Summary
The microphthalmia transcription factor MITF plays important roles in several cell lineages including retinal and neural crest-derived pigment cells. Previous reports have shown that besides its regulation at the transcriptional level, MITF is also regulated post-translationally by phosphorylation and ubiquitination which affect the protein's activity and stability. Here we demonstrate that in addition, MITF is modified in melanoma cells by small ubiquitin-like modifier (SUMO). In vitro assays further show that sumoylation occurs at two lysine residues, K182 and K316, and depends on SUMO E1 activating enzyme (SAE I/SAE II) and E2 conjugating enzyme (Ubc9). Interestingly, MITF with double lysine 182/316 to arginine mutations, although displaying normal DNA binding, stability and nuclear localization, shows a substantial increase in the transcriptional stimulation of promoters containing multiple but not single MITF binding sites. MITF containing the double lysine-to-arginine substitution also shows enhanced cooperation with Sox10 on the Dct promoter. We conclude that SUMO modification of MITF regulates the protein's transcriptional activity especially with respect to synergistic activation. The results suggest that sumoylation plays a significant role among the multiple mechanisms that regulate MITF during development and in adulthood.
Keywords: melanocyte, transcription regulation, post-translational regulation, cooperation, Sox10
Introduction
The microphthalmia-associated transcription factor (MITF) is a basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor that is expressed in a number of cell types including melanocytes, mast cells, osteoclasts, and heart and skeletal muscle cells (Hodgkinson et al., 1993; Nakayama et al., 1998; Steingrimsson et al., 1994). MITF plays an important role in the development and function of neural crest-derived melanocytes and optic neuroepithelium-derived retinal pigment epithelium (RPE) cells. Mutations at the mouse Mitf locus lead to coat color dilution, white spotting or complete loss of pigmentation, deafness, reduced eye size, and can be associated with phenotypes in other Mitf-expressing tissues (Arnheiter et al., in press; Hodgkinson et al., 1993). Similarly, mutations in MITF, the human homolog of mouse Mitf, are associated with abnormal pigmentation and deafness as observed in Waardenburg syndrome type IIa and Tietz syndrome (Arnheiter et al., in press; Steingrimsson et al., 2004; Tassabehji et al., 1994; Vance and Goding, 2004).
The MITF protein is composed of several functional domains: amino- and carboxyl (C)-terminal transactivation domains, a basic domain required for DNA binding, and a helix-loop-helix-leucine zipper (HLH-Zip) domain required for dimerization and interaction with a variety of other factors. The basic domain of MITF recognizes Ephrussi (E)-box motifs with a preference for CATGTG, flanked by A or T, and CACATG motifs (Aksan and Goding, 1998). MITF strongly stimulates pigment cell, mast cell, and osteoclast specific promoters (Bentley et al., 1994; Morii et al., 1996; Weilbaecher et al., 2001; Yasumoto et al., 1997).
The activity of MITF is in part regulated by the levels of accumulation of its own mRNA, the availability of interacting factors, and modifications of MITF protein itself. Previous studies showed, for instance, that MITF becomes phosphorylated at multiple distinct serine residues following the activation of specific kinases. Phosphorylation of Ser73 by ERK2 (Hemesath et al., 1998) enables recruitment of the transcriptional activator p300/CBP through the amino terminal transactivation domain of MITF (Price et al., 1998; Sato et al., 1997). Erk2 also activates p90RSK which has been reported to phosphorylate Ser409 of MITF (Wu et al., 2000). Moreover, phosphorylation of Ser73 and Ser409 have been shown to promote proteasome-mediated degradation of MITF following polyubiquitination (Wu et al., 2000; Xu et al., 2000). Other phosphorylation sites include Ser298, thought to be phosphorylated by GSK3b (Takeda et al., 2000) and Ser307, phosphorylated by p38(Mansky et al., 2002). While phosphorylation of MITF is known to modulate MITF activity in cultured cells, however, information on the role these modifications play in living organisms and during melanoma formation is still sparse.
Recently, a number of ubiquitin-like proteins including small ubiquitin-like modifier (SUMO), Nedd8, and Apg12 have been described that become covalently linked to the side chain of lysine residues (Kamitani et al., 1997; Mahajan et al., 1997; Mizushima et al., 1998). SUMO proteins, of which there are three mammalian members (SUMO-1, -2, -3), are structurally homologous to ubiquitin (Lapenta et al., 1997; Qi et al., 1998). Sumoylation occurs by a pathway similar to ubiquitination although the enzymes involved are distinct. SUMO is first activated by a heterodimeric E1 enzyme (SAE I-SAE II, also known as Aos-Uba2) that catalyzes the ATP-dependent formation of an isopeptide linkage between the C-terminus of SUMO and a cystein residue in SAE II. In a transesterification reaction, SUMO is then transferred from SAE II to the E2 SUMO-conjugating enzyme Ubc9. Subsequently, the thioester-linked Ubc9-SUMO conjugate catalyzes the formation of an isopeptide linkage between the C-terminus of SUMO and the e-amino group of a lysine embedded in W-K-X-E consensus sequences in target proteins (W marks hydrophobic residue, X stands for any residue) (Desterro et al., 1999). Recently, the PIAS family of proteins, Ran-BP2, and Polycomb 2(Pc2) have been reported to function as SUMO-E3 ligases that promote the transfer of SUMO from E2 to the substrate protein (Kagey et al., 2003; Kahyo et al., 2001; Sachdev et al., 2001).
A number of proteins, both cytoplasmic and nuclear, have been found to be modified by sumoylation, including Ran-GAP1, PML, IjB, and p53 (Desterro et al., 1998; Kahyo et al., 2001; Mahajan et al., 1997; Muller et al., 1998). Notably, a large number of transcription factors are sumoylated whereby sumoylation usually attenuates transcriptional activity, for instance because of sequestration into distinct subnuclear domains. Recently, it has been recognized, however, that sumoylation can also affect promoter-bound transcriptional complexes. For example, studies of the glucocorticoid receptor (GR) and C/EBPa have identified a 'synergy control' motif whose hallmark is the W-K-X-E sumoylation signature. Interestingly, in GR, for instance, mutations in this motif that prevent sumoylation lead to an increase in the activity of GR from promoters bearing multiple but not single binding sites (Iniguez-Lluhi and Pearce, 2000).
In order to further understand the molecular mechanisms of MITF's transcriptional role, we here explore the modification of MITF with ubiquitin-like proteins through post-translational modifications. In this study, we show that MITF can be modified by SUMO in vitro, is SUMO-modified in melanoma cells, and that non-sumoylatable mutant MITF displays a substantial increase in activity on promoters containing multiple but not single binding sites. This enhanced activity extends to cooperation with heterologous factors such as SOX10.
