retention time of the primer extension products with those of the products of the sequencing reactions. (A) Primer extension products from total RNA of strain FIBX6 (top) and plasmid DNA of pIB30 (bottom) run on an A.L.F. sequencer. (B) B1, primer extension products from total RNA of strain FIBX5 grown in heparin-only medium; B2, primer extension products from total RNA of strain FIBX5 grown in glucose-only medium; and B3, primer extension products from total RNA of wild-type F. heparinum grown in heparin-only medium. Bottom, primer extension products from plasmid pIB17.
Purification and characterization of the GAG lyases from the transconjugant strains.
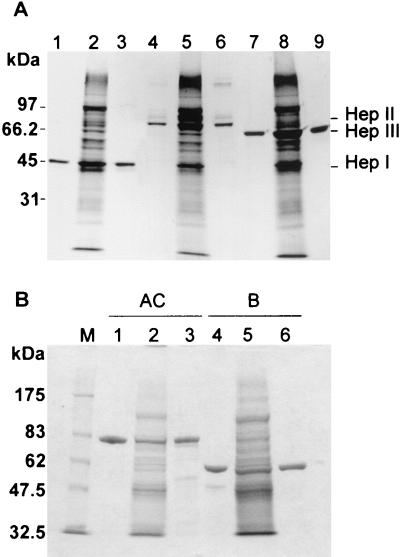
It was shown that the expression levels of HepI, HepII, HepIII, ChnA, and ChnB in strains FIBX5, FIBX3, FIBX4, FIBX6, and FIBX7, respectively, were very high. However, whether these enzymes displayed biochemical characteristics similar to their wild-type counterparts was not known. The three heparinases and two chondroitinases expressed recombinantly, named tHepI, tHepII, tHepIII, tChnA, and tChnB, were purified from FIBX5, FIBX3, FIBX4, FIBX6, and FIBX7 cultures, respectively, and further characterized. As shown in Fig. 5, tHepI, tHepII, tHepIII, tChnA, and tChnB were purified to appear as a single band on an SDS-PAGE gel stained with Coomassie blue. In addition, the apparent molecular weights, as judged by migration on an SDS-PAGE gel, of both recombinant and native GAG lyases were similar. The specific activities of heparinases and chondroitinases from wild-type F. heparinum and the transconjugant strains were shown to be similar, as seen in Table 4 and Table 5. The molecular weights of the recombinant heparinases were determined by electron mass spectroscopy to be 42,514, 85,763, and 73,217 Da for tHepI, tHepII, and tHepIII, respectively, and are in close agreement with those of their wild-type counterparts. Similar crystal structures and glycosylation patterns were observed for tChnA and tChnB and their counterparts from wild-type F. heparinum (7, 13). Together, these analyses indicate that these five GAG lyases, whether synthesized from wild-type F. heparinum or the transconjugant strains, share the same biochemical characteristics.
FIG. 5.
Coomassie blue R-250 analysis of HepI, -II, and -III and ChnA and -B purified from either the native or transconjugant F. heparinum strain. (A) SDS-10% PAGE Coomassie blue R-250-stained gel. Lane 1, 1 μg of purified native HepI; lane 4, native HepII; and lane 7, native HepIII. Lane 2, 0.1 OD600 units of soluble FIBX5; lane 5, same amount of FIBX3; and lane 8, FIBX4 cell extract. Lane 3, 1 μg of purified tHepI; lane 6, same amount of tHepII; and lane 9, same amount of tHepIII. (B) SDS-7.5% PAGE Coomassie blue R-250-stained gel. Lane M, molecular weight marker; lane 1, 1 μg of purified native ChnA; lane 4, 1 μg of native ChnB; lane 2, 0.1 OD600 units of soluble FIBX6; lane 5, same amount of FIBX7; lane 3, 1 μg of purified tChnA; and lane 6, same amount of tChnB.
TABLE 4.
Specific activity of HepI, -II, and -III from F. heparinum for heparin and heparan sulfate
| Substrate | Sp act (IU mg−1)
|
|||||
|---|---|---|---|---|---|---|
| HepI
|
HepII
|
HepIII
|
||||
| na | tb | n | t | n | t | |
| Heparin | 110 | 105 | 4.3 | 6.1 | 0.65 | 0.5 |
| Heparan sulfate | 1.9 | 2.0 | 8.1 | 9.9 | 56.3 | 54.8 |
Enzyme purified from the native F. heparinum strain.
