Abstract
Depletion of the SlpA protein from the bacterial surface greatly reduced the adhesion of Lactobacillus brevis ATCC 8287 to the human intestinal cell lines Caco-2 and Intestine 407, the endothelial cell line EA-hy926, and the urinary bladder cell line T24, as well as immobilized fibronectin. For functional analysis of the SlpA surface protein, different regions of the slpA gene were expressed as internal in-frame fusions in the variable region of the fliCH7 gene of Escherichia coli. The resulting chimeric flagella carried inserts up to 275 amino acids long from the mature S-layer protein, which is 435 amino acids in size. The expression of the SlpA fragments on the chimeric flagella was assessed by immunoelectron microscopy and Western blotting using anti-SlpA antibodies, and their binding to human cells was assessed by indirect immunofluorescence. Chimeric flagella harboring inserts that represented the N-terminal part of the S-layer protein bound to the epithelial cell lines, whereas the C-terminal part of the S-layer protein did not confer binding on the flagella. The shortest S-layer peptide capable of detectable binding was 81 amino acid residues in size and represented residues 96 through 176 in the unprocessed S-layer protein. The bacteria and the chimeric flagella did not show detectable binding to erythrocytes, whereas the SlpA-expressing ATCC 8287 cells as well as the chimeric SlpA 96-245/FliC flagella bound to immobilized fibronectin. The N-terminal SlpA peptide 96-176 or 96-200 fused to FliC was not recognized in Western blotting or immunoelectron microscopy by a polyclonal serum raised against the S-layer protein; the antiserum, however, reacted in immunofluorescence with the ATCC 8287 cells. In contrast, an antiserum raised against the His-tagged peptide 96-245 of SlpA bound to the hybrid flagella with the N-terminal SlpA inserts but did not react with ATCC 8287 cells. The results identify the S-layer of L. brevis ATCC 8287 as an adhesin with affinity for human epithelial cells and fibronectin and locate the receptor-binding region within a fragment of 81 amino acids in the N-terminal part of the molecule, which in native S-layer seems inaccessible to antibodies.
Bacterial adhesion to epithelial and subepithelial tissue is an important initial event in successful colonization of the mammalian intestine and other tissue sites. Several adhesion molecules have been characterized for bacterial species that cause infectious diseases in humans or animals (18, 29, 44). This is in sharp contrast to our limited knowledge of the adhesins present on the mammalian commensal genus Lactobacillus. Species of Lactobacillus are major members of the indigenous bacterial microbiota in the gastrointestinal and genital tracts of humans and animals. Lactobacilli are considered beneficial to their host organism and have a long history of use in humans and animals to prevent or cure various minor illnesses. As lactobacilli are members of the normal intestinal microbiota and are food-grade organisms, their possible application as carriers of oral vaccine antigens (28) or other medically important effector molecules (39) in the intestine has aroused interest. Isolates of lactobacilli have been found to adhere to the intestinal epithelial cell lines derived from their mammalian hosts (9, 12, 16, 34), intestinal or gastric mucus (17), extracellular matrix components (30, 41), and human platelets (14) as well as uncharacterized mannoconjugates on intestinal cells (1). Early studies suggested a critical role of lactobacillar adhesiveness in determining host specificity of bacterial colonization in the chicken (13), but later studies failed to demonstrate such a role for lactobacilli isolated from humans (2). The mechanisms by which lactobacilli adhere to and colonize human tissues have remained poorly characterized; to date, two adhesion proteins of lactobacilli have been characterized on a genetic level, an ABC transporter protein of Lactobacillus reuteri (30) and the CbsA S-layer protein of Lactobacillus crispatus (36), both of which bind to collagens of pericellular tissue.
S-layers are paracrystalline surface protein arrays that are commonly expressed by species of Eubacteria and Archaebacteria (3, 33). Most S-layers are composed of a single protein species, the S-layer protein, greatly varying in size in different bacterial genera. S-layers are hydrophobic and crystallize to form a two-dimensional layer on the bacterial surface. The genes encoding S-layers are efficiently transcribed, and the S-layer protein is the dominant protein species, representing 10 to 20% of the total protein mass in the bacterial cell (7). S-layers are commonly expressed by species of the genus Lactobacillus (27). Their role in bacterial adhesiveness to chicken epithelium has been suggested previously (35), and we recently described a collagen-binding S-layer protein of L. crispatus (36), but overall, the functions of lactobacillar S-layers have remained poorly characterized. The primary structures of the few lactobacillar S-layer genes that have been determined predict proteins of 43 to 46 kDa with considerable sequence variability in the N-terminal half of the proteins (4, 5, 8, 43), which could suggest differing functions and antigenic variation for these proteins (36). In lactobacilli, S-layer proteins apparently possess important cellular functions, as several laboratories, including ours (6, 26; M. Kahala and A. Palva, unpublished results), have failed to construct viable null mutants of the S-layer genes. Thus, molecular display methods are a method of choice for genetic analyses of S-layer functions.
Most S-layer proteins can reassemble in physiological buffers to form a regular, insoluble array. Their adhesive properties have been analyzed by solid-phase assays using a soluble ligand, which has remained a severe limitation in the functional analysis of these abundant surface proteins of eubacteria and archaea. Flagella of Salmonella enterica serovar Typhimurium or Escherichia coli have successfully been applied to express heterologous peptides in thousands of copies on the surface of the filament, mainly for vaccination purposes (reviewed in reference 46). We have introduced a flagellum display system in which adhesive peptides up to 300 amino acids in size are expressed in a functional form (47). The flagellum display also offers a system to express large hydrophobic adhesive peptides in a soluble form, and we have applied it to analyze the collagen-binding region in the S-layer-like YadA protein of Yersinia enterocolitica (47). We report here the use of the flagellum display to demonstrate and characterize an adhesive property in the S-layer protein SlpA of Lactobacillus brevis, whose primary structure is known (43).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strain ATCC 8287 of L. brevis and its slpA gene have been described previously (43). The nucleotide sequence for the slpA gene of L. brevis ATCC 8287 has been deposited in the GenBank database under GenBank accession no. Z14250, and the amino acid sequence of this protein can be accessed through the Swiss Protein Database under SwissProt accession no. Q05044. The bacteria were grown overnight at 37°C in 20 ml of static MRS broth. The extraction of the S-layer from the bacterial surface was performed with 2 M guanidine hydrochloride (27), and the control cells were treated with phosphate-buffered saline (PBS), pH 7.1, alone. For adhesion assays, the bacterial cells were suspended in the epithelial cell culture medium or PBS (see below). To analyze the removal of the S-layer protein from the bacterial surface, guanidine hydrochloride-treated and control cells were suspended in an equal volume of the sample buffer used for sodium dodecyl sulfate (SDS) gel electrophoresis (24), the suspensions were boiled for 10 min, and the solubilized proteins were analyzed by SDS-polyacrylamide gel electrophoresis in 12% slab gels. This treatment did not cause detectable cell lysis.