Results
SUMO modification of MITF
We first examined whether MITF is modified by ubiquitin-like proteins in mammalian cells. As schematically depicted in Figure 1A, Flag-tagged SUMO-1, SUMO-2, Nedd8 and ubiquitin were coexpressed with V5-tagged MITF in HEK293 cells and the modification was studied using immunoprecipitation assays. For these experiments, we used two splice variants of MITF, +6 MITF which contains the six residues ACIFPT located upstream of the basic domain and encoded by exon 6a, and)6 MITF, which lacks this sequence, has a slightly reduced activity, and is the only form present in mice carrying the hypomorphic Mitfmi-sp (microphthalmia-spotted) allele (Hemesath et al., 1994; Steingrimsson et al., 1994). SUMO modification is a dynamic and reversible reaction, and under the standard cell lysis condition, endogenous isopeptidases readily catalyze the release of SUMO from target proteins. Hence, we used a combination of the isopeptidase inhibitors N-ethyl-maleimide (NEM) and iodoacetamide for preparation of cell lysates (Suzuki et al., 1999). When ubiquitin or ubiquitin-like proteins were expressed in HEK293 cells, many proteins became modified by the respective modifiers as indicated from a shift in electrophoretic mobility (Figure 1B, right panel). This suggests that HEK293 cells express all enzymes required for ubiquitination, neddylation, and sumoylation. We then immunoprecipitated whole cell extracts with anti-V5 antibody and analyzed the precipitates by immunoblotting with horseradish peroxidase-labeled anti-V5 antibody. As shown in Figure 1B (left panel), the blots revealed two slower migrating forms of MITF (in this case + 6 MITF), with Mr = 80 000 and 100 000, respectively, after co-expression of Flag-SUMO1 (Mr = approximately 11 500) but not after co-expression of Flag-Nedd8 (Mr = approximately 8000). This result suggested that either one or two SUMO proteins are covalently attached to MITF. To confirm that the slower migrating forms are indeed MITF/SUMO-1 conjugates, we immunoprecipitated extracts with anti-V5 or anti-Flag antibody and prepared immunoblots using either anti-V5 or anti-Flag antibodies. Figure 1C shows that the anti-V5-precipitated, slower migrating forms of +6 MITF were also detected by anti Flag-immunoblotting. Furthermore, two slower migrating forms of similar electrophoretic mobility were also seen after anti-Flag immunoprecipitation followed by anti-V5 immunoblotting. Similar results were obtained with)6 MITF (Figure 1C, right-most panel). The results suggest that the slower migrating forms of MITF were indeed SUMO-1 modified, and that there are at least two sumoylation sites in MITF, regardless of which of the two isoforms were used.
Figure 1.
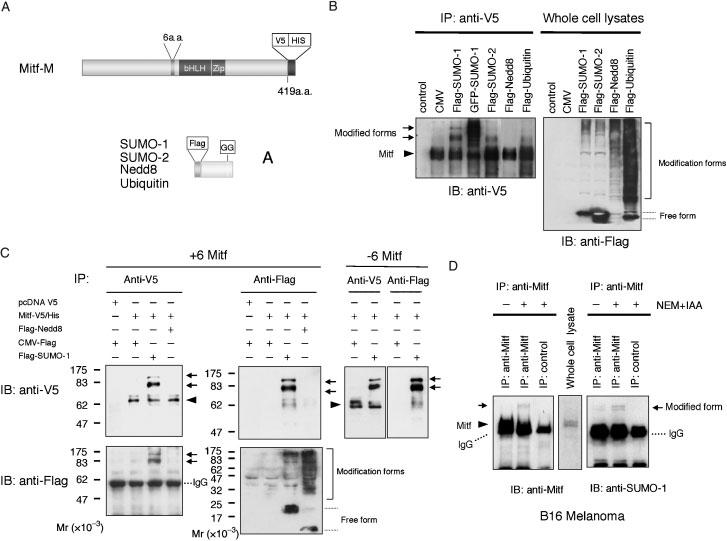
MITF is modified by SUMO in mammalian cells. (A) Schematic representation of V5-His tagged MITF and Flag-tagged ubiquitin and ubiquitin-like proteins used in this study. (B) HEK293 cells were transfected with the plasmid expressing V5-tagged Mitf together with the plasmids expressing Flag-tagged SUMO-1, SUMO-2, Nedd8, ubiquitin and GFP-tagged SUMO-1. Thirty-six hours after transfection, cell lysates were prepared and immunoprecipitated with anti-V5 antibody. The immunoprecipitates were subjected to SDS-PAGE and analyzed by immunoblotting with anti-V5 antibody (left panel). Whole cell lysates were subjected to SDS-PAGE and analyzed by immunoblot with anti-Flag antibody (right panel). The V5-His tagged MITF (several isoforms representing different degrees of phosphorylation as previously described (Wu et al., 2000) is indicated by an arrowhead and its corresponding conjugated forms by arrows. (C) Flag-tagged SUMO-1 or Nedd8 were co-expressed with V5-tagged MITF with or without exon 6a (+6 MITF or)6 MITF) in HEK293 cells. The cell lysates were immunoprecipitated with anti-V5 or anti-Flag antibodies and the immunoprecipitates were subjected to SDS-PAGE and analyzed by immunoblotting with anti-V5 or anti-Flag antibodies. (D) Endogenous MITF is modified by SUMO-1. Whole cell lysates from B16 melanoma cells were prepared either in the presence or absence of isopeptidase inhibitors and then immunoprecipitated with anti-MITF. The precipitates were analyzed by immunoblotting with horseradish peroxidase coupled anti-MITF or anti-SUMO-1 antibodies. Note that in all figures, MITF proteins are indicated by an arrowhead, and their corresponding conjugated forms by arrows.
Next we investigated whether endogenous MITF can be post-translationally modified by SUMO-1. Cell lysates from B16 melanoma cells which express MITF and SUMO-1 endogenously were prepared either in the presence or absence of isopeptidase inhibitors and subjected to immunoblotting using anti-MITF or anti-SUMO-1 antibody. As shown in Figure 1D, there were slower migrating forms which could be detected with both anti-MITF and anti-SUMO-1 antibodies after immunoprecipition with anti-MITF antibody, provided the isopetidase inhibitors were present. In the absence of isopeptidase inhibitors, however, these slower migrating forms were barely visible. The results provide evidence that at least a fraction of endogenous MITF is post-translationally modified by SUMO-1 in melanoma cells.
Lys182 and Lys316 are major sumoylation sites in Mitf
Recent studies have identified a consensus sequence for sumoylation (I/L/V)KXE (Rodriguez et al., 2001). We searched for the presence of consensus SUMO modification motifs in MITF and identified two putative sites at lysine 182 and lysine 316 (Figure 2A). To determine if these lysines were indeed modified by SUMO, we prepared MITF plasmid constructs containing lysine to arginine mutations at position 182 (K182R), 316 (K316R), or both (2KR). Given that +6 and)6 MITF were equally sumoylatable (see Figure 1C), all subsequent experiments were done with +6 MITF, henceforth referred to as MITF. Protein extracts from cells transfected with constructs encoding either V5-tagged wild-type or mutant MITF along with Flag-SUMO-1 were immunoprecipitated with anti-V5 antibody. Western blot analysis using anti-V5 antibody showed that the two slower migrating forms of SUMO-modified wild type MITF (Figure 2B, lane 3) were not detected in precipitates from cells with double mutant MITF (2KR) (Figure 2B, lane 5). Further, we could not detect the slowest form of Mr = 100 000 in precipitates with mutation of K182R or K316R alone (lane 6 or 8). In contrast, mutation of the reported site of ubiquitination, K201, to arginine did not abrogate SUMO-1 modification (lane 7). These experiments suggest that both K182 and K316 are functional sumoylation sites and confirmed the above suggestion that the upper SUMO-modified band corresponds to doubly sumoylated MITF.
Figure 2.
