Abstract
Biofilm formation was repressed by glucose in several species of Enterobacteriaceae. In Escherichia coli, this effect was mediated at least in part by cyclic AMP (cAMP)-cAMP receptor protein. A temporal role for cAMP in biofilm development was indicated by the finding that glucose addition after ∼24 h failed to repress and generally activated biofilm formation.
In the natural environment, bacteria predominantly exist in matrix-enclosed, sessile communities referred to as biofilms (4). Biofilms protect cells from deleterious conditions, such as attack by the mammalian immune system (5). Biofilms are complex assemblages of cells which exhibit channels and pillars that are thought to permit the exchange of nutrients and wastes. A recent model for biofilm development proposes that it is initiated by the attachment of individual cells to a surface, followed by their migration and replication to form microcolonies that eventually produce the mature biofilm (20, 22). A variety of extracellular molecules and surface organelles participate in E. coli biofilm formation (6, 7, 23, 33).
Central carbon flux and its regulation may represent key features of bacterial biofilm development. We recently reported that the RNA binding protein CsrA of Escherichia coli represses biofilm formation and activates biofilm dispersal (13). The effect of CsrA on biofilm formation is mediated largely through its regulatory role in central carbon flux and intracellular glycogen synthesis and catabolism (17, 18, 24, 25, 28, 34). The influence of CsrA is substantially greater than that of other regulators of E. coli biofilm formation, OmpR, RpoS, or the Cpx two-component system (1, 8, 33). Studies with other species have revealed that the global regulator Crc (catabolite repression control) of Pseudomonas aeruginosa activates biofilm formation (21), and the expression of the staphylococcal biofilm polysaccharide PIA (polysaccharide intracellular adhesin) requires a functional glucose phosphoenolpyruvate:sugar phosphotransferase system (15).
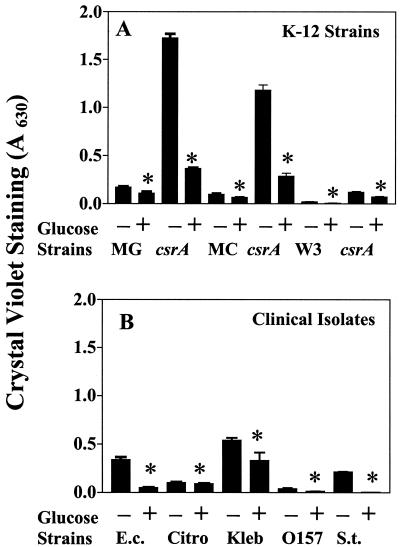
During studies of biofilm formation, we noted that the addition of glucose to media was inhibitory. To substantiate this observation, E. coli K-12 parental strains MG1655, MC4100, and W3110 and their isogenic csrA mutants (Table 1) were grown in microtiter wells in colony-forming antigen (CFA) medium (9) with or without glucose (0.2% wt/vol), and biofilm was quantitated after 24 h of growth using crystal violet staining (A630), as described previously (13) (Fig. 1A). Essentially identical results were observed in Luria-Bertani (LB) medium (19) (data not shown). These and other biofilm experiments described in this article were performed at least in triplicate with three samples per experiment, and data were analyzed by Tukey multigroup analysis (Stat View; SAS Institute Inc., Cary, N.C.). Glucose caused a statistically significant decrease in biofilm formation in every case, which varied from ∼30 to 95% reduction, depending primarily on the strain background but also on the medium. Biofilm formation by related clinical isolates, including urinary catheter isolates of E. coli, Citrobacter freundii, and Klebsiella pneumoniae and the intestinal pathogens Salmonella enterica Typhimurium and E. coli O157:H7, was also repressed by glucose in CFA medium (Fig. 1B) or LB medium (data not shown). These effects generally varied from ∼50 to 75%. The three urinary catheter isolates exhibited similar repression by glucose in artificial urine medium (13), which mimics the urinary tract environment (data not shown).
TABLE 1.
Bacterial strains or phage used in this study
| Strain or phage | Relevant genotype or description | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| MG1655 | F− λ− | Michael Cashel |
| CAG18642 | zhf-331::Tn10; 57.5 min | 30 |
| MC4100 | F Δ(argF-lac)U169 rpsL relA flhD deoC ptsF rbsR | 11 |
| ML2a | met gal hsdKRsupE supF Δcya::Kanr | 10 |
| SA2777a | F rpsL relA Δcrp::cam | S. Garges and S. Adhya |
| W3110 | F− λ−mcrA mcrB IN(rrnD-rrnE)1 | Richard E. Wolf, Jr. |
| TR1-5BW3414a | csrA::Kanr | 25 |
| Clinical strains | ||
| P5 | C. freundii | 14 |
| P18 | E. coli | 14 |
| P30 | K. pneumoniae | 14 |
| EF302 | E. coli O157:H7 | 16 |
| ATCC 14028 | S. enterica serovar Typhi- urium | 2 |
| Bacteriophage P1vir | Strictly lytic P1; forms clear plaques | 30 |
Mutant alleles Δcya::kanR, Δcrp::cam, and csrA::kanR were moved among strains by P1vir transduction or cotransduction. The strain prefix TR indicates the presence of the csrA::Kanr allele.