Bacterial adhesion assays.
The human Intestine 407 (ATCC CCL6), the Caco-2 (ATCC HTB-37), and the EA-hy926 (11) cell lines were cultivated to confluence in RPMI 1640 medium (Life Technologies, Paisley, Scotland) supplemented with fetal calf serum (PAA Laboratories GmbH, Linz, Austria), l-glutamine, nonessential amino acids (for Caco-2 and Intestine 407 cells; Life Technologies), and gentamicin. The T24 (ATCC HTB-4) cells were cultivated in McCoy's 5A medium with the same supplements. The Intestine 407, EA-hy926 and T24 cells were cultured for 2 days on diagnostic glass slides (Knittel Glassbearbeitungs GmbH, Braunschweig, Germany), and the Caco-2 cells were cultured for 17 to 21 days in Lab-Tek eight-well chamber slides (Nunc, Roskilde, Denmark). Before the adhesion assays, the cells were washed once with PBS. The bacteria were suspended in the culture medium without supplements at cell densities ranging from 5 × 107 to 1 × 109 cells/ml, 40 μl (300 μl for Caco-2 cells) of the suspension per well was added to the epithelial cells, and the slides were incubated for 1 h at 37°C in a moist chamber. The slides were washed five times at room temperature with PBS for 5 min each and fixed for 10 min with methanol. The epithelial cells with adherent bacteria were then examined in a Carl Zeiss Jenaval microscope (Jena, Germany) directly by Nomarski interference optics (for photography), or for quantitative analysis, the bacteria were stained for 5 min with 10% (vol/vol) Giemsa stain (Oy Reagena Ltd., Kuopio, Finland) or, in the case of Caco-2 cells, with crystal violet and iodine and were analyzed by light microscopy. Because of difficulties in visualizing individual adherent bacteria on the Caco-2 cell surface, we used fluorescein-tagged bacteria with these epithelial cells, and the quantitation of adhesion was done as described previously (20). The mean number of adherent bacteria ± standard deviation was quantitated from 20 epithelial cells or, in the case of Caco-2 cells, from 12 microscopic fields (field, 2.16 × 104 μm2). In inhibition assays, the SlpA 96-245/FliC or the ΔFliCH7 flagella were added at the concentration of 120 μg of FliC/ml (see below) with the bacteria (5 × 108 cells/ml) onto the Intestine 407 cells.
Bacterial adhesion to human plasma fibronectin (Becton Dickinson, Bedford, Mass.) and type IV collagen (Sigma Chemical Co., St. Louis, Mo.) immobilized on glass was tested essentially as described previously (45): the target proteins were used at the surface concentration of 2.5 pmol, the slides were blocked with 5% (wt/vol) skim milk (Valio, Helsinki, Finland) in PBS, the incubation time with bacteria in PBS was 2 h, and washing was performed with 0.05% skim milk in PBS.
Hemagglutination assays with bacteria as well as treatment of human erythrocytes with endo-β-galactosidase, neuraminidase, trypsin, or pronase were conducted by routine procedures (22). We used human 01 and Resolve Panel B (Ortho Diagnostic Systems Inc., Raritan, N.J.) red blood cells. In addition, sheep, calf, goat, horse, dog, rabbit, mouse, cat, and pig erythrocytes were tested. The chimeric flagella were tested at the concentration of 200 μg/ml, and the agglutination was read by naked eye as well as under the microscope.
Flagellum display of SlpA fragments and production of His-SlpA 96-245.
The construction of chimeric SlpA/FliC flagella and their testing were performed essentially as described previously (47). Fragments representing different parts of the slpA gene were amplified by PCR with Pfu polymerase and with chromosomal DNA from the L. brevis strain ATCC 8287 as the template. The primers were designed on the basis of the nucleotide sequence of slpA (43) and contained an AccI restriction site at the 5′ terminus. The primers amplified regions of slpA encoding the peptide fragment 31-300, 31-245, 96-370, 96-245, 96-200, 96-176, 126-156, or 239-447, where the residue numbers include the 30-amino-acid-long signal sequence of SlpA. The slpA fragments were cloned into the AccI site in the plasmid pFliCH7Δ deleted for 174 bp in the variable region of fliC, and the resulting plasmids were expressed in E. coli strain JT1, which is a fliC::Tn 10 and fimA::cat derivative of E. coli C600 (47). The ΔFliCH7 flagella lacking an insert were available from previous work, and the chimeric SlpA/FliC flagellar filaments were isolated as described previously (47).
DNA isolation and manipulation were performed by routine procedures (32). To express SlpA 96-245 as a His-tagged peptide, the corresponding region in slpA was PCR amplified and cloned in the pQE-30 expression vector. The primers were designed on the basis of the nucleotide sequence of slpA (43) and contained in the forward primer a 5′ BamHI restriction site and in the reverse primer a HindIII restriction site and the stop codon. The fusion protein was expressed and purified by affinity chromatography as described in the manufacturer's instructions (Qiagen GmbH, Hilden, Germany).
Immunological methods.
The flagellar or SlpA proteins were separated by SDS gel electrophoresis (27) in 10% (wt/vol) slab gels and subjected to Western blotting (42) with polyclonal antibodies against the H7 flagella (47), the SlpA protein of L. brevis ATCC 8287 (43), or the His-SlpA 96-245 peptide as primary antibodies and alkaline phosphatase-conjugated secondary antibodies. For immunoelectron microscopy (IEM), bacterial cells expressing flagellar constructs were suspended in Luria broth and immobilized on copper grids coated with Pioloform and carbon. After being washed with PBS containing 1 mg of bovine serum albumin (BSA) in PBS/ml, the anti-H7, the anti-SlpA, or the anti-His SlpA 96-245 antiserum (diluted 1:300 in PBS containing 10 mg of BSA/ml) was added and left to react with the flagella for 90 min at 20°C. The grids were washed in PBS with 1 mg of BSA/ml, and bound antibodies were detected with AuroprobeEM protein A conjugate (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England; diluted 1:40). Bacteria were negatively stained with 1% potassium tungstic acid, pH 6.5, for 1 min, and the grids were examined in a JEOL JEM-100EX transmission electron microscope at an operating voltage of 60 kV.