Enzyme purified from the transconjugant F. heparinum strain.
TABLE 5.
Specific activity of ChnA and -B from F. heparinum for various chondroitin sulfates
| Substrate | Sp act (IU/mg)
|
|||
|---|---|---|---|---|
| ChnA
|
ChnB
|
|||
| na | tb | n | t | |
| Chondroitin sulfate A | 278 | 271 | ND | ND |
| Chondroitin sulfate C | 168 | 168 | ND | ND |
| Dermatan sulfate | NDc | ND | 104 | 96 |
Enzyme purified from the native F. heparinum strain.
Enzyme purified from the transconjugant F. heparinum strain.
ND, not determined.
DISCUSSION
We describe here a system for a high level of gene expression in F. heparinum. This system was used to express hepA as well as hepB, hepC, cslA, and cslB under the control of the hepA regulatory region using the F. heparinum conjugation-integration plasmid pIBXF1. The transconjugant strains expressed approximately five times more HepI and HepII and 10 times more HepIII than did wild-type F. heparinum grown in MH medium, as well as 20- and 15-fold more ChnA and ChnB, respectively, than did wild-type F. heparinum grown in MA medium, as judged by SDS-PAGE-Western analysis and by the corresponding activity assay. These results demonstrated that this system, i.e., the gene expression cassette composed of the hepA regulatory region within the integrative plasmid system, can be used to reach high levels of protein expression in F. heparinum.
Several hypotheses were considered in explaining why such high levels of expression were obtained in F. heparinum with the addition of a second copy of the hepA gene or other GAG lyase genes under the control of the hepA upstream region. It was first suspected that the increase of hepB and hepC expression levels in strains FIBX3 and FIBX4, respectively, or that high levels of cslA and cslB expression in strains FIBX6 and FIBX7, respectively, were the result of a cloning manipulation that modified the ATG context by adding an NdeI restriction site in hepBHU, hepCHU, cslAHU, and cslBHU expression gene cassettes. This possibility was ruled out, since the hepA DNA fragment was cloned without such a modification and still showed an unexpectedly high level of HepI expression. It was then suspected that, if the orientation of the expression gene cassette lay in the same orientation as the HU Tpr gene cassette, an antibiotic-resistant gene (dhfrII) from R751 fused to the HU region of hepA (35), the strength of hepA promoter function might have been amplified. This possibility was also ruled out, since the strains with the hepCHU and the cslAHU expression gene cassettes were inserted at the XbaI site of plasmid pIBXF1 in opposing orientations and still displayed similar activity profiles (data not shown). The possibility that the hepA downstream region may code for a certain instability of the translated mRNA, which may be reflected by low expression levels of HepI in wild-type F. heparinum, was raised. A possible RNA hairpin secondary structure that may serve as a transcription termination signal (nucleotides 1329 to 1359 of the published hepA sequence [27]) was identified and located to 1 nucleotide after the termination codon. This sequence was included in the construction of strain FIBX5, which still yielded fivefold more heparin-degrading activity than the wild-type strain. Consideration was given to the possibility that, during the selection process, transconjugants displaying a mutation that deregulated hepA gene expression lead to a higher level of trimethoprim resistance for the selection of a few transconjugant strains, as discussed previously (35). This hypothesis was also ruled out, since no increase in heparin-degrading activity was seen in these transconjugant strains. It was then suggested that the site of the integration of plasmid pIBXF1 might influence GAG lyase gene expression. However, this was highly unlikely, since strains FIBX8 and FIBX9, which carry the cslAHU and cslBHU cassettes in plasmid pIBXF2, respectively, showed expression profiles similar to those of strains FIBX6 and FIBX7 grown in MH medium. Furthermore, when the hepCHU gene cassette was cloned into an integration plasmid carrying a 10-kbp EcoRI DNA fragment from plasmid pIB10 (34) corresponding to the hepA loci, the resulting transconjugant strain displayed a similar heparan sulfate-degrading activity when grown in MH medium (data not shown). It was also suggested that the upstream region of hepA at its loci may somehow repress hepA expression in wild-type F. heparinum. This possibility was also discarded by that fact that strain FIBX10 harboring a 3.5-kbp hepA upstream region showed chondroitin sulfate A-degrading activity similar to that of strain FIBX6, in which only an 830-bp hepA upstream region was fused to cslA, when grown in MH medium. Finally, the transcriptional start sites were shown to be identical for the hepA in wild-type F. heparinum and cslAHU in strain FIBX6, ruling out the possibility of modified promoter elements. The reason(s) underlying these drastic increases in GAG lyase expression observed with our conjugative-integrative plasmid system could not be unveiled.