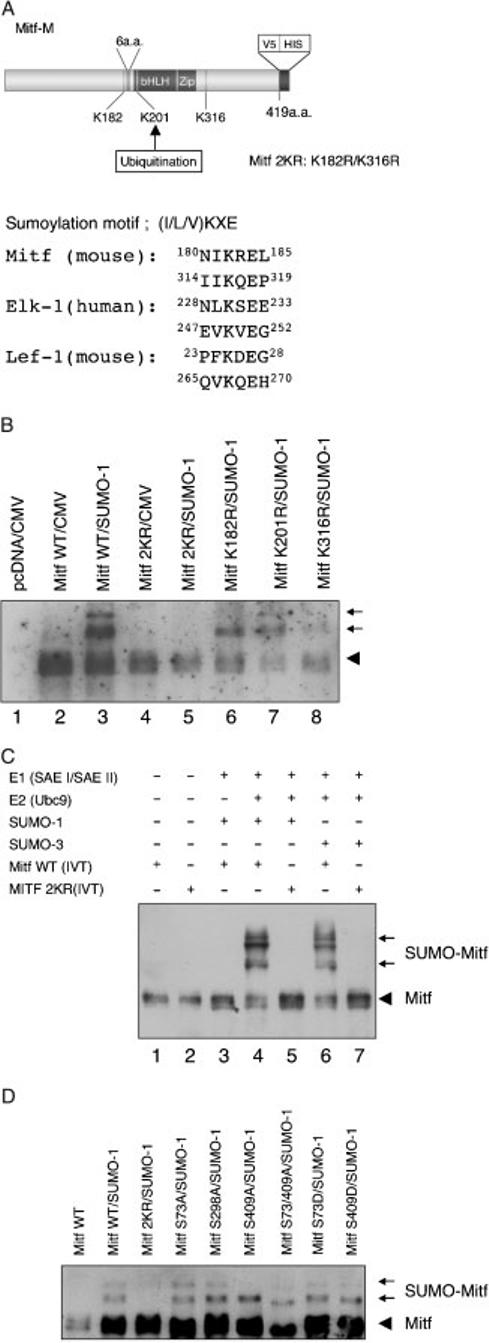
Lysine 182 and lysine 316 are the sites of SUMO-conjugation in MITF in vitro and in vivo. (A) Schematic representation of MITF. Two possible sumoylation sites are present at position 182 and 316. The putative acceptor sites and the alignment of sumoylation motifs in MITF and reference proteins are shown on the right. (B) Protein extracts from cells co-transfected with constructs encoding wild type MITF or MITF with the indicated mutations [K182R, K316R, both K182/316R (2KR) or K201R], and flag-tagged SUMO-1 were immunoprecipitated with anti-V5 antibody. The precipitates were analyzed by immunoblotting with horseradish peroxidase-coupled anti-V5 antibody. The V5-His tagged Mitf is indicated by an arrowhead and its corresponding conjugated forms by arrows. (C) In vitro sumoylation of MITF. In vitro translated wild type and mutant MITF were subjected to in vitro sumoylation using purified recombinant E1 (SAE I/SAE II) enzyme, E2 (Ubc9) enzyme, and recombinant SUMO-1 or SUMO-3 in the presence of ATP. The reaction mixes were subjected to SDS-PAGE and immunoblotted with anti-V5 antibody. Note that in vitro reactions can result in additional MITF forms not detected in transfected cells which may reflect conformational isomers. (D) MITF with mutations in phosphorylation sites are normally sumoylated. The indicated plasmids encoding wild type and mutant MITF were transfected into HEK293 cells along with the SUMO-1 expression plasmid, and cellular extracts subjected to electrophoresis and immunoblotting. Note that only 2KR mutant MITF shows no SUMO-modified forms. Also note that the S73A/409A double mutant, modified or not, shows a faster migration than the other mutants, consistent with previous results (Wu et al., 2000) and independent unpublished observations (Arnheiter et al., in press). Longer exposures (not shown) clearly revealed both mono- and doubly sumoylated forms in wild type MITF and all mutants except 2KR-MITF.
In order to delineate which enzymes are required for MITF sumoylation, we then established an in vitro sumoylation assay using purified recombinant E1 (SAE I/SAE II) and E2 (Ubc9) enzymes and recombinant SUMO-1 or SUMO-3. As substrates, we used in vitro translated V5-tagged wild type MITF or double mutant-MITF (2KR). As shown in Figure 2C, wild type MITF, but not 2KR mutant MITF, was covalently conjugated with both SUMO-1 or SUMO-3 in the presence of E1 enzyme and Ubc9. This result confirms that lysines at position 182 and 316 are major sites for sumoylation, and suggests that sumoylation depends at least on E1 and E2 enzymes.
The fact that in vitro translated MITF can be sumoylated in vitro suggests that other post-translational modifications that occur in vivo, such as phosphorylation and ubiquitination, are not prerequisite for sumoylation. To confirm this notion, we also tested whether MITF mutated at either of three phosphorylation sites can be sumoylated like wild type MITF. For these experiments, we used cells transfected with expression vectors encoding MITFs with the following mutations: S73A, S298A, S409A and S73/409A double mutant. In addition, we tested S73D and S409D, each prepared to mimic the charge of the respective phosphorylated serines. Figure 2D shows that as predicted on the basis of the above in vitro reactions, none of these mutations seemed to interfere with sumoylation, except that in some case, such as with S409A, the doubly sumoylated form became clearly visible only after prolonged exposure (not shown). It remains to be shown, however, whether sumoylated MITF can become phosphorylated at any or all of these sites.
Sumoylation of MITF is not altered by PIAS family members
Besides the SUMO-conjugating E2-enzyme Ubc9, the SUMO E3 ligase PIAS3 was also previously shown to associate with MITF (Levy et al., 2002; Xu et al., 2000). This suggested that PIAS-3 and perhaps other PIAS family members might influence E1/Ubc9-mediated sumoylation. To explore this possibility, we first confirmed that MITF binds these proteins, using an in vitro binding assay. Escherichia coli derived GST-UBC9 was immobilized on glutathione-agarose beads and in vitro translated V5-tagged MITF was then tested for retention and elution from the respective beads. In similar ways, E. coli-derived GST-MITF was first attached to beads and HEK293-expressed Flag-tagged PIAS-1, PIAS-3 or PIAS Xα were then tested. As shown in Figure 3A, MITF is retained, and can be eluted, from GST-Ubc9, and each of the three PIAS proteins is retained, and can be eluted, from GST-MITF.
Figure 3.
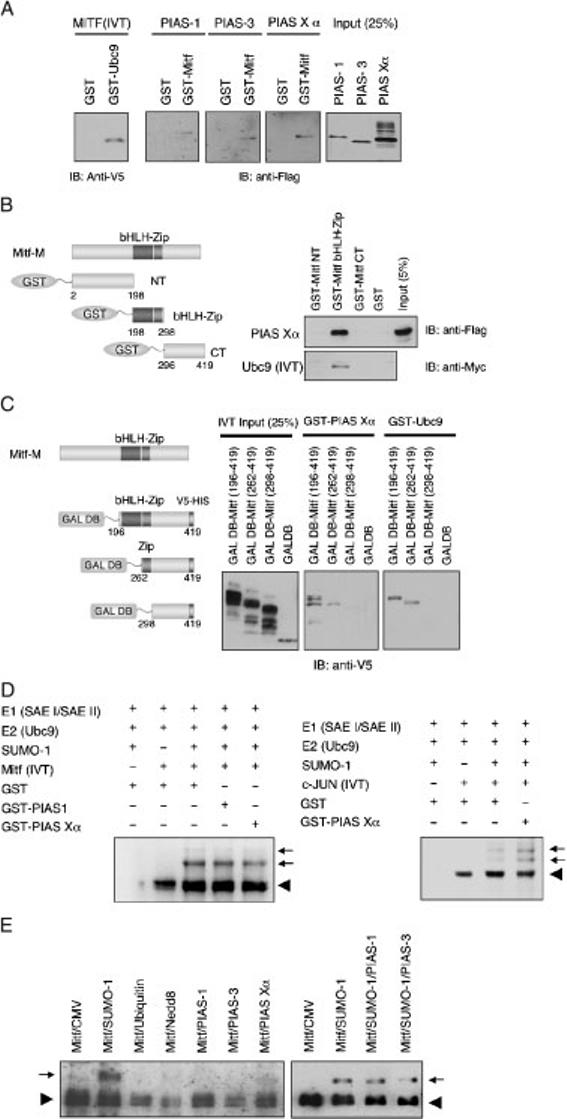
PIAS family members do not alter in vitro MITF sumoylation. (A) Ubc9 and PIAS interact with MITF in vitro. In vitro translated V5-tagged MITF was incubated with GST or GST-Ubc9 fusion protein immobilized on glutathione-agarose beads. Bound proteins were separated on 10% SDS-PAGE and subjected to immunoblotting with anti-V5 antibody (left panel). The lysates from HEK293 cells expressing Flag-tagged PIAS-1, -3, or -Xα were incubated with GST-MITF immobilized on glutathione-agarose beads. Bound proteins were detected with anti-Flag antibody. (B) Schematic representation of GST-MITF fusion proteins used in this study (left panel). In vitro translated myc-tagged Ubc9 or lysates of HEK293 cells expressing Flag-tagged PIAS Xα protein were incubated with the designated GST-MITF fusion proteins that were immobilized on glutathione agarose beads. Bound proteins were detected with anti-Flag or anti-myc antibody respectively. (C) Schematic representation of GAL4 DNA binding domain (GAL DB)-MITF fusion proteins used in this study. In vitro translated GAL DB fusion proteins were incubated with GST-PIAS Xα or GST-Ubc9. Bound proteins were detected by anti-V5 antibody. (D) PIAS does not enhance the sumoylation of MITF in vitro. In vitro translated V5-tagged MITF or V5-tagged c-JUN were subjected to in vitro sumoylation in the presence of GST-PIAS-1, Xα, or GST. The reaction mixtures contain the indicated proteins. Anti-V5 western blotting was used to detect free and SUMO-1 conjugated MITF. (E) PIAS proteins do not alter sumoylation in transfected cells. HEK293 cells were cotransfected with the indicated plasmids and subjected electrophoresis and western blotting as indicated above. Note that neither PIAS-1 nor PIAS-3 altered the extent of sumoylation after co-expression of SUMO-1. Also note that under the conditions employed, the doubly sumoylated MITF form was not detected.