Binding tests with chimeric flagella.
The binding of the chimeric flagella to the epithelial cells was assessed by indirect immunofluorescence essentially as described previously (47). Briefly, the epithelial cells were washed at room temperature with PBS, fixed with methanol for 10 min at −20°C, and then washed with PBS at room temperature. The FliC concentration of each extract was adjusted to 20 μg/ml with the Tina (version 2.0) image analysis program (Raytest Isotopenmessgeräte GmbH, Straubenhardt, Germany) and Coomassie blue-stained SDS-polyacrylamide gels with the YadA 26-202/FliC chimeric flagella (47) or BSA as internal standard. The flagellar extracts (40 μl, or 300 μl for Caco-2 cells) were added to the cells, and the slides were kept for 5 h at 4°C. After washing and a second fixing with methanol, the bound flagella were visualized by staining with immunoglobulin G molecules from an anti-FliCH7 rabbit antiserum and with fluorescein isothiocyanate-labeled secondary antibodies as detailed previously (47). The control assays included staining of the epithelial cells as described above but with the ΔFliCH7 flagella lacking an insert, or with omission of the flagellar extract or the flagellar extract and the immunoglobulins.
Binding of chimeric flagella to immobilized plasma fibronectin (Becton Dickinson) was tested by an enzyme-linked immunosorbent assay as described in reference 47. The coating of microwells was performed with fibronectin (11 μg/ml) or 1% (wt/vol) skim milk in PBS, quenching was performed for 2 h with 1% skim milk, and flagellar extracts were incubated overnight at 4°C at 0.8 to 25 μg/ml in 0.05% skim milk in PBS.
Homology searches.
Protein and DNA sequence homology searches were performed with the BLASTP2, the FASTA, and the Lalign programs available at the website http://www.dna.affrc.go.jp/htdocs/Blast/fasta.html, as well as with the ClustalW multiple sequence alignment program at http://pbil.ibcp.fr/cgi-bin/align_clustalw.pl. The grand average hydropathy values were calculated by using the program ProtParam at http://www.expasy.ch/cgi-bin/protparam.
RESULTS
Expression of slpA fragments as fusions to fliC.
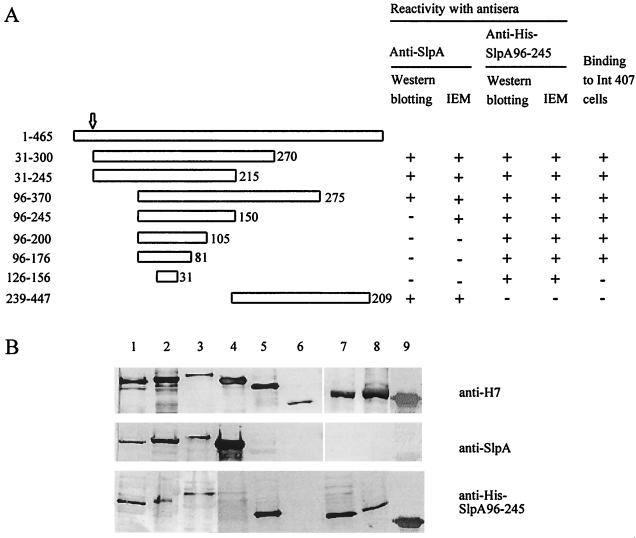
After an initial screening of various lactobacillar isolates for adhesiveness to human small intestinal cell lines, we chose the strain L. brevis ATCC 8287 on the basis that it showed highly efficient adhesion to various human epithelial cell lines. Cells of L. brevis ATCC 8287 were known to express the S-layer protein SlpA as their major cell surface protein (43). We observed that treatment of ATCC 8287 cells with guanidine hydrochloride, which is the routine procedure to deplete cells of the S-layer (38), completely abolished bacterial adhesion to the human small intestinal cell line Intestine 407 (see below). We used the flagellum display technology (47) to analyze the possible adhesive function of SlpA. Fragments of slpA were cloned into the AccI site in the plasmid pFliCH7Δ, which contains fliC with a 174-bp deletion in the variable region, and the chimeric flagella were prepared from the surface of the complemented derivatives of E. coli JT1, whose genotype is fliC::Tn10 andfimA::cat. The fragments of SlpA that were expressed as a FliC fusion are schematically presented in Fig. 1; the numbering of amino acid residues refers to the unprocessed SlpA peptide, which has a signal sequence of 30 amino acids. To confirm the expression of the SlpA fragments in the FliC protein, the chimeric flagella were analyzed by Western blotting using polyclonal anti-SlpA or anti-FliCH7 antibodies (Fig. 1B). The apparent size of the chimeric flagellins, as estimated from the Western blots stained with anti-FliCH7 antibodies, corresponded to those predicted from the nucleotide sequence. The ΔFliCH7 flagella lacking an insert did not react with the anti-SlpA antibodies (lane 6 in Fig. 1B), whereas the SlpA 31-245/FliC, the SlpA 31-300/FliC, the SlpA 96-370/FliC, and the SlpA 239-447/FliC flagella were recognized by the anti-SlpA antibodies (lanes 1 through 4 in Fig. 1B). These chimeric flagella carried an insertion of 215, 270, 275, or 209 amino acid residues, respectively. In contrast, the chimeric flagella SlpA 96-245/FliC, SlpA 96-200/FliC, SlpA 96-176/FliC, and SlpA 126-156/FliC, which carried SlpA inserts ranging in size between 150 and 31 amino acid residues and covering part of the N-terminal region of the SlpA molecule, did not detectably react with the anti-SlpA antibodies (lanes 5 and 7 through 9 in Fig. 1B). We also raised rabbit antibodies against His-tagged peptide 96-245 of SlpA purified from the cytoplasm of recombinant E. coli. These antibodies reacted in Western blotting with the chimeric flagella carrying the N-terminal SlpA inserts but not with the ΔFliCH7 or the SlpA 239-447/FliC flagella (Fig. 1B), which indicates that the chimeras indeed expressed the shorter N-terminal SlpA inserts.
