Abstract
The entomopathogenic bacterium Bacillus thuringiensis is known to secrete a zinc metalloprotease (InhA) that specifically cleaves antibacterial peptides produced by insect hosts. We identified a second copy of the inhA gene, named inhA2, in B. thuringiensis strain 407 Cry−. The inhA2 gene encodes a putative polypeptide showing 66.2% overall identity with the InhA protein and harboring the zinc-binding domain (HEXXH), which is characteristic of the zinc-requiring metalloproteases. We used a transcriptional inhA2′-lacZ fusion to show that inhA2 expression is induced at the onset of the stationary phase and is overexpressed in a Spo0A minus background. The presence of a reverse Spo0A box in the promoter region of inhA2 suggests that Spo0A directly regulates the transcription of inhA2. To determine the role of the InhA and InhA2 metalloproteases in pathogenesis, we used allelic exchange to isolate single and double mutant strains for the two genes. Spores and vegetative cells of the mutant strains were as virulent as those of the parental strain in immunized Bombyx mori larvae infected by the intrahemocoelic route. Exponential phase cells of all the strains displayed the same in vitro potential for colonizing the vaccinated hemocoel. We investigated the synergistic effect of the mutant strain spores on the toxicity of Cry1C proteins against Galleria mellonella larvae infected via the oral pathway. The spores of ΔinhA2 mutant strain were ineffective in providing synergism whereas those of the ΔinhA mutant strain were not. These results indicate that the B. thuringiensis InhA2 zinc metalloprotease has a vital role in virulence when the host is infected via the oral route.
Bacillus thuringiensis is a gram-positive, spore-forming bacterium that is a member of the Bacillus cereus group (Bacillus anthracis, B. cereus, Bacillus mycoides, and B. thuringiensis). B. thuringiensis is well known for its insecticidal activity, which is mainly due to the crystallized δ-endotoxins produced upon spore formation. After ingestion by susceptible insects, the δ-endotoxins (also termed Cry proteins) are dissolved. They are then activated in the insect's gut before binding to epithelial cell-specific receptors, causing cell lysis and death (49). B. thuringiensis spores and vegetative cells also have insecticidal activity (12, 21, 58). Indeed, mutants lacking δ-endotoxins are still pathogenic when injected into certain Lepidopteran larvae. Moreover, the addition of B. thuringiensis and B. cereus spores or vegetative cells strongly increases the insecticidal activity of Cry toxins against some insect species that are weakly susceptible following the ingestion of crystals alone (26, 32, 46). The appearance of bacteria in the hemocoel when spores are added to the crystals suggests that the synergism is due to septicemia (36). The PlcR regulon (46) and the Vip3A toxin (10) are involved in this synergism.
Pathogenic bacteria express a myriad of virulence or metabolic genes in their hosts. The factors produced by these genes allow the pathogen to survive in the hostile environment of the host, to escape the immune system, and to establish a biotope where they can proliferate. We do not know how B. thuringiensis cells or spores manage to invade and to kill insects after hemocoelic inoculation. However, B. thuringiensis is highly resistant to the humoral defense system of the host, especially to cecropins and attacins, which are the main classes of inducible antibacterial peptides in various lepidopterans and dipterans (5, 23, 24). A zinc metalloprotease secreted by B. thuringiensis, termed InhA or InA, specifically hydrolyzes cecropins and attacins in the immune hemolymph of Hyalophora cecropia in vitro (8, 12).
Although the degradation of cecropins and attacins by InhA may partly explain the success of the bacterium in invading hemocoel, the importance of this protease in virulence has been debated (8, 33, 51). Indeed, the role of InhA in resistance to the humoral defense system is not consistent with the time course of InhA production. Steiner (53) studied the kinetics of InhA production in the hemolymph of H. cecropia infected with B. thuringiensis cells and found that the concentration of this protease was maximal long after insect death. Moreover, a chromosomal transcriptional inhA′-lacZ fusion showed that inhA expression starts at the onset of sporulation and is indirectly activated by Spo0A (18). This late expression of inhA is not consistent with the production of the antibacterial peptide, which is an initial host defense reaction (12, 40, 57). Another putative role is suggested by the fact that purified InhA has a lethal effect when injected to the insect host. The symptoms associated with the administration of InhA are typical of toxemia and not bacterial septicemia (33, 51).