To determine which domain of MITF is required for interaction with Ubc9 or PIAS proteins, we then used various truncated mutants of GST-MITF for binding assays. Figure 3B shows that MITF interacts with Ubc9 and PIAS Xα through the bHLH-Zip domain. We also generated constructs of MITF deletion mutants fused to the GAL4 DNA binding domain (Figure 3C). The proteins were synthesized by in vitro transcription/translation and were mixed with GST-Ubc9 or GST-PIAS Xα on glutathione-agarose beads. GST-Ubc9 and GST-PIAS Xα bound to the deletion mutant which retains the bHLH-Zip or the leucine zipper domain, but not to the C-terminal domain lacking the leucine zipper. These results suggest that Ubc9 and PIAS Xα bind MITF through the leucine zipper domain directly.
Given the interaction between MITF and PIAS family members, we then investigated whether sumoylation of MITF might be stimulated by PIAS Xα or PIAS-1. Figure 3D shows that neither PIAS-1 nor PIAS Xα enhanced in vitro sumoylation of MITF compared with control (GST). Serving as a positive control, sumoylation of c-JUN was enhanced by GST-PIAS Xα (Figure 3D, right panel), consistent with a previous report (Kotaja et al., 2002). To confirm this result in living cells, we also transfected HEK293 cells with the constructs expressing MITF and PIAS family members, with or without the SUMO-1 expression construct. Likewise, under these conditions, we did not observe enhancement of MITF sumoylation by PIAS-1, 3, and Xα (Figure 3E).
Taken together, these results suggest that the presence of E1 enzyme along with Ubc9 is sufficient for MITF sumoylation and that, at least in vitro, PIAS proteins seem to be dispensable for this reaction.
SUMO modification of Mitf does not alter DNA binding
As shown above, there are two sumoylation sites in MITF, one of them at residue 182, i.e. immediately upstream of the basic (DNA-binding) domain. It was conceivable, therefore, that conjugation of SUMO to K182 would modify binding of MITF to DNA. To address this question, electrophoretic mobility shift assays (EMSAs) with wild type and mutant MITF were performed, using as probe a double-stranded oligonucleotide with a central E-box. Purified GST-MITF was subjected to in vitro sumoylation under conditions assuring a high degree of sumoylation, and the reaction was subsequently used for EMSA.
The sumoylation status of MITF used in the reaction was verified by western blot analysis (Figure 4A). EMSA showed similar DNA binding abilities between wild type MITF, double mutant MITF, and SUMO1-conjugated wild type MITF. As would be predicted from the increased molecular weight of SUMO-MITF, SUMO-MITF/DNA complexes migrated slightly slower than their non-sumoylated counterparts (Figure 4B). These results suggest that SUMO modification of MITF does not significantly alter its DNA binding ability although we have not formally ruled out that only monosumoylated MITF (or heterodimers between mono- and doubly sumoylated MITF) bind DNA normally.
Figure 4.
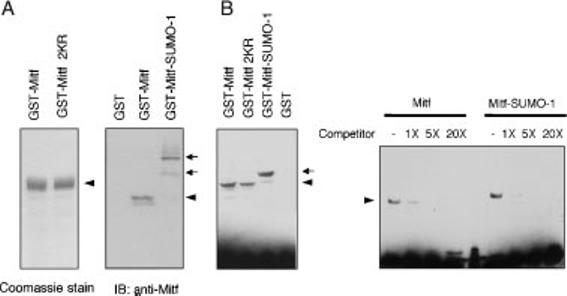
Sumoylation does not affect DNA binding of MITF. (A) The purified GST-wild type MITF or mutant MITF (K182R/K316R) were subjected to SDS-PAGE and the proteins were detected by Coomassie stain (left panel). GST-MITF fusion protein was subjected to in vitro sumoylation in the presence or absence of Ubc9. The reaction mixtures were subjected to SDS-PAGE and immunoblotted with anti-MITF antibody (right panel). (B) The reaction mixtures were subsequently analyzed by EMSA. For competition assays, all DNA-binding reactions contained 4 fmol Biotin-labeled double-stranded oligonucleotide and various amounts of unlabeled double-stranded oligonucleotide.
In results not shown, we also tested whether the sumoylation mutants accumulated normally in the nuclei of transfected cells. In fact, no change in nuclear distribution was seen with any of the single or double lysine 182/316 mutant forms of MITF. This was not unexpected, however, as only a minor fraction of MITF is normally sumoylated in living cells, even in the presence of isopeptidase inhibitors (see Figure 1D).
SUMO modulates the transcriptional activity of Mitf
Although sumoylation did not affect DNA binding of MITF, it was possible that it still altered its transcriptional activity. To address this question, we compared the activity of wild type MITF with that of single or double mutant MITF, using a luciferase reporter assay. The first reporter plasmid employed contained four copies of an M-Box appended to a minimal simian virus 40 (SV40) promoter. B16 melanoma cells and NIH3T3 cells were cotransfected with this reporter plasmid and either empty vector or vectors encoding wild type or mutant MITF. Cell extracts were then analyzed by a luciferase assay. The results (Figure 5A) showed that wild type MITF as well as K182R, K201R and K316R mutant MITF activated the reporter approximately 2-3-fold above background. In contrast, K182/316R doubly mutant MITF showed activations of 6-8-fold, suggesting that prevention of sumoylation leads to an increased transcriptional activity on this artificial promoter.
Figure 5.