FIG. 1.
(A) Schematic presentation of the SlpA fragments expressed as fusions to FliC. On top is shown the entire SlpA polypeptide of 465 amino acids; the arrow indicates the cleavage site of the signal sequence. The bars indicate the fragments expressed as fusions in FliC, and the numbers on the left refer to N- and C-terminal amino acids in the SlpA peptide. The size of the amino acid insert in FliC is given after each construct. Reactivity of each chimeric flagellum with the anti-SlpA as well as the anti-His-SlpA 96-245 peptide antibody was determined by Western blotting (see below) and by IEM (Fig. 2). The binding of the chimeric flagella to the human Intestine 407 cells was tested by indirect immunofluorescence (Fig. 3) and is indicated on the right. (B) Western blotting of the chimeric flagella with anti-FliCH7, anti-SlpA, and anti-His-SlpA 96-245 peptide polyclonal antibodies. The flagella were SlpA 31-245/FliC (lane 1), SlpA 31-300/FliC (lane 2), SlpA 96-370/FliC (lane 3), SlpA 239-447/FliC (lane 4), SlpA 96-245/FliC (lane 5), ΔFliC lacking an insert (lane 6), SlpA 96-176/FliC (lane 7), SlpA 96-200/FliC (lane 8), and SlpA 126-156/FliC (lane 9).
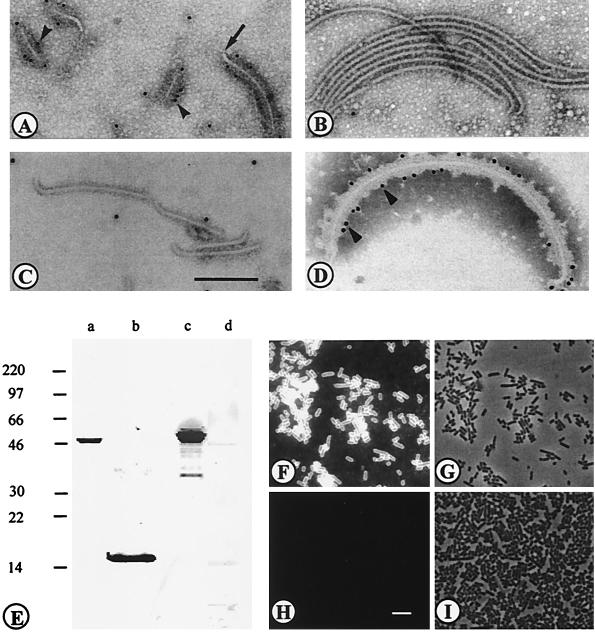
IEM with anti-SlpA antibodies and protein A-gold was also used to analyze the chimeric flagella. As an example, Fig. 2 shows IEM of the SlpA 96-245/FliC, the SlpA 96-176/FliC, and the ΔFliCH7 flagella with anti-SlpA antibodies as well as the reactivity of the SlpA 96-245/FliC with anti-FliCH7 antibodies. The SlpA 96-245/FliC flagella bound anti-SlpA antibodies (Fig. 2A), whereas the ΔFliCH7 and the SlpA 96-176 flagella were unreactive (Fig. 2B and C). The specificity of the reactivity was further illustrated by a lack of the anti-SlpA antibody binding to flagellar hooks encoded by the flgE gene (arrow in Fig. 2A). The chimeric flagella SlpA 31-300/FliC, SlpA 31-245/FliC, SlpA 96-370/FliC, SlpA 96-245/FliC, and SlpA 239-447/FliC reacted with the anti-SlpA antibodies, whereas the other chimeric flagella were unreactive. All the flagella reacted with the anti-FliCH7 antibodies; Fig. 2D shows reactivity of the SlpA 96-245/FliC as an example, and the results are summarized in Fig. 1A.
FIG. 2.
(A to D) IEM of chimeric flagella. The staining was done with anti-SlpA (A to C) or anti-FliCH7 (D) antibodies and with protein A-gold. The flagella were SlpA 96-245/FliC (A), ΔFliC lacking an insert (B), SlpA 96-176/FliC (C), and SlpA 96-245/FliC (D). Arrowheads indicate binding of protein A-gold, and the arrow in panel A indicates the flagellar hook encoded by flgE. (E to I) Reactivities of antisera with SlpA and His-SlpA 96-245 proteins, L. brevis ATCC 8287 cells, and recombinant flagella. (E) Western blotting of the SlpA protein from ATCC cells (lanes a and c) and the His-SlpA 96-245 peptide from recombinant E. coli (lanes b and d) with anti-His-SlpA 96-245 peptide (lanes a and b) and anti-SlpA (lanes c and d) antibodies. Numbers at left indicate protein sizes in kilodaltons. (F) Indirect immunofluorescence staining of ATCC 8287 cells with anti-SlpA antibodies. (H) Staining with anti-His-SlpA 96-245 antibodies. (G and I) Fields corresponding to panels F and H, respectively, under light microscopy. Bars, 200 nm (A to D) and 5 μm (F to I).
Binding of the SlpA/FliC flagella to Intestine 407 cells.
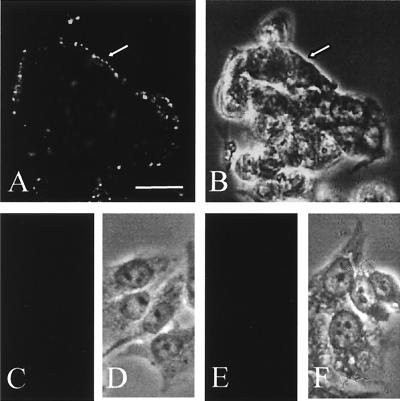
We initially tested the binding of chimeric flagella to the Intestine 407 cells because bacterial adhesion (see below) and the flagellar binding to nonconfluent cell culture were easily evaluated. The reactivity of the chimeric flagella with human Intestine 407 cells was assessed by indirect immunofluorescence using purified anti-FliCH7 immunoglobulin G and fluorescein isothiocyanate-conjugated secondary antibodies. Representative examples of the assays are shown in Fig. 3. The SlpA 31-300/FliC (data not shown) and the SlpA 96-245/FliC flagella efficiently bound to the Intestine 407 cells (Fig. 3A), whereas the construct SlpA 239-447/FliC, carrying the C-terminal part of SlpA, failed to bind (Fig. 3C). No binding was observed with the ΔFliCH7 flagella lacking an insert (Fig. 3E). The granular pattern of staining by the chimeric flagella probably results from their organelle-like structure, and no significant aggregation of the chimeric filaments was observed by electron microscopy (data not shown).