The hepA transcriptional start site was mapped to 26 bp upstream of the translational start site using a cslA expression strain, FIBX6. When we attempted to confirm this in the wild-type F. heparinum and hepA transconjugant strains, two additional transcriptional signals were seen. It was demonstrated that these two signals were not related to HepI expression, because the signals presented even when the strains were grown in glucose minimal medium, where HepI synthesis is repressed. The absence of one of the nonspecific peaks in the cslA expression strain FIBX6 can be explained by the disruption of the nucleotide sequence by the introduction of a restriction site for cloning. In addition, the absence of this peak may offer a clue to understand why the other four GAG lyase expression strains expressed levels of GAG lyases higher than the hepI in strain FIBX5.
GAG lyases synthesized in F. heparinum are posttranslationally processed to remove their N-terminal signal sequences (34, 37) and translocated to the periplasmic space, and HepI, HepII, ChnA, and ChnB are further modified by the addition of carbohydrate molecule(s) [Laliberte et al., 10th Symp. Protein Soc., Protein Sci. 5(Suppl. 1):435s, 1996]. The host cell machinery involved in these processes was revealed not to be a limiting factor in achieving a high level of GAG lyase production in F. heparinum. Electrospray mass spectroscopy analysis revealed that the heparinases produced from either the transconjugant or wild-type strains were similarly processed and glycosylated. Taken together with the fact that the crystal structures of native and recombinant chondroitinases were identical (7, 13), it is believed that F. heparinum can efficiently produce large quantities of fully matured proteins. Therefore, this system offers an alternative to E. coli for efficient expression of secreted proteins.
The productivity of the F. heparinum transconjugants for their respective GAG lyases was better than that obtained with the recombinant E. coli strains. It was calculated that HepI and HepIII levels reached approximately 2.8 and 6.0% of total soluble protein (Table 2; the total soluble proteins were 0.25 mg per ml−1 OD600−1) in strains FIBX5 and FIBX4, respectively. The ChnA and ChnB expression levels reached approximately 3.5 and 3.7%, in strains FIBX6 and FIBX7 grown in heparin-only medium, respectively. In E. coli, the recombinant HepI level reached 250 mg/liter when expressed as inclusion bodies or 150 mg/liter in a soluble form in a small-scale fermentation setting (5). In a 3-day FIBX5 fermentation, the productivity of HepI averaged 500 mg/liter but could reach a much higher level when the fermentation time was extended (B. Eggimann and H. Su, personal communication). The HepIII production level from FIBX4 was reported at 844 mIU ml−1 OD600−1, while E. coli yielded only 280 mIUml−1 OD600−1 (5). Although it is possible to double the biomass with E. coli in a high-cell-density fermentation process, its production would still be 35% lower than with FIBX4, and purification would be hindered by the significant presence of degradation products of recombinant HepIII. The HepIII production levels in strain FIBX4 fermentation reached more than 1 g/liter (Eggimann and Su, personal communication). The production of recombinant ChnA and recombinant ChnB was about 50- and 15-fold lower in E. coli, respectively, than in transconjugant strains FIBX6 and FIBX7 (37). Previously, the methods of chondroitinase production from wild-type F. heparinum used chondroitin sulfate A medium and proved to be very expensive with a low productivity due to limited biomass accumulation. Strains FIBX6 and FIBX7 were used for ChnA and ChnB production, and productivity was in the levels of grams/liter (Eggimann and Su, personal communication). In summary, the FIBX5, FIBX3, FIBX4, FIBX6, and FIBX7 transconjugant strains offer a very effective system for HepI, HepII, HepIII, ChnA, and ChnB production, respectively. The enzymes can be purified using preestablished procedures and possess identical biophysical and biochemical characteristics as their native counterparts. In addition, the expression system offers the possibility of conducting structural and functional studies of these GAG lyases in their native environment.
Acknowledgments
The first two authors, Francoise Blain and A. Lydia Tkalec, contributed equally to this work.
We thank Philippe Marchessault (IBEX Pharmaceuticals Inc.) for the preparation of Fig. 4. We acknowledge Peter Lau (Biotechnology Research Institute, Montreal, Canada) for helpful discussion and Elain B. Newman (Concordia University, Montreal, Canada) for her support and encouragement during the course of this study.