FIG. 1.
Effects of glucose on biofilm formation by E. coli K-12 strains and their csrA mutants (A) and by enteric pathogens (B) in CFA medium. The K-12 parent strains were MG1655 (MG), MC4100 (MC), and W3110 (W). Clinical strains were E. coli P18 (E.c.), C. freundii P5 (Citro), K. pneumoniae P30 (Kleb), E. coli O157:H7 EF302 (O157), and S. enterica Typhimurium ATCC 14028 (S.t.). Biofilm was determined after 24 h of growth at 26°C in the presence or absence of 0.2% glucose. Each bar shows the average and standard error of three separate experiments (P < 0.0001). ∗, significant difference with respect to cultures lacking glucose. Essentially identical effects of glucose were observed in LB medium (not shown).
The glucose effect, or catabolite repression, is mediated in part by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in E. coli (reviewed in references 3 and 29). In classical catabolite repression, transport of glucose leads to dephosphorylation of IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system, which prevents this protein from activating membrane-bound adenylate cyclase (Cya). The binding of cAMP to CRP forms a complex that interacts specifically and with high affinity to its cis elements in the promoter regions of cAMP-regulated genes and thereby regulates transcription. CRP levels also decline during catabolite repression (12, 32). Through these mechanisms, glucose affects the expression of genes located throughout the genome.
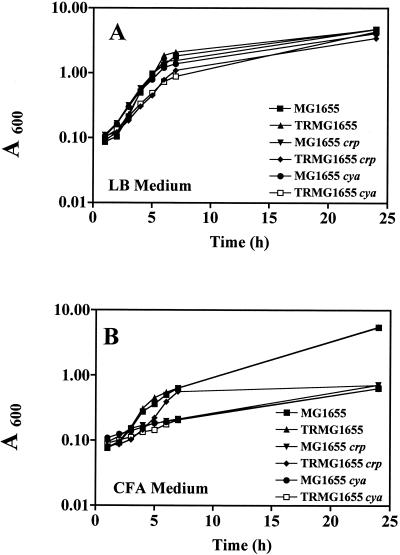
To determine whether biofilm formation was subject to classical catabolite repression by cAMP and CRP, the effects of crp and cya deletions and exogenous cAMP on biofilm formation by MG1655 and its csrA mutant were examined. Because cAMP and CRP may have pleiotropic effects on growth, the growth curves of these strains were compared in LB medium (with 0.2% glucose) or CFA medium (lacking glucose) at 26°C with shaking at 280 rpm. Growth rates in LB medium containing 0.2% glucose were unaffected by cya or crp mutations in MG1655 and very slightly decreased in the csrA mutant background (Fig. 2A). However, all of the cya and crp mutants exhibited substantial growth defects in CFA medium (Fig. 2B). Because of these effects, biofilm formation was corrected for total cell protein to yield specific biofilm values (A630 per milligram of protein) in experiments with cya and crp mutants. Protein assays on cultures containing planktonic and sessile cells were conducted as described previously (13).
FIG. 2.
Growth curves of MG1655, its isogenic csrA mutant, TRMG1655, and their crp and cya derivatives. Cultures were grown at 26°C in LB medium containing 0.2% glucose (A) or CFA medium (B) and sampled at the indicated times, and growth was determined (A600).