FIG. 3.
Binding of the chimeric flagella to the human Intestine 407 cells. The bound flagella were detected by indirect immunofluorescence using anti-FliCH7 immunoglobulin G and fluorochrome-conjugated secondary antibodies. (A) Binding of the SlpA 96-245/FliC chimeric flagella. (B) The corresponding field under light microscopy. (C) Binding of SlpA 239-447/FliC. (E) Binding of the ΔFliC flagella lacking an insert. (D and F) Light microscopic fields corresponding to panels C and E, respectively. Arrows indicate cell contours. Bar, 20 μm.
The results shown in Fig. 3 suggested that the receptor-binding region is located within the N-terminal part of the SlpA molecule. In order to localize the binding site in more detail, the fragments covering the N-terminal half of the SlpA molecule were expressed in FliC and tested for binding. The shortest SlpA fragment conferring detectable binding to Intestine 407 cells was in the construct SlpA 96-176/FliC, which contained an 81-amino-acid-long insert from SlpA (Fig. 1A). The construct SlpA 126-156/FliC with an insert of 31 amino acids did not exhibit detectable adhesiveness. When flagella were tested at the concentration of 120 μg of FliC/ml and with 5 × 108 bacteria/ml, no inhibition of bacterial adhesion to Intestine 407 cells was detected with the SlpA 96-245/FliC or the ΔFliCH7 flagella.
Comparison of the reactivities of the anti-SlpA and anti-His-SlpA 96-245 peptide antibodies.
The His-SlpA 96-245 peptide was aggregated and unsuited for binding tests; we therefore raised antipeptide antibodies in order to test their effect on the adhesiveness of the strain ATCC 8287. We could not detect any significant inhibition of bacterial adhesion by Fab fragments prepared from immunoglobulins against the peptide or the entire SlpA molecule (data not shown). The anti-SlpA antibodies reacted in Western blotting or IEM with chimeric flagella expressing the larger N-terminal regions of SlpA (summarized in Fig. 1) but not with the His-SlpA 96-245 peptide (Fig. 2E). The antibodies reacted with the ATCC 8287 cells in indirect immunofluorescence (Fig. 2F and G). The anti-His-SlpA 96-245 peptide antibodies, in contrast, recognized the hybrid flagella with N-terminal SlpA inserts in Western blotting and in IEM (Fig. 1), and no binding of the antibody to the SlpA 239-447/FliC flagella or the ΔFliCH7 flagella (Fig. 1B) was detected. However, the anti-His-SlpA 96-245 peptide antibodies did not bind to ATCC 8287 cells (Fig. 2H and I).
Adhesion of S-layer-expressing and S-layer-depleted bacteria to human cells.
SlpA represents the single major protein in ATCC 8287 cells (43), and we have not been able to produce a viable slpA mutant strain. On the other hand, the peripheral S-layer proteins can be extracted from the lactobacillar cell surface with guanidine hydrochloride or SDS without disrupting the bacterial cells (38). Our results above suggested that removal of SlpA from the ATCC 8287 cell surface should abolish bacterial adhesiveness, and we next analyzed the adhesion of native and guanidine hydrochloride-treated bacterial cells as well as the binding of the SlpA 96-245/FliC, the SlpA 239-447/FliC, and the ΔFliCH7 flagella to human cell types. The strain ATCC 8287 adhered to the Intestine 407, human bladder T24, and endothelial EA-hy926 cells, as well as polarized large intestinal Caco-2 cells (Table 1). For Intestine 407, T24, and EA-hy926 cells, guanidine hydrochloride treatment of L. brevis cells abolished bacterial adhesion, whereas only a partial decrease in bacterial adhesion to Caco-2 cells was detected. The SlpA 96-245/FliC flagella bound to all four epithelial cell lines, whereas no binding of the SlpA 239-447/FliC or the ΔFliCH7 flagella was detected (Table 1).
TABLE 1.
Adhesion of L. brevis cells and binding of chimeric SlpA/FliC flagella to human epithelial cellsa
| Human cell line | Adhesion (no. of bacteria/human cell)
|
Binding of flagellum type:
|
|||
|---|---|---|---|---|---|
| − GnHCl | + GnHCl | SlpA 96-245/FliC | SlpA 239-447/FliC | ΔFliC | |
| Intestine 407 | 264 ± 66 | 4 ± 2 | + | − | − |
| Caco-2 | 297 ± 90b | 175 ± 34b | + | − | − |
| T24 | 56 ± 18 | 3 ± 2 | + | − | − |
| EA-hy926 | 110 ± 35 | 8 ± 3 | + | − | − |
Bacterial adhesion was assessed before and after removal of the S-layer by treatment with 2 M guanidine hydrochloride (GnHCl). The results are given as means and standard deviations of values for adherent bacteria on 20 epithelial cells. The bacteria were tested at 5 × 108 cells/ml. The flagellar binding was tested by indirect immunofluorescence (Fig. 3).
For the confluent Caco-2 cell culture, the results are given as means ± standard deviations for 12 microscopic fields (field, 2.16 × 104 μm2) of confluent cells.
The L. brevis ATCC 8287 cells did not agglutinate human 01 erythrocytes or those in a commercial blood group typing panel, nor did they react with erythrocytes from nine animal species. Bacterial binding to the erythrocytes was not induced by treatment of human or animal erythrocytes with endo-β-galactoside, neuraminidase, trypsin, or pronase, each of which has been used to expose cryptic target sites on erythrocytes for bacterial adhesion proteins (22). We did not detect any binding of the SlpA 96-245/FliC or the ΔFliCH7 flagella to human 01 erythrocytes.
Bacterial adherence and binding of chimeric flagella to fibronectin.