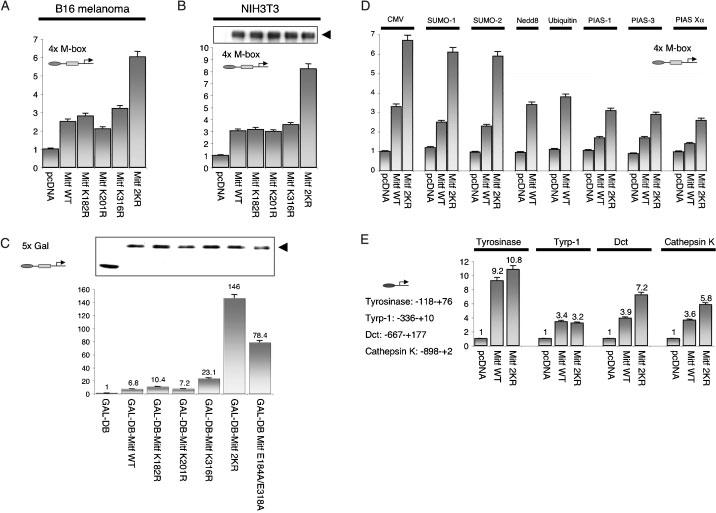
Effect of SUMO modification on transcriptional activity of MITF. (A) B16 melanoma cells; or (B) NIH3T3 cells were transiently transfected with the 4X M-box-luciferase reporter construct and phRL-TK carrying Renilla luciferase gene together with the expression plasmids carrying wild type MITF, MITF-K182R, MITF-K201R, MITF-K316R, MITF-K182R/K316R or control (empty) plasmid. Gel electrophoresis and MITF immunoblottting indicated that similar amounts of MITF were expressed in NIH3T3 cells [upper panel in (B), arrow head marking MITF protein]. Luciferase activity was measured 24 h after transfection and normalized using the activities of the Renilla luciferase. Results represent averages from at least three independent experiments. (C) NIH3T3 cells were transfected with pFR-luc and phRL-TK together with pcDNAGAL4 DB-wild type MITF, mutants, or pcDNAGAL4DB and luciferase activities were analyzed as in (A). The upper panel shows the expression levels of GAL4 DNA binding domain and fusion proteins, as revealed by immunoblotting. (D) NIH3T3 cells were cotransfected with the 4X M-box-luciferase reporter construct, phRL-TK, and the plasmids expressing wild-type MITF or mutant together with the plasmids expressing SUMO-1, SUMO-2, Nedd8, ubiquitin, PIAS family members, or with the empty plasmid (pcDNA), and luciferase activity was measured. (D) NIH3T3 cells were co-transfected with pGL-tyrosinase-, tyrp-1, Dct, or cathepsin K promoter as well as phRL-Tk and the plasmids expressing wild-type or mutant MITF, or empty plasmid (pcDNA).
Mutant MITF protein was then compared with wild type MITF as GAL4 fusion proteins to allow interaction through a heterologous DNA-binding motif. As shown in Figure 5B, wild type GAL4-MITF, GAL4-MITF(K182R) and GAL4-MITF(K201R) activated a reporter plasmid containing five GAL4 binding sites 6.8-10.4-fold. GAL4-MITF(K316R) activated this reporter 23.1-fold, and GAL4-MITF(2KR) 146-fold. In addition, GAL4-MITF(E184A/E318A), in which alanines were substituted for the glutamic acid residues in the respective sumoylation motifs, activated the reporter 78.4-fold. This latter mutant shows a substantially decreased SUMO modification (data not shown). Thus, the effect of the double sumoylation mutation seen on E-box containing promoters extends to heterologous DNA binding motifs and hence is not restricted to E-boxes.
Given the increased activity of the non-(or hypo) sumoylatable mutants, we then tested whether forced increase in wild type MITF modification by overexpression of SUMO, Nedd8, ubiquitin, PIAS-1, -3, Xα, would be consistent with the observations made with mutant MITF. To this end, wild type MITF, and, to control for effects not dependent on direct MITF sumoylation, its 2KR mutant, were expressed along with the 4xM-box reporter and appropriate expression vectors for the above modifiers. As shown in Figure 5B, the transcriptional activity of wild type MITF was decreased when SUMO-1 or SUMO-2 were coexpressed. Although this decrease was relatively weak (30-35%), it was still stronger than the relative repression seen on the 2KR mutant MITF where it was only approximately 15%. This suggested that at least part of the repression induced by SUMO-overexpression is mediated through the sumoylation of MITF. Interestingly, overexpression of Nedd8 or ubiquitin had no effect on the transcriptional activity of wild type MITF and so was not further tested on mutant MITF. Overexpression of PIAS family members also repressed the transcriptional activity of MITF but to a comparable degree, regardless of whether wild type or 2KR mutant MITF was used (approximately 50% in both cases). The results suggested that forced increase of sumoylation of wild type MITF represses the protein's activity. PIAS proteins, although repressing MITF's activity as well, apparently do so independent of K182/316 sumoylation and likely act through the modification of other proteins. This latter observation is consistent with the above notion that PIAS proteins have no direct stimulatory effect on MITF sumoylation (see Figure 3D).
All reporter plasmids used above were artificial constructs containing multiple MITF or GAL4 binding sites. To test whether the effect of MITF sumoylation can be extended to more natural promoters, we then used several reporter constructs containing the indicated promoter regions of the MITF-responsive genes tyrosinase, tyrp-1, Dct, and cathepsin K. Interestingly, compared to wild type, 2KR mutant MITF displayed an increased transcriptional activity on the Dct and cathepsin K promoters, but modest or no changes on the tyrosinase and tyrp-1 promoters. Intriguingly, increased activity was associated with higher numbers of MITF binding sites. The respective promoter fragment of tyrp-1 contained one, that of tyrosinase two, that of Dct three, and that of cathepsin K four E boxes (of these latter four, three are positively regulated by MITF and one negatively) (Motyckova et al., 2001). Thus, the enhanced transcriptional stimulation of promoters by 2KR mutant MITF depended on the promoter context and was positively correlated with the number of MITF binding sites.
Sumoylation regulates synergistic transcriptional activity of MITF
It thus appeared that sumoylation affected the transcriptional activity of MITF in a manner dependent on the number of binding sites in a promoter. This suggested that sumoylation may in fact regulate the cooperation of individual MITF molecules on promoter DNA. Interestingly, recent studies described a synergy control (SC) motif in the GR and other transcription factors that regulates synergistic transcriptional activity. The SC consensus motif contains a sumoylation sequence (W-K-X-E) as well as the presence of a proline residue within 1-4 residues flanking either side (or both) of the core sumoylation sequence (Iniguez-Lluhi and Pearce, 2000). In fact, proline residues found around the K182 and K316 sumoylation motifs in MITF (see Figure 7) suggest that these motifs may act as SC motifs. Thus, we used reporter genes containing one, three or five GAL4 binding sites (schematically shown in Figure 6A) and tested them in cotransfection assays with expression vectors for GAL4-MITF (wild type) or GAL4-MITF(2KR). As shown in Figure 6B, C, the 2KR mutant showed a substantially enhanced transcriptional activity only from the 3X GAL4 and 5X GAL4 reporters and not from the 1X GAL4 reporter. This result is consistent with the above observation that the effect of sumoylation-mutations are more pronounced on promoters with multiple E boxes and suggests that MITF sumoylation regulates cooperative activation on reporter DNA.
Figure 7.
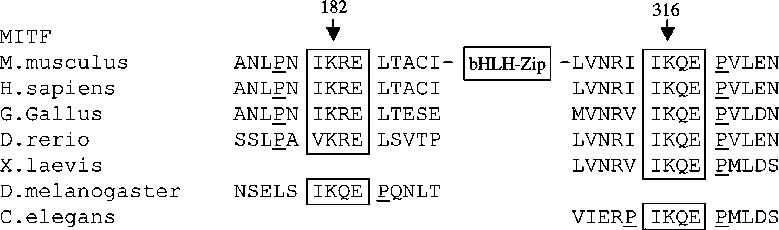
Sumoylation sites and neighboring proline residues suggesting their function as ‘synergy control’ motifs are conserved throughout evolution. Note that D. melanogaster (Hallsson et al., 2004) and C. elegans (Rehli et al., 1999) each have only one gene most closely related to MITF while D. rerio and X. laevis each have two and G. gallus, M. musculus and H. sapiens each have one MITF gene. In addition, vertebrates have several more distantly MITF-related genes (Tfe genes) that were not analyzed in this study. For each species, only one exemplary MITF (or closely MITF-related) sequence is shown.
Figure 6.