Southern blot analyses have shown that multiple copies of inhA-like genes are present in some B. thuringiensis strains (33). Here, we report the characterization of a gene encoding a putative metalloprotease that is very similar to InhA in B. thuringiensis strain 407 Cry−. This gene was designated inhA2. The transcription start site of inhA2 was determined, and the regulation of inhA2 expression in different B. thuringiensis genetic backgrounds was studied. Allelic exchange was used to construct single and double mutants for the two inhA genes, and these mutants were studied to determine the respective roles of the two genes in pathogenesis. Two different infection models were used: Bombyx mori larvae for the injection experiments and Galleria mellonella larvae for the force-feeding assays.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli K-12 strain TG1 [Δ(lac-proAB) supE thi hsd-5 (F′ traD36 proA+ proB+ lacIq lacZΔM15)] (15) was used as a host for cloning experiments. E. coli strain SCS110 [rpsL (Strr) thr leu endA thi-1 lacY galK galT ara tonA tsx dam dcm supE44 Δ(lac-proAB) (F′ traD36 proA+ proB+ lacIq lacZΔM15)] (Stratagene, La Jolla, Calif.) was used to prepare DNA, which was then transformed into B. thuringiensis. Alternatively, E. coli strain ET 12567 (F− dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1) was used. Cells of E. coli strains TG1, SCS110, and ET12567 were grown in LB (Luria broth) with vigorous shaking at 37°C. Conventional CaCl2 or electroporation procedures were used to transform E. coli (11), and the transformants were selected on LB plates supplemented with ampicillin (100 μg ml−1) or kanamycin (20 μg ml−1).
The acrystalliferous B. thuringiensis wild-type strain (407 Cry−) belonging to serotype 1 (31) and the asporogenic and acrystalliferous 407 Cry− Δspo0A mutant strain (29) were used throughout this study. B. thuringiensis 407 Cry− carrying a disrupted inhA gene (407−[inhA′-lacZ]) has been described previously (18). In this study, this strain was designated 407 Cry− ΔinhA. B. thuringiensis cells were transformed by electroporation as described previously (31). Transformants were selected on LB plates supplemented with kanamycin (200 μg ml−1), erythromycin (10 μg ml−1), or kanamycin (200 μg ml−1) plus erythromycin (3 μg ml−1).
Spores of the various B. thuringiensis strains were obtained by culturing cells in 10 ml of sporulation-specific (HCT) medium (28) at 30°C for 3 days. Spores were harvested by centrifugation (16,000 × g; 2 min), washed twice with distilled water (twice, each time with 2 ml), and finally resuspended in 2 ml of sterile distilled water. The concentrations of the spore preparations were estimated by plating them onto LB agar plates containing appropriate antibiotics.
The Cry1C toxins were prepared from the asporogenic strain 407 ΔsigK (6) transformed with pHT1C (47) as described by Gominet and colleagues (16).
Nucleic acid manipulations.
Plasmid DNA was extracted from E. coli by a standard alkaline lysis procedure using QIA prep spin columns (Qiagen). Chromosomal DNA was extracted from B. thuringiensis cells harvested in mid-log phase as described previously (42). Restriction enzymes and T4 DNA ligase were used as recommended by the manufacturer (Biolabs New England). Oligonucleotide primers (Table 1) were synthesized by Genset (Paris, France). PCRs were performed in a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer). Amplified DNA fragments were purified using the QIAquick PCR purification Kit (Qiagen) and migrated on 0.7% agarose gels after digestion. Digested DNA fragments were extracted from agarose electrophoresis gels using a centrifugal filter device (Ultrafree-DA; Amicon Laboratories). The double-stranded DNA was then cloned and sequenced by the dideoxy-chain termination method (48) using the T7 sequencing kit from Pharmacia P-L and [α-35S]dATP (15 TBq; Amersham, Little Chalfont, United Kingdom). Some nucleotide sequences were determined by Genome Express (Paris, France).
TABLE 1.