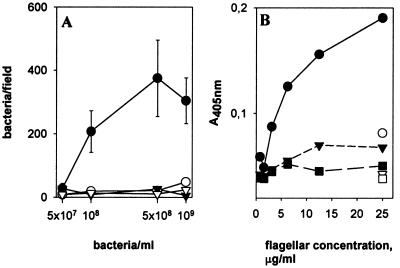
Fibronectins have been shown previously to mediate bacterial adherence to a number of epithelial cell types (reviewed in reference 19), and we therefore tested whether SlpA binds to human fibronectin. ATCC 8287 cells with intact SlpA adhered to fibronectin, whereas removal of SlpA from the cell surface abolished adherence (Fig. 4A). No adherence to type IV collagen was detected. The SlpA 96-245/FliC flagella bound to immobilized fibronectin, whereas binding of the SlpA 239-447/FliC or the ΔFliC flagella was close to background level (Fig. 4B).
FIG. 4.
Binding of SlpA to immobilized fibronectin. (A) Adhesiveness of L. brevis ATCC 8287 to fibronectin (circles) or type IV collagen (inverted triangles) is shown before (closed symbols) and after (open symbols) removal of SlpA by guanidine hydrochloride. Means and standard deviations of bacterial numbers in 20 randomly chosen microscopic fields of 4.8 × 103 μm2 are shown. (B) Binding of chimeric flagella to fibronectin (closed symbols) and skim milk proteins (open symbols) is shown. The flagella were SlpA 96-245/FliC (circles), SlpA 239-447/FliC (inverted triangles), and ΔFliC (squares).
Sequence analysis of SlpA.
We found in the GenBank, EMBL, SwissProt, or PIR databases only sequences with a low level of homology to the DNA sequence encoding the SlpA 96-176 region or to the corresponding amino acid sequence. The identity of the SlpA sequence with other sequenced lactobacillar S-layer proteins was 19 to 23%, with no regions showing significant local identity. The grand hydropathy values of the processed form of SlpA, the fragment SlpA 126-156, the fragment SlpA 96-176, the fragment SlpA 96-245, and the fragment SlpA 239-447 were −0.383, −0.300, −0.621, −0.489, and −0.341, respectively.
DISCUSSION
Lactobacilli are important bacterial colonizers of intestinal surfaces. Despite the high importance of lactobacilli as members of intestinal normal microbiota and their potential for biotechnological applications, the adhesion proteins of lactobacilli have remained poorly characterized. Our present and previous (36, 41) results establish S-layers as a class of lactobacillar adhesins that exhibit affinity for different tissue targets. We used the flagellum display technology to obtain direct evidence for the adhesive function of SlpA and expressed fragments of slpA as a fusion to the fliCH7Δ gene of E. coli. The N-terminal region of SlpA conferred adhesiveness on the chimeric flagella SlpA 31-300/FliC, whereas no binding was observed with the SlpA 239-447/FliC chimera expressing a C-terminal SlpA insert or with the truncated ΔFliCH7 flagella lacking an insert. Our finding that the adhesive character of SlpA of L. brevis resides in the N-terminal region of the S-layer protein is in accordance with the situation in the collagen-binding S-layer protein CbsA of L. crispatus (36). This region in lactobacillar S-layer proteins exhibits sequence variability (36), which suggests functional diversity in lactobacillar S-layer proteins, and indeed we did not observe any collagen binding by SlpA.
Expression of the N-teminal SlpA fragments by the flagellum display technology showed that the 81-amino-acid insert in SlpA 96-176/FliC conferred adhesiveness on the flagella, whereas the 31-amino-acid insert in SlpA 126-156/FliC failed to do so. This indicates that the core epithelium-binding region in SlpA is less than 100 amino acids in size. We found no epithelium binding with the SlpA 239-447/FliC flagella, which, according to the grand average hydropathy value, is more hydrophobic than are the N-terminal SlpA regions supporting adhesion. Our finding that SlpA of L. brevis binds to various human epithelial cell types but not to erythrocytes is in agreement with the tissue distribution of fibronectins and fibronectin receptors. Fibronectins are present on epithelial cell surfaces as well as in a soluble form in circulation. They bind to human β1 integrins that are present on numerous epithelial cells (15). On the other hand, fibronectins bind to a number of bacterial species and mediate bacterial adhesion to epithelial cells by direct or bridging mechanisms (reviewed in reference 19). While this study was in progress, we became aware of the report by Kapczynski et al. (21) showing that adherence of Lactobacillus to Intestine 407 cells correlates with fibronectin binding. Our results agree with their data and identify ATCC 8287 S-layer as a surface structure mediating fibronectin binding by lactobacilli. It seems apparent that other fibronectin-binding structures must also exist on lactobacilli (25, 37), and their molecular natures remain to be established.
The crystalline S-layers bear morphological similarity to the regular envelope structure of animal viruses, where structural and immunological studies have demonstrated the so-called canyon hypothesis (31). The receptor-binding pocket on the viral surface is located within a conserved concave structure surrounded by variable regions and is inaccessible to the antigen-binding region of antibodies. Our results suggest a similar organization in the S-layer of L. brevis ATCC 8287. The chimeric SlpA/FliC flagella reacted with anti-FliCH7 as well as anti-His-SlpA 96-245 peptide antibodies in IEM and Western blotting, whereas the N-terminal fragments of SlpA of 150 amino acids or less failed to give a detectable reaction with the polyclonal anti-SlpA antibodies. This region in SlpA expressed the epithelium-binding activity. The anti-SlpA antibodies that we used recognize SlpA on the surface of L. brevis ATCC 8287 cells as well as in Western blots (43) (Fig. 2). In contrast, antibodies raised against the His-SlpA 96-245 peptide failed to bind to ATCC 8287 cells (Fig. 2H and I) but recognized the SlpA epitope in the denatured form in Western blotting of chimeric flagella and the His-tagged peptide (Fig. 1 and 2), as well as in a nondenatured conformation in IEM of chimeric flagella. These findings indicate that in the polymerized S-layer the region 96-245 of SlpA is poorly accessible to the antigen-binding regions of antibodies, whereas the region is exposed to antibody binding in the chimeric flagella. Failure of anti-SlpA antibodies to react with SlpA 96-245 and chimeric flagella with shorter inserts also indicates that this region was poorly immunogenic when complete S-layer protein was used as an immunogen. Another explanation for the behavior of the two antisera could be that the flagellum inserts fold into a different conformation than does the intact protein. Resolution of the question whether the receptor-binding region in the L. brevis SlpA indeed is in a concave pocket requires a structural model of SlpA not currently available.