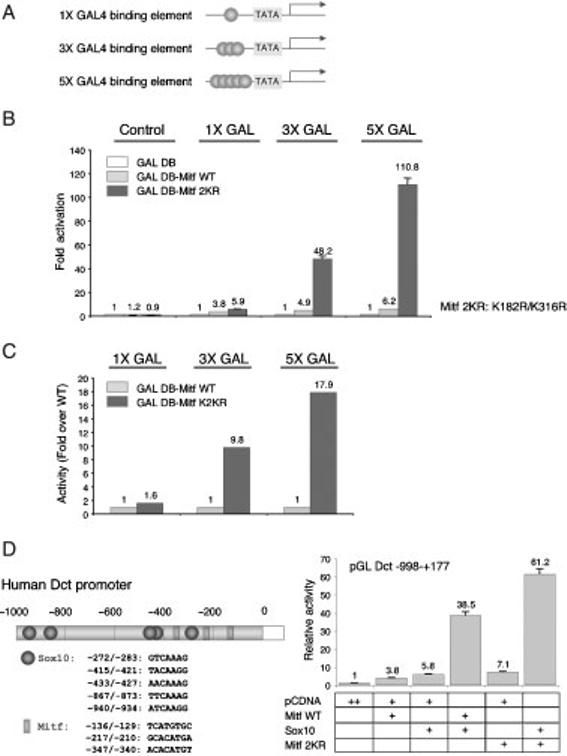
Synergistic transcriptional activities are enhanced by sumoylation site mutations of MITF. (A) Schematic illustration of GAL4-responsive reporter genes used in this study. (B) Expression plasmids for GAL4 DNA binding domain fused with the wild type and 2KR mutant of MITF were transfected into NIH3T3 cells with a luciferase reporter gene containing one (1X GAL), three (3X GAL) or five (5X GAL) GAL4 binding sites upstream of the TATA box and phRL. (C) Ratio of mutant over WT activity measured in Figure 5B. (D) Schematic representation of promoter fragment of the human Dct gene. Effects of the 2KR mutation on the synergistic transcription between MITF and SOX10. Expression plasmids for wild type or 2KR mutant MITF, wild type or SOX10, a luciferase reporter gene containing the DCT promoter, and phRL-TK were cotransfected into NIH3T3 cells. Luciferase activity was measured as described for Figure 5.
It is well established that promoters are efficiently activated by the cooperation of multiple distinct transcription factors. To further test whether the effect of MITF sumoylation on transcription factor cooperation would extend to other factors, we then analyzed the Dct promoter in the context of its activation by both MITF and SOX10. The chosen human Dct promoter fragment contains five potential SOX10 binding sites, in addition to its three MITF sites, and synergisms between MITF and SOX10 have been reported (Jiao et al., 2004; Ludwig et al., 2004; Yasumoto et al., 2002). As shown in Figure 6D, wild type MITF activated the promoter 3.8-fold, SOX10 5.8-fold, and MITF and SOX10 together 38.5-fold, confirming previous results (Ludwig et al., 2004). The 2KR mutant MITF, while displaying an intrinsic 7.1-fold acivation of the reporter, cooperated with SOX10 to give a 61.5-fold activation. These results are consistent with the notion that MITF sumoylation not only affects MITF/MITF cooperation, but also MITF/SOX10 cooperation.
Discussion
In this study, we present a series of experiments that show that the melanocyte transcription factor MITF can be covalently modified by SUMO both in vitro and in living cells, and that a fraction of MITF, although a minor one, is constitutively SUMO-modified in melanoma cells. The analysis of mutants whose sumoylation is reduced or totally abolished allows us to conclude that sumoylation has a functional consequence - the reduction of transcriptional activity on some but not other MITF-responsive promoters. In fact, MITF's activity is modulated by sumoylation on promoters with multiple MITF binding sites but not on promoters with single binding sites. This suggests that MITF's transcriptional output is subtly regulated by the extent of sumoylation. It will be important in the future, therefore, to determine whether sumoylation is physiologically regulated in vivo, for instance in a cell type- and spatio-temporal manner during development, in processes that lead to cellular pathology including vitiligo or melanoma formation, or during normal cellular turn-over as it occurs with melanocytes during hair regeneration and physiological aging.
MITF can be sumoylated at two distinct lysine residues, K182 and K316, while lysine 201, reported to be a site for ubiquitination (Xu et al., 2000), and other lysines are not targets of sumoylation. In vitro reactions indicate that sumoylation of MITF critically depends on the E1 activating enzyme dimer SAEI/SAEII and the E2 conjugating enzyme Ubc9, both of which previously implicated in sumoylation. In fact, in contrast to ubiquitination, which can be catalized by a large number of different E2 enzymes, sumoylation only utilizes Ubc9 which itself is incapable of forming thioester conjugates with ubiquitine (Desterro et al., 1997; Gong et al., 1997). In fact we found that Ubc9 directly interacts with MITF, thus confirming earlier observations (Xu et al., 2000), and delineate this interaction to occur via MITF's leucine zipper domain. In contrast to our results, however, Xu et al. (2000) described that Ubc9 serves as a ubiquitin and not a SUMO conjugating enzyme and that MITF/Ubc9 interactions lead to MITF degradation following ubiquitination. Additional studies will be needed to provide an explanation for this discrepancy.
Since previous results have shown that PIAS-3, an E3 ligase, binds MITF (Levy et al., 2002, 2003), we also tested whether MITF sumoylation is modulated by PIAS proteins. E3 ligases normally promote the transfer of SUMO from Ubc9 to substrate proteins. Since MITF interacts with Ubc9 directly, however, there may not be an absolute requirement for E3 ligases, similar to previous observations with other proteins (Mao et al., 2000; Shiio and Eisenman, 2003). In fact, we found that in vitro MITF sumoylation was not stimulated by PIAS family members, notwithstanding the facts that the respective PIAS proteins interacted with MITF and that overexpression of PIAS family members suppressed MITF activity as described for PIAS-3 (Levy et al., 2003). In support of the notion that PIAS-mediated suppression of MITF activity may not act through MITF sumoylation, we also observed that a similar suppressive PIAS effect was seen with the non-sumoylatable 2KR mutant MITF. Furthermore, we found that MITF with a mutation of serine 409 → aspartate is normally sumoylatable (see Figure 2D). This mutant was reported not to bind PIAS-3, and not to be inhibited by PIAS-3 (Levy et al., 2003). In similar ways, PIASy-mediated repression of transcriptional activity of the androgen receptor (AR) does also not require sumoylation (Gross et al., 2004). We conclude, therefore, that MITF sumoylation- and PIAS-mediated suppression of MITF activity are parallel but independent events.
One of the most interesting part of our study concerns the fact that MITF sumoylation has differential effects on distinct MITF-responsive promoters. The 2KR mutant MITF showed enhanced transcriptional stimulation of reporter plasmids containing multiple E box binding sites, such as the Dct promoter or the cathepsin K promoter, while no or only modest enhancement was seen with promoters containing just one (tyrp-1) or two (tyrosinase) E boxes. Further analyses indicated that enhanced stimulation extended to the cooperation with heterologous transcription factors, such as seen with respect to the synergistic interaction between MITF and SOX10 on the Dct promoter. Likewise, cooperation was observed when GAL4-MITF proteins were allowed to interact through heterologous GAL4 binding sites on artificial promoters, indicating that MITF/E-box interactions are not prerequisite to reveal the effect of sumoylation. Similarly, a poorly sumoylatable mutant of the GR showed increased transcriptional activity on some target promoters but not others, depending on the number of functional binding sites (Poukka et al., 2000; Tian et al., 2002). These and other data indicate that SUMO modification may specifically affect the ability of transcription factors to function synergistically.