REFERENCES
- 1.Baug, R. F., and J. J. Zimmermann. 1993. Heparinase in the activated clotting time assay: monitoring heparin-independent alternations in coagulation function. Perfusion Rev. 1:14-28. [Google Scholar]
- 2.Boorstein, W. R., and E. A. Craig. 1989. Primer extension analysis from RNA. Methods Enzymol. 180:347-369. [DOI] [PubMed] [Google Scholar]
- 3.Christensen, P. 1980. Description and taxonomic status of Cytophaga heparina (Payza and Korn) comb. nov. (basionym: Flavobacterium heparinum Payza and Korn 1956). Int. J. Syst. Bacteriol. 30:473-475. [Google Scholar]
- 4.Denholm, E. M., E. Cauchon, C. Poulin, and P. J. Silver. 2000. Inhibition of human dermal fibroblast proliferation by removal of dermatan sulfate. Eur. J. Pharmacol. 400:145-153. [DOI] [PubMed] [Google Scholar]
- 5.Ernst, S., O. A. Garro, S. Winkler, G. Venkataraman, R. Langer, C. L. Cooney, and R. Sasisekharan. 1997. Process simulation for recombinant protein production: cost estimation and sensitivity analysis for heparinase I expressed in Escherichia coli. Biotechnol. Bioeng. 53:575-582. [DOI] [PubMed] [Google Scholar]
- 6.Ernst, S., R. Langer, C. L. Cooney, and R. Sasisekharan. 1995. Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 30:387-444. [DOI] [PubMed] [Google Scholar]
- 7.Fethiere, J., B. Eggimann, and M. Cygler. 1999. Crystal structure of chondroitinase AC: a glycosaminoglycan lyase with a new fold. J. Mol. Biol. 288:635-647. [DOI] [PubMed] [Google Scholar]
- 8.Fethiere, J., B. H. Shilton, Y. Li, M. Allaire, M. Laliberte, B. Eggimann, and M. Cygler. 1998. Crystallization and preliminary analysis of chondroitinase AC from Flavobacterium heparinum. Acta Crystallogr. Sect. D 54:279-280. [DOI] [PubMed] [Google Scholar]
- 9.Galliher, P. M., C. L. Cooney, R. S. Langer, and R. J. Linhardt. 1981. Heparinase production by Flavobacterium heparinum. Appl. Environ. Microbiol. 41:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godavarti, R., and R. Sasisekharan. 1998. Heparinase I from Flavobacterium heparinum, role of positive charge in enzymatic activity. J. Biol. Chem. 273:248-255. [DOI] [PubMed] [Google Scholar]
- 11.Gu, K., R. J. Linhardt, M. Laliberté, K. Gu, and J. Zimmermann. 1995. Purification, characterization and specificity of chondroitin lyases and glycuronidase from Flavobacterium heparinum. Biochem. J. 312:569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, L., H. Van Halbeek, B. Eggimann, and J. J. Zimmermann. 1995. Structural characterization of the noval O-linked carbohydrate chain of heparinase I from Flavobacterium heparinum. Glycobiology 5:712. [Google Scholar]
- 13.Huang, W., A. Matte, Y. Li, Y. S. Kim, R. J. Linhardt, H. Su, and M. Cygler. 1999. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J. Mol. Biol. 294:1257-1269. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, R. L., S. J. Busch, and A. D. Cardin. 1991. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol. Rev. 71:481-539. [DOI] [PubMed] [Google Scholar]
- 15.Kjellen, L., and U. Lindahl. 1991. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 60:401-475. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y., A. Matte, H. Su, and M. Cygler. 1998. Crystallization and preliminary X-ray analysis of chondroitinase B from Flavobacterium heparinum. Acta Crystallogr. Sect. D 55:1055-1057. [DOI] [PubMed] [Google Scholar]
- 17.Linhardt, R. J., P. M. Galliher, and C. L. Cooney. 1986. Review: polysaccharide lyases. Appl. Biochem. Biotechnol. 12:135-177. [DOI] [PubMed] [Google Scholar]
- 18.Liu, D., Z. Shriver, R. Godavarti, G. Venkataraman, and R. Sasisekharan. 1999. The calcium-binding sites of heparinase I from Flavobacterium heparinum are essential for enzymatic activity. J. Biol. Chem. 274:4089-4095. [DOI] [PubMed] [Google Scholar]
- 19.Lohse, D. L., and R. J. Linhardt. 1992. Purification and characterization of heparin lyases from Flavobacterium heparinum. J. Biol. Chem. 267:24347-24355. [PubMed] [Google Scholar]