The adhesive chimeric flagella did not assemble into an S-layer-like structure. We have recently observed that collagen binding by CbsA of L. crispatus is optimal with regular polymeric S-layers (36) but can also be observed in a display system where S-layer sheets are not formed (26). Our results indicate that the binding region in SlpA represents a conformational domain that is not critically dependent on a polymerized S-layer. SlpA resembles the fibronectin-binding S-layer protein A of Aeromonas salmonicida, where a soluble 35-kDa N-terminal peptide from the 51-kDa A-layer protein retains the adhesiveness of the S-layer but does not assemble into a tetragonal array (10, 40). Our attempts to inhibit the adhesion of L. brevis to Intestine 407 cells with the chimeric flagella failed. This was not surprising in light of the highly efficient adhesion of the ATCC 8287 cells and the fairly low concentration of the chimeric flagella that we were able to use in the assays. Furthermore, we hypothesize that the efficiency of binding is lower with the SlpA peptide than with the complete SlpA molecule or with the intact S-layer on the surface of L. brevis cells, where the binding epitope is exposed in thousands of regularly and closely arranged copies.
We found that L. brevis ATCC 8287 carrying SlpA expressed affinity for human intestinal, urinary bladder, and endothelial cells but not for human or animal erythrocytes. Adhesion of ATCC 8287 cells to Intestine 407, T24, and EA-hy926 cells was abolished after removal of S-layer by guanidine hydrochloride treatment of the bacteria. Extraction with guanidine hydrochloride or SDS, as used in this study, is a routine procedure to remove S-layer proteins (38) and did not cause detectable lysis of L. brevis cells. It is obvious that minor amounts of other cell wall proteins also were removed by the extraction; the poor adhesiveness of S-layer-depleted ATCC 8287 cells, however, is in accordance with the notion that SlpA is an adhesin. The adhesion of L. brevis ATCC 8287 to Caco-2 cells was reduced but not abolished after the guanidine hydrochloride treatment, although the SlpA 96-245/FliC flagella also bound to this cell type. Caco-2 cells are recognized by a number of lactobacillar isolates, including strains that do not express an S-layer protein (12), and our ongoing work (37) has shown that a number of S-layer-expressing lactobacillar isolates express adhesiveness to Caco-2 cells after S-layer removal by guanidine hydrochloride. It is obvious that surface structures other than the S-layer protein mediate adhesion of such strains. Such an adhesin(s) has not, however, been identified so far. Expression of multiple adhesins that bind to epithelial or nonepithelial surfaces is common in bacterial pathogens (18, 23, 44). It is likely that L. brevis ATCC 8287 expresses, in addition to SlpA, another adhesin with affinity for Caco-2 cells and that this adhesin is not removed from the cell surface or denatured by the guanidine hydrochloride extraction. The importance of the SlpA-mediated adhesion for lactobacilli awaits analysis of their non-S-protein adhesins as well as further characterization of their receptor-binding specificities.
Acknowledgments
We thank Ilkka Palva for discussion and critical comments on the manuscript and Anna Pirkola, the Finnish Red Cross Blood Transfusion Center, for the erythrocytes. Electron microscopy was performed at the Electron Microscopy Unit, the Institute of Biotechnology, University of Helsinki.
This study was supported by the Academy of Finland (grant no. 40836, 44600, 44602, and 44168) and by the University of Helsinki.
REFERENCES
- 1.Adlerberth, I., S. Ahrné, M.-L. Johansson, G. Molin, L. Å. Hanson, and A. E. Wold. 1996. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to human colonic cell line HT-29. Appl. Environ. Microbiol. 62:2244-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrné, S., S. Nobaek, B. Jeppson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88-94. [DOI] [PubMed] [Google Scholar]
- 3.Beveridge, T. J. 1994. Bacterial S-layers. Curr. Opin. Struct. Biol. 4:204-212. [Google Scholar]
- 4.Boot, H. J., C. P. A. M. Kolen, J. M. van Noort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J. Bacteriol. 175:6089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boot, H. J., C. P. A. M. Kolen, and P. H. Pouwels. 1995. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 177:7222-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot, H. J., P. A. M. Kolen, and P. J. Pouwels. 1996. Interchange of the active site and silent S-protein genes of Lactobacillus acidophilus by inversion of the chromosomal slp gene. Mol. Microbiol. 21:799-809. [DOI] [PubMed] [Google Scholar]
- 7.Boot, H. J., and P. H. Pouwels. 1996. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol. Microbiol. 21:1117-1123. [DOI] [PubMed] [Google Scholar]
- 8.Callegari, M. L., B. Riboli, J. W. Sanders, P. S. Cocconelli, J. Kok, G. Venema, and L. Morelli. 1998. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology 144:719-726. [DOI] [PubMed] [Google Scholar]
- 9.Coconnier, M.-H., T. R. Klaenhammer, S. Kerneis, M.-F. Bernet, and A. L. Servin. 1992. Protein-mediated adhesion of Lactobacillus acidophilus BG2FO4 on human enterocyte and mucus-secreting cell lines in culture. Appl. Environ. Microbiol. 58:2034-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doig, P., L. Emödy, and T. J. Trust. 1992. Binding of laminin and fibronectin by the trypsin-resistant major structural domain of the crystalline virulence surface array protein of Aeromonas salmonicida. J. Biol. Chem. 267:43-49. [PubMed] [Google Scholar]
- 11.Edgell, C.-J. S., G. C. McDonald, and J. B. Graham. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. USA 80:3734-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elo, S., M. Saxélin, and S. Salminen. 1981. Attachment of Lactobacillus casei strain GG to human colon carcinoma cell line Caco-2. Lett. Appl. Microbiol. 13:154-156. [Google Scholar]
- 13.Fuller, R. 1973. Ecological studies on the Lactobacillus flora associated with the crop epithelium of the fowl. J. Appl. Bacteriol. 36:131-139. [Google Scholar]
- 14.Harty, D. W. S., M. Patrikakis, and K. W. Know. 1993. Identification of Lactobacillus strain isolates from patients with infective endocarditis and comparison of surface-associated properties with those of other strains of the same species. Microb. Ecol. Health Dis. 6:191-201. [Google Scholar]
- 15.Hemler, M. 1993. Integrins, p. 143-145. In T. Kreis and R. Vale (ed.), Guidebook to the extracellular matrix and adhesion proteins, 1st ed. Oxford University Press, Oxford, United Kingdom.