Support for a SUMO-mediated modulation of transcription factors acting synergistically comes from recent studies that have identified a negatively acting protein domain referred to as the 'synergy control (SC) motif'. This motif is defined by a common core sequence, I/L-K-X-E, reminiscent of the sumoylation signature, and the presence of proline residues within one to four positions from either or both ends of the core (Iniguez-Lluhi and Pearce, 2000). Synergy control motifs are present in a number of transcription factors and co-factors and were shown to be modified by SUMO (Holmstrom et al., 2003). Interestingly, both sumoylation sites of MITF also contain the tell-tale proline residues within three aminoacids from either end of the core sumoylation sequence, and this feature seems to be conserved throughout the evolution of MITF from C. elegans to man (Figure 7). The fact that non-sumoylatable mouse MITF with mutations at lysines 182 and 316 shows enhanced transcriptional stimulation of promoters with compound but not with single binding sites strongly suggests that these MITF motifs function as SC motifs.
How might sumoylation modulate MITF's cooperative transcriptional activity? Although it has been observed with some factors that sumoylation can alter cytoplasmic/nuclear transport or subnuclear localization, we have no evidence for such mechanisms regulating MITF, as immunofluorescent assays showed no alterations in nuclear localization between wild type and 2KR mutant MITF, or between wild type MITF in cells with normal and that in cells with artificially high levels of SUMO-1. Moreover, sumoylation did also not appear to disrupt DNA binding of MITF, nor was there evidence for a change in protein stability that might have explained a suppression of transcriptional activity. More likely, therefore, is the possibility that MITF sumoylation has effects on the recruitment of transcriptional co-factors, along the line of observations made with the Elk-1 transcription factor which, when sumoylated, preferentially recruits the histone deacetylase HDAC2 (Yang and Sharrocks, 2004). HDAC2 recruitment leads to reduced histone acetylation and hence transcriptional repression. It is conceivable that if sumoylation of MITF indeed plays a role in the recruitment of co-repressors, such recruitment may be enhanced on complex promoters containing multiple binding sites compared to those with single binding sites. Nevertheless, we recognize that sumoylation is a subtle regulator, but we anticipate that it is precisely the subtlety of distinct promoter regulations that may finally explain how MITF exerts its multiple developmental and adult functions in such a variety of cell types.
Materials and methods
Plasmid constructs
Wild-type mouse melanocyte (M)-Mitf cDNA, containing or lacking the alternatively spliced 18 bp sequence encoding ACIFPT, was introduced into pcDNA3.1. V5/HIS expression plasmid (Invitrogen, Carlsbad, CA, USA) or pcDNA3.1 GAL DB-V5/HIS expression plasmids were as described previously (Shimono et al., 2000). Mouse SUMO-1, SUMO-2, Nedd8, ubiquitin, Ubc9, PIAS-3, human PIAS-1 and PIAS Xα were amplified by PCR and introduced into the pFLAG-CMV2 expression vector (Sigma, St Louis, MO, USA) or pcDNA3.1 myc/HIS (Invitrogen), thereby fusing these cDNAs with the FLAG sequence or myc/HIS sequence. All Mitf mutations were introduced into double-stranded DNA by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the instructions of the manufacturer. The sequences of all constructs were confirmed. A CMV-SOX10 expression plasmid was provided by William Pavan, and Tyrosinase, Tyrp-1 and Dct promoter fragments were from Vincent Hearing. The 1xGal4 and 3xGal4 reporter plasmids were constructed by inserting double-stranded oligonucleotides in pGL3 vector (Promega, Madison, WI, USA). The inserts are as follows: 1xGal4: ctcgagCGGAGTACTGTCCTCCGaggggagactcagaggTATA TAatggaattccccatccagctt; 3xGal4: ctcgagCGGAGTACTGTCCTCC GagCGGAGTACTGTCCTCCGagCGGAGTACTGTCCTCCGagcggagactc tagaggTATATAatggaattccccatccagctt; 5xGal4 (pFR-Luc) was from Stratagene.
Cell culture and transfection
HEK293 cells were cultured in RPMI supplemented with 10% fetal calf serum. NIH3T3 mouse fibroblast and B16 melanoma cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS. Cells were transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Antibodies
Anti-Flag monoclonal antibody (M2) was purchased from Sigma and horseradish peroxidase-coupled, or uncoupled, anti-V5 antibody and anti-myc antibody were from Invitrogen. Anti-MITF (C5) monoclonal antibody was from Lab Vision Corp. (Fremount, CA, USA), and anti-sentrin monoclonal antibody was from Zymed (San Francisco, CA, USA).
Immunoprecipitation and immunoblotting
Cells were grown to subconfluency in 35-mm dishes. They were then lysed in lysis buffer which consisted of 20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; 1 mM sodium orthovanadate; 20 mM N-ethylmaleimide (Pierce); 200 lM iodoacetamide (Sigma) supplemented with Halt™ protease inhibitor cocktail (Pierce, Rockford, IL, USA) and 1 mM phenylmethylsulfonyl fluoride (PMSF). The lysates were clarified by centrifugation (15 000 x g) for 30 min, and the supernatants were incubated with antibodies for 3 h at 4°C. The resulting immunocomplexes were collected with Protein G-Sepharose (Invitrogen) and washed four times with lysis buffer. The complexes were eluted in sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl, pH 6.8; 5 mM EDTA; 2% SDS; 10% glycerol; 1 mg/ml bromphenol blue; 80 mM dithiothreitol) by boiling for 10 min and subjected to SDS-polyacrylamide gel electrophoresis. Separated proteins were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) and reacted with the respective antibodies. The reaction was examined by enhanced chemiluminescence detection kit (Pierce) according to the instructions of the manufacturer.
Pull-down assay using glutathione S-transferase fusion proteins
To generate glutathione S-transferase (GST) fusion proteins with each domain of MITF, cDNA fragments of MITF, Ubc9, PIAS-1, and PIAS Xα were amplified by PCR and cloned into the pGEX-KG vector. These constructs were used to transform B21 E. coli (Amersham, Piscataway, NJ, USA) to produce GST fusion proteins that were purified using glutathione-agarose beads as described previously (Murakami et al., 2002).
The lysates of HEK293 cells expressing PIAS family members or in vitro translated proteins were incubated with 2 lg of immobilized GST or GST fusion proteins for 3 h at 4°C and washed with lysis buffer four times. The proteins bound to GST fusion proteins were eluted by boiling in SDS-sample buffer, separated by SDS-polyacrylamide gel electrophoresis, and immunoblotted with anti-V5 anti-FLAG or anti-myc antibody.
In vitro translated proteins were synthesized using the TnT quick coupled transcription/translation system with T7 RNA polymerase according to the manufacturer's instructions (Promega).
In vitro sumoylation assay
In vitro sumoylation was performed in a 20 μl reaction mixture containing 20 mM Hepes (pH7.5), 2 μl of in vitro translated MITF (either wild type or K182/316R), 5 mM MgCl2, 2 mM ATP, 500 ng SUMO-1 or SUMO-3, 50 ng human SAEI/SAEII, 200 ng Ubc9 and 500 ng GST fusion proteins. Purified SUMO-1, SUMO-3, SAE-I/SAE-II and Ubc9 were purchased from LAE lnternational (Rockville, MD, USA). After incubation for 1 h at 30°C, the reaction mixture was subjected to SDS-PAGE and immunoblotting with an anti-V5 antibody.
Electrophoretic gel mobility shift assays
About 1 μg of purified GST-Mitf protein was subjected to in vitro sumoylation assay. Reaction mixtures were incubated for 3 h at 30°C. Subsequently, electrophoretic gel mobility shift assays (EMSAs) were performed using LightShift™ Chemiluminescent EMSA kit (Pierce) that uses a non-isotopic method to detect DNA-protein interactions according to the manufacture's instructions. Oligonucleotides, 5′-CCTTGTGGAGATCATGTGACTTCCTGATTC-3 and 5′-GAATCAGGAAGTCACAATGATCTCCACAAGG-3′ containing an E-box binding site for MITF were labeled with biotin for chemiluminescence using a biotin 3′ end DNA labeling kit (Pierce). After labeling, the oligonucleotides were annealed for 1 h at 37°C to form the double-stranded probe. Gel mobility shift assays were performed by incubating GST-MITF protein along with labeled and unlabeled competing oligonucleotides in binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 5 mM MgCl2, 50 mM KCl and 0.2 lg salmon sperm DNA) at room temperature for 30 min. Mixtures were size-fractionated on a non-denaturing 4% polyacrylamide gel followed by transfer to nitrocellulose membranes and detection by streptavidin-HRP/chemiluminescence for biotin-labeled probes.