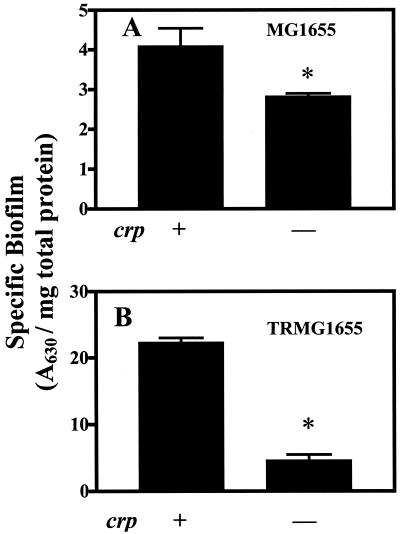
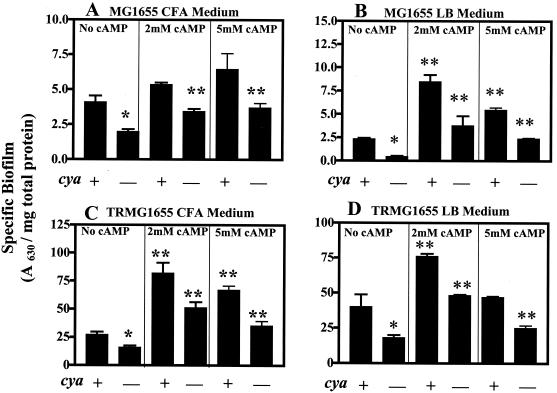
The disruption of crp in MG1655 or its csrA mutant significantly decreased specific biofilm formation (Fig. 3). The effect of crp was ∼30% in MG1655 and ∼75% in the csrA mutant both in CFA medium (Fig. 3) and in LB medium containing 0.2% glucose (data not shown). The magnitudes of these effects were comparable to those of the glucose effects on these strains (Fig. 1). Disruption of cya also decreased biofilm formation in these strains (Fig. 4). The addition of cAMP (2 or 5 mM) to the growth medium of cya mutants significantly increased specific biofilm formation (about two- to fivefold) in all experiments, and in most cases it increased biofilm formation by cya wild-type strains. Taken together, these results reveal that glucose effects on biofilm formation in E. coli are mediated at least in part by the classical catabolite repression system, i.e., cAMP and CRP.
FIG. 3.
Effects of crp on specific biofilm formation by MG1655 and its isogenic csrA mutant TRMG1655. Cultures of crp wild type or isogenic mutant strains were grown for 24 h in CFA medium, and biofilm was determined after 24 h at 26°C. Each bar shows the average and standard error of three experiments (P < 0.0001). ∗, significant difference between the crp mutant and its parent strain. Results were essentially identical in LB medium containing 0.2% glucose (not shown)
FIG. 4.
Effects of cya and exogenous cAMP on specific biofilm formation by MG1655 and its csrA mutant TRMG1655. Cultures were grown for 24 h in LB medium plus 0.2% glucose or CFA medium in the presence of 0, 2, or 5 mM cAMP. Each bar shows the average and standard error of three experiments. ∗, that cya disruption significantly decreased biofilm formation (P < 0.0001); ∗∗, addition of cAMP to the culture resulted in a significant increase in biofilm (P < 0.0001).
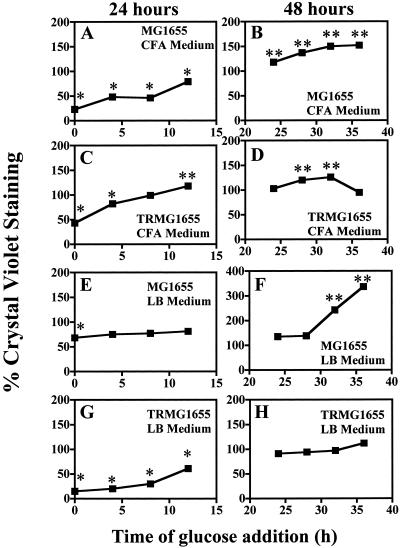
We previously evaluated the time courses of biofilm formation by MG1655 and its csrA mutant, TRMG1655, and observed that biofilm accumulated considerably beyond 24 h in both strains (13). Therefore, a final study was conducted to assess the temporal effects of catabolite repression on biofilm formed by MG1655 and its csrA mutant, TRMG1655 (Fig. 5). In this experiment, 0.2% glucose (wt/vol, final concentration) was added to cultures at various times during growth, and biofilm was assayed at 24 or 48 h. The presence of glucose at the time of inoculation led to statistically significant inhibition in every case (Fig. 5A, C, E, and G). Thereafter, glucose effects became progressively weaker. One of the 24-h biofilms, that of TRMG1655 in CFA medium, no longer was inhibited but exhibited a modest yet statistically significant increase when glucose was added at 12 h (Fig. 5C). The addition of glucose after 24 h invariably failed to inhibit biofilm formation at 48 h. In fact, glucose addition after 24 h tended to increase biofilm formation, with the exception of TRMG1655 growing in LB medium. These observations suggest that the cAMP-dependent steps of biofilm development are completed by 24 h of growth under these conditions.
FIG. 5.