- 20.Meyer, R. J., and J. A. Shapiro. 1980. Genetic organization of the broad-host-range IncP-1 plasmid R751. J. Bacteriol. 143:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullis, K., F. Faloona, S. Scharf, R. Saiki, G. Horn, and H. Erlich. 1986. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp. Quant. Biol. 51:263-269. [DOI] [PubMed] [Google Scholar]
- 22.Myöhänen, S., and J. Wahlfors. 1993. Automated fluorescent primer extension. BioTechniques 14:16-17. [PubMed] [Google Scholar]
- 23.Payza, A. N., and E. D. Korn. 1956. Bacterial degradation of heparin. Nature (London) 177:88-89. [DOI] [PubMed] [Google Scholar]
- 24.Plummer, T. H. J., A. L. Tarentino, and C. R. Hauer. 1995. Novel, specific O-glycosylation of secreted Flavobacterium meningosepticum proteins. Asp-Ser and Asp-Thr-Thr consensus sites. J. Biol. Chem. 270:13192-13196. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasisekharan, R., M. Bulmer, K. W. Moremen, C. L. Cooney, and R. Langer. 1993. Cloning and expression of heparinase I gene from Flavobacterium heparinum. Proc. Natl. Acad. Sci. USA 90:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shriver, Z., Y. Hu, and R. Sasisekharan. 1998. Heparinase II from Flavobacterium heparinum. Role of histidine residues in enzymatic activity as probed by chemical modification and site-directed mutagenesis. J. Biol. Chem. 273:10160-10167. [DOI] [PubMed] [Google Scholar]
- 29.Shriver, Z., D. Liu, Y. Hu, and R. Sasisekharan. 1999. Biochemical investigations and mapping of the calcium-binding sites of heparinase I from Flavobacterium heparinum. J. Biol. Chem. 274:4082-4088. [DOI] [PubMed] [Google Scholar]
- 30.Silver, P. J. 1998. IBT 9302 (heparinase III): a novel enzyme for the management of reperfusion injury-related vascular damage, restenosis and wound healing. Expert Opin. Investig. Drugs 7:1003-1014. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 2:784-791. [Google Scholar]
- 32.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 33.Steyn, P. L., P. Segers, M. Vancanneyt, P. Sandra, K. Kersters, and J. J. Joubert. 1998. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int. J. Syst. Bacteriol. 48:165-177. [DOI] [PubMed] [Google Scholar]
- 34.Su, H., F. Blain, R. A. Musil, J. J. F. Zimmermann, K. Gu, and D. C. Bennett. 1996. Isolation and expression in Escherichia coli of hepB and hepC, genes coding for the glycosaminoglycan-degrading enzymes heparinase II and heparinase III, respectively, from Flavobacterium heparinum. Appl. Environ. Microbiol. 62:2723-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su, H., Z. Shao, A. L. Tkalec, F. Blain, and J. J. Zimmermann. 2001. Development of a genetic system for the transfer of DNA into Flavobacterium heparinum. Microbiology 147:581-589. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi, M., and A. Yokota. 1992. Proposals of Shingobacterium faecium sp. nov., Sphingobacterium piscium sp. nov., Sphingobacterium heparinum comb. nov., Sphingobacterium thalpophilum comb. nov. and two genospecies of genus Sphingobacterium, and synonymy of Flavobacterium yabuuchiae and Sphingobacterium spiritivorum. J. Gen. Appl. Microbiol. 38:465-482. [Google Scholar]
- 37.Tkalec, A. L., D. Fink, F. Blain, G. Zhang-Sun, M. Laliberte, D. C. Bennett, K. Gu, J. J. F. Zimmermann, and H. Su. 2000. Isolation and expression in Escherichia coli of cslA and cslB, genes coding for the chondroitin sulfate-degrading enzymes chondroitinase AC and chondroitinase B, respectively, from Flavobacterium heparinum. Appl. Environ. Microbiol. 66:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 39.Yang, V. C., R. J. Linhardt, H. Bernstein, C. L. Cooney, and R. Langer. 1985. Purification and characterization of heparinase from Flavobacterium heparinum. J. Biol. Chem. 260:1849-1857. [PubMed] [Google Scholar]
- 40.Zimmermann, J. J., R. Langer, and C. L. Cooney. 1990. Specific plate assay for bacterial heparinase. Appl. Environ. Microbiol. 56:3593-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]