- 16.Henriksson, A., R. Szewzyk, and P. L. Conway. 1991. Characteristics of the adhesive determinants of Lactobacillus fermentum 104. Appl. Environ. Microbiol. 57:499-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriksson, A., and P. L. Conway. 1996. Adhesion of Lactobacillus fermentum 104-S to porcine stomach mucus. Curr. Microbiol. 33:31-34. [DOI] [PubMed] [Google Scholar]
- 18.Hultgren, S. J., S. N. Abraham, and S. Normark. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383-415. [DOI] [PubMed] [Google Scholar]
- 19.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Höök. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18:211-223. [DOI] [PubMed] [Google Scholar]
- 20.Källenius, G., T. K. Korhonen, V. Väisänen-Rhen, and S. B. Svenson. 1985. Adherence assays, p. 321-332. In T. K. Korhonen, E. A. Dawes, and P. H. Mäkelä (ed.), Enterobacterial surface antigens: methods for molecular characterization. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 21.Kapczynski, D. R., R. J. Meinersmann, and M. D. Lee. 2000. Adherence of Lactobacillus to intestinal 407 cells in culture correlates with fibronectin binding. Curr. Microbiol. 41:136-141. [DOI] [PubMed] [Google Scholar]
- 22.Korhonen, T. K., and J. Finne. 1985. Agglutination assays for detecting bacterial binding specificities, p. 301-313. In T. K. Korhonen, T. Hovi, and P. H. Mäkelä (ed.), Enterobacterial surface antigens: methods for molecular characterization. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 23.Korhonen, T. K., R. Virkola, B. Westerlund, H. Holthöfer, and J. Parkkinen. 1990. Tissue tropism of Escherichia coli adhesins in human extraintestinal infections. Curr. Top. Microbiol. Immunol. 151:115-127. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lorca, G., M. I. Torino, G. F. de Valdez, and Å. Ljung. 2002. Lactobacilli express cell surface proteins which mediate binding of immobilized collagen and fibronectin. FEMS Microbiol. Lett. 206:31-37. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, B., J. Sillanpää, E. Smit, T. K. Korhonen, and P. H. Pouwels. 2000. Expression of cbsA encoding the collagen-binding S-protein of Lactobacillus crispatus JCM5810 in Lactobacillus casei ATCC 393T. J. Bacteriol. 182:6857-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda, K., and T. Kawata. 1983. Distribution and chemical characterization of regular arrays in the cell wall of strains of the genus Lactobacillus. FEMS Microbiol. Lett. 20:145-150. [Google Scholar]
- 28.Mercenier, A., H. Müller-Alouf, and C. Grangette. 2000. Lactic acid bacteria as live vaccines. Curr. Issues Mol. Biol. 2:17-25. [PubMed] [Google Scholar]
- 29.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 30.Roos, S., P. Aleljung, N. Robert, B. Lee, T. Wadström, M. Lindberg, and H. Jonsson. 1996. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system. FEMS Microbiol. Lett. 144:33-38. [DOI] [PubMed] [Google Scholar]
- 31.Rossman, M. G. 1989. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J. Biol. Chem. 264:14587-14590. [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Sara, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182: 859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savage, D. C. 1984. Adherence of the normal flora, p. 3-10. In E. C. Boedeker (ed.), Attachment of organisms to the gut mucosa, vol. 1. CRC Press, Boca Raton, Fla. [Google Scholar]
- 35.Schneitz, C., L. Nuotio, and K. Lounatmaa. 1993. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface layer (S-layer). J. Appl. Bacteriol. 74:290-294. [DOI] [PubMed] [Google Scholar]
- 36.Sillanpää, J., B. Martinez, J. Antikainen, T. Toba, N. Kalkkinen, S. Tankka, K. Lounatmaa, J. Keränen, M. Höök, B. Westerlund-Wikström, P. H. Pouwels, and T. K. Korhonen. 2000. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182:6440-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sillanpää, J. V. 2001. Ph.D. thesis. University of Helsinki, Helsinki, Finland.
- 38.Sleytr, U. B., and M. Sára. 1997. Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol. 15:20-26. [DOI] [PubMed] [Google Scholar]
- 39.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, S., J. W. Austin, W. D. McCubbin, C. M. Kay, and T. J. Trust. 1992. Roles of structural domains in the morphology and surface anchoring of the tetragonal paracrystalline array of Aeromonas hydrophila. Biochemical characterization of the major structural domain. J. Mol. Biol. 228:652-661. [DOI] [PubMed] [Google Scholar]
- 41.Toba, T., R. Virkola, B. Westerlund, Y. Björkman, J. Sillanpää, T. Vartio, N. Kalkkinen, and T. K. Korhonen. 1995. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl. Environ. Microbiol. 61:2467-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidgrén, G., I. Palva, R. Pakkanen, K. Lounatmaa, and A. Palva. 1992. S-layer protein gene of Lactobacillus brevis: cloning by polymerase chain reaction and determination of the nucleotide sequence. J. Bacteriol. 74:7419-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687-694. [DOI] [PubMed] [Google Scholar]
- 45.Westerlund, B., P. Kuusela, T. Vartio, I. van Die, and T. K. Korhonen. 1989. A novel lectin-independent interaction of P fimbriae of Escherichia coli with immobilized fibronectin. FEBS Lett. 243:199-204. [DOI] [PubMed] [Google Scholar]
- 46.Westerlund-Wikström, B. 2000. Peptide display on bacterial flagella: principles and applications. Int. J. Med. Microbiol. 290:223-230. [DOI] [PubMed] [Google Scholar]
- 47.Westerlund-Wikström, B., J. Tanskanen, R. Virkola, J. Hacker, M. Lindberg, M. Skurnik, and T. K. Korhonen. 1997. Functional expression of adhesive peptides as fusions to Escherichia coli flagellin. Protein Eng. 11:1319-1326. [DOI] [PubMed] [Google Scholar]