Luciferase assay
For reporter-gene assays, NIH3T3, B16 melanoma, or HEK293 cells were cultured in 24-well tissue culture plates for 20-24 h prior to transfection. The cells were then co-transfected with the following plasmids: (1) 400 ng of wild-type or mutant MITF expression constructs; (2) 400 ng of one of the following: 4X M-box, tyrosinase, tyrp-1, Dct, or cathepsin K reporter plasmid; and (3) 25 ng of phRL-TK plasmid carrying the Renilla luciferase gene (Promega). The cells were cultured for 24 h after transfection and then lysed. The resulting extracts were assayed for the luciferase activity using the Dual-Luciferase reporter Assay System (Promega). To investigate GAL4 fusion protein mediated gene expression in NIH3T3 cells, cells were transfected with 400 ng luciferase reporter plasmid containing GAL4 DNA binding elements, 25 ng of phRL-TK plasmid, 400 ng of pcDNA-GAL4 DNA binding domain-MITF fusion constructs (pcDNA-GAL4DB-MITF). The cells were harvested 24 h after transfection and assayed for luciferase activity.
To investigate the effect of expression of ubiquitin-like proteins or PIAS family members on luciferase activity, NIH3T3 cells were co-transfected with 300 ng of the 4X M-box containing reporter plasmid, 25 ng of phRL-TK, and 300 ng of MITF expression constructs and 300 ng constructs expressing ubiquitin-like proteins or PIAS family members. The total amounts of transfected DNAs were kept constant by including the appropriate empty vector where needed. Twenty-four hours after transfection, luciferase assays were performed. All experiments were done in duplicates or triplicates, were repeated three times, and calculated as fold stimulation ± SD.
Acknowledgements
We thank Drs Vincent Hearing and William Pavan for promoter and expression constructs, and the NINDS sequencing facility for excellent services.
Footnotes
Note added in proofRecently, an independent study reaching similar conclusions was published by Miller et al. (A. J. Miller, C. Levy, I. J. Davis, E. Razin, D. E. Fisher (2005) J. Biol. Chem. 280, 146-155).
References
- Aksan I, Goding CR. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheiter H, Hou L, Nguyen M-TT, Bismuth K, Csermely T, Murakami H, Skuntz S, Liu W, Bharti K.Mitf—a matter of life and death for the developing melanocyte Melanocytes to Melanoma: The Progression to Malignancy Humana Press; Totowa, NJ: in pressin press [Google Scholar]
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol. Cell. Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IjBa inhibits NF-jB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- Gross M, Yang R, Top I, Gasper C, Shuai K. PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene. 2004;23:3059–3066. doi: 10.1038/sj.onc.1207443. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Stivers C, Odenwald W, Arnheiter H, Pignoni F, Steingrimsson E. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics. 2004;167:233–241. doi: 10.1534/genetics.167.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, Mcgill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Holmstrom S, VanAntwerp ME, Iniguez-Lluhi JA. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl Acad. Sci. U. S. A. 2003;100:15758–15763. doi: 10.1073/pnas.2136933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez-Lluhi JA, Pearce D. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 2000;20:6040–6050. doi: 10.1128/mcb.20.16.6040-6050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Z, Mollaaghababa R, Pavan WJ, Antonellis A, Green ED, Hornyak TJ. Direct interaction of Sox10 with the promoter of murine Dopachrome Tautomerase (Dct) and synergistic activation of Dct expression with Mitf. Pigment Cell Res. 2004;17:352–362. doi: 10.1111/j.1600-0749.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapenta V, Chiurazzi P, VanDerSpek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21 qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- Levy C, Nechushtan H, Razin E. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 2002;277:1962–1966. doi: 10.1074/jbc.M109236200. [DOI] [PubMed] [Google Scholar]
- Levy C, Sonnenblick A, Razin E. Role played by microphthalmia transcription factor phosphorylation and its Zip domain in its transcriptional inhibition by PIAS3. Mol. Cell. Biol. 2003;23:9073–9080. doi: 10.1128/MCB.23.24.9073-9080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Rehberg S, Wegner M. Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett. 2004;556:236–244. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J. Biol. Chem. 2002;277:11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl Acad. Sci. U. S. A. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Morii E, Tsujimura T, Jippo T, Hashimoto K, Takebayashi K, Tsujino K, Nomura S, Yamamoto M, Kitamura Y. Regulation of mouse mast cell protease 6 gene expression by transcription factor encoded by the mi locus. Blood. 1996;88:2488–2494. [PubMed] [Google Scholar]
- Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, Fisher DE. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc. Natl Acad. Sci. U. S. A. 2001;98:5798–5803. doi: 10.1073/pnas.091479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. Embo. J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Yamamura Y, Shimono Y, Kawai K, Kurokawa K, Takahashi M. Role of Dok1 in cell signaling mediated by RET tyrosine kinase. J. Biol. Chem. 2002;277:32781–32790. doi: 10.1074/jbc.M202336200. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev. 1998;70:155–166. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl Acad. Sci. U. S. A. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, Hemesath TJ, Fisher DE. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J. Biol. Chem. 1998;273:17983–17986. doi: 10.1074/jbc.273.29.17983. [DOI] [PubMed] [Google Scholar]
- Qi F, Ridpath JF, Berry ES. Insertion of a bovine SMT3B gene in NS4B and duplication of NS3 in a bovine viral diarrhea virus genome correlate with the cytopathogenicity of the virus. Virus Res. 1998;57:1–9. doi: 10.1016/s0168-1702(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Rehli M, DenElzen N, Cassady AI, Ostrowski MC, Hume DA. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999;56:111–120. doi: 10.1006/geno.1998.5588. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding CR. CBP/p300 as a co-factor for the Microphthalmia transcription factor. Oncogene. 1997;14:3083–3092. doi: 10.1038/sj.onc.1201298. [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. U. S. A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Murakami H, Hasegawa Y, Takahashi M. RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J. Biol. Chem. 2000;275:39411–39419. doi: 10.1074/jbc.M006585200. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ichiyama A, Saitoh H, Kawakami T, Omata M, Chung CH, Kimura M, Shimbara N, Tanaka K. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J. Biol. Chem. 1999;274:31131–31134. doi: 10.1074/jbc.274.44.31131. [DOI] [PubMed] [Google Scholar]
- Takeda K, Takemoto C, Kobayashi I, Watanabe A, Nobukuni Y, Fisher DE, Tachibana M. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Hum. Mol. Genet. 2000;9:125–132. doi: 10.1093/hmg/9.1.125. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- Tian S, Poukka H, Palvimo JJ, Janne OA. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002;367:907–911. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004;17:318–325. doi: 10.1111/j.1600-0749.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Motyckova G, Huber WE, Takemoto CM, Hemesath TJ, Xu Y, Hershey CL, Dowland NR, Wells AG, Fisher DE. Linkage of M-CSF signaling to Mitf, TFE3, and the osteoclast defect in Mitf(mi/mi) mice. Mol. Cell. 2001;8:749–758. doi: 10.1016/s1097-2765(01)00360-4. [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzymeh UBC9. Exp. Cell. Res. 2000;255:135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Takeda K, Saito H, Watanabe K, Takahashi K, Shibahara S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. Embo. J. 2002;21:2703–2714. doi: 10.1093/emboj/21.11.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]