Temporal effects of glucose on biofilm formation at 24 or 48 h of growth. Glucose (0.2% [wt/vol], final concentration) was added at the indicated times after inoculation of MG1655 or its csrA mutant, TRMG1655, into CFA or LB medium. Crystal violet staining was measured at A630 at either 24 or 48 h of growth, and results were calculated as percentages of values for the control cultures that lacked glucose. Each point is the average and standard error of three separate experiments. ∗ and ∗∗, significant decrease or increase (P < 0.0001), respectively, in biofilm formation relative to controls lacking glucose.
Conclusions.
Several global regulatory factors influence biofilm development in E. coli. The most striking effects to have been recognized thus far are those of CsrA (13), while those of cAMP-CRP and the other regulators (1, 8, 33) are more modest. Previous studies demonstrated that cAMP-CRP activates the expression of certain operons involved in biofilm formation, flhDC, which encodes the activator protein for the flagellar cascade of gene expression (23, 31), and glgCAP, which encodes glycogen biosynthetic and degradative enzymes (13, 26, 27). However, the relative contributions of these or other genes to the observed catabolite repression remain to be determined. Six E. coli K-12 strains and five pathogens, representing four genera of Enterobacteriaceae, exhibited glucose inhibition of biofilm formation, suggesting that catabolite repression of biofilm development is a common theme in this family of bacteria. While cAMP exhibits a relatively broad phylogenetic distribution (3), its role in biofilm formation is unknown outside of the Enterobacteriaceae. Interestingly, the cAMP-independent catabolite repression control protein Crc of P. aeruginosa is essential for biofilm formation (21). Perhaps Crc and cAMP-CRP represent evolutionarily convergent regulatory features of biofilm development in the Pseudomonadaceae and the Enterobacteriaceae, respectively.
Acknowledgments
We gratefully acknowledge gifts of strains from M. Cashel, C. Gross, M. Hammar, S. Garges, R. E. Wolf, Jr., J. R. Johnson, and J. W. Foster and manuscript critique by M. E. Hart.
This study was supported by a grant from the National Science Foundation (MCB-9726197).
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Danese, P., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese, P., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 8.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D. G., D. J. Evans, Jr., and W. Tjoa. 1977. Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect. Immun. 18:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerinot, M. L., and B. K. Chelm. 1984. Isolation and expression of the Bradyrhizobium japonicum adenylate cyclase gene (cya) in Escherichia coli. J. Bacteriol. 159:1068-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammar, M., A. Arnquivist, Z. Bian, A. Olsén, and S. Nomark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 12.Ishizuka, H., A. Hanamura, T. Kunimura, and H. Aiba. 1993. A lowered concentration of cAMP receptor protein caused by glucose is an important determinant for catabolite repression in Escherichia coli. Mol. Microbiol. 10:341-350. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, D. W., K. Suzushi, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, J. R., P. Delvari, and M. Azar. 1999. Activities of a nitrofurazone-containing urinary catheter and a silver hydrogel catheter against multidrug-resistant bacteria characteristic of catheter-associated urinary tract infection. Antimicrob. Agents Chemother. 43:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiel, K., H. Rohde, J. K. M. Knobloch, and D. Mach. 2002. Overexpression of PIA in Staphylococcus epidermidis is inhibited by inactivation of PTS glucose permease GlcA or its regulating antiterminator GlcT. Int. J. Med. Microbiol. 291(Suppl. 32.):V88. [Google Scholar]
- 16.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, M. Y., and T. Romeo. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179:4639-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, M. Y., H. Yang, and T. Romeo. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177:2663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pratt, L. A., and R. Kolter. 1999. Genetic analyses of bacterial biofilm formation. Curr. Opin. Microbiol. 2:598-603. [DOI] [PubMed] [Google Scholar]
- 23.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 24.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 25.Romeo, T., M. Gong, M. Y. Liu, and A.-M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romeo, T., J. Black, and J. Preiss. 1990. Genetic regulation of glycogen synthesis in Escherichia coli: in vivo effects of the catabolite repression and stringent response systems in glg gene expression. Curr. Microbiol. 21:131-137. [Google Scholar]
- 27.Romeo, T., and J. Preiss. 1989. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5′-diphosphate 3′-diphosphate and analysis of in vivo transcripts. J. Bacteriol. 171:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabnis, N., H. Yang, and T. Romeo. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 270:29096-29104. [DOI] [PubMed] [Google Scholar]
- 29.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1343. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 30.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tagami, H., T. Inada, T. Kunimura, and H. Aiba. 1995. Glucose lowers CRP* levels resulting in repression of the lac operon in cells lacking cAMP. Mol. Microbiol. 17:251-258. [DOI] [PubMed] [Google Scholar]
- 33.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, H., M. Y. Liu, and T. Romeo. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]