Primer sequences used in this study
| Name | Nucleotide sequencea | Restric- tion site |
|---|---|---|
| InB1 | 5′-CGCGGATCCATGCAATTTTGCATATTGTC-3′ | BamHI |
| InB2 | 5′-CCGGAATTCACTAATCACGTGCGCC-3′ | EcoRI |
| InB3 | 5′-CGGAATTCGGAGTTTGGAAAGCATGC-3′ | EcoRI |
| InB4 | 5′-CCCAAGCTTTTGTGTATAGCCAAGTAAC-3′ | HindIII |
| InB8 | 5′-AAACTGCAGCATGCAATTTTGCATATTGTC-3′ | PstI |
| InB9 | 5′-CGCGGATCCCTCTTTTGCTGGCGTTTCTGC-3′ | BamHI |
| Bext.1 | 5′-CCCTCCAGTTTTCTGGTCAGACCCTCTATC-3′ | |
| B6sqBt | 5′-AAACTGCAGCCCAGCAAACGTAATTGCTTC-3′ | PstI |
| B9sqBt | 5′-CGCGGATCCCTCTTTTGCTGGCGTTTCTGC-3′ | BamHI |
| Bex2 | 5′-AAACTGCAGGTTTAATGTTTATAGAATTATGTC-3′ | PstI |
| B2sq | 5′-CGTTTTGATTCCCATCCCCG-3′ | |
| inB2sq | 5′-GGTTAACTGTTCCAGGAAAAGC-3′ | |
| inB3sq | 5′-GCATGCTTTCCAAACTCCGG-3′ | |
| inB4sq | 5′-GCGTGGTGTAGGAGTTCCTAC-3′ |
Restriction sites are underlined.
Plasmid and mutant strain constructions.
The inhA2 gene in B. thuringiensis 407 Cry− and 407 Cry− ΔinhA was disrupted as follows. First, 1,066-bp BamHI-EcoRI and 1,183-bp EcoRI-HindIII DNA fragments, corresponding to the chromosomal DNA regions upstream and downstream of the inhA2 gene, respectively, were generated by PCR using the oligonucleotide pairs InB1-InB2 and InB3-InB4 (Table 1). A kanamycin resistance (Kmr) cassette was purified from pDG783 as a 1.5-kb EcoRI fragment carrying the aphA3 gene from Enterococcus faecalis (54). The amplified DNA fragments and the Kmr cassette were digested by appropriate enzymes and cloned between the HindIII and the BamHI sites of the thermosensitive plasmid pRN5101, conferring erythromycin resistance to gram-positive hosts and ampicillin resistance to E. coli (55). The resulting plasmid was verified by restriction mapping and used to transform the wild-type strain 407 Cry− and the mutant strain 407 Cry− ΔinhA. Strains that were resistant to kanamycin and sensitive to erythromycin arose through a double-crossover event in which the chromosomal wild-type copy of inhA2 was deleted and replaced with the Kmr cassette (29). The chromosomal allele exchange was checked by PCR using appropriate oligonucleotide primers. The corresponding mutant strains were designated 407 Cry− ΔinhA2 and 407 Cry− ΔinhA ΔinhA2.
A transcriptional inhA2′-lacZ fusion was constructed using a PstI-BamHI DNA fragment corresponding to the upstream region of inhA2 that was generated by PCR using the primer pair InB8 and InB9 (Table 1). The PCR fragment was digested by appropriate enzymes, purified as a 438-bp fragment, and ligated between the BamHI and PstI sites of pHT304-18′Z (2). The recombinant plasmid, designated pHT304-inhA2′Z, was introduced into the B. thuringiensis wild-type strain (407 Cry−) and the mutant strain (407 Cry− Δspo0A) by electroporation. Transformants were named 407 Cry− [pHT304ΩinhA2′Z] and 407 Cry− Δspo0A [pHT304ΩinhA2′Z], respectively.
Mapping of mRNA start site by primer extension.
Total RNA was extracted from wild-type B. thuringiensis (407 Cry−) cells grown in LB at 30°C with shaking. The inhA2 transcription start site was determined by primer extension as described previously (3) using a synthetic 30-mer oligonucleotide, Bext.1 (Table 1), which is complementary to the DNA sequence at positions −132 to −162 with respect to the translational start site of inhA2 gene. DNA sequencing was performed by the dideoxy chain termination method with the primer Bext.1 and using the double-stranded pHT304ΩinhA2′Z as the template.
β-Galactosidase assay.
The B. thuringiensis strains harboring lacZ transcriptional fusions were cultured in LB or in sporulation-specific medium (HCT) at 30°C. β-Galactosidase specific activities were measured as described previously (42). The specific activities are expressed in units of β-galactosidase milligram−1 of protein (Miller units).
Determination of B. thuringiensis 407 Cry− inhA2 nucleotide sequence.
Four pairs of oligonucleotides (B1 [B6sqBt-B9sqBt], B2 [Bex2-B2sq], B3 [inB2sq-inB3sq], and B4 [inB4sq-InB4]) (Table 1) were designed based on the available B. anthracis nucleotide sequence. These primers were used to amplify four fragments of 595, 1,025, 765, and 1,290 bp, respectively, using 407 Cry− chromosomal DNA as a template. The four fragments are overlapping and cover the entire inhA2 region from position −482 to +240 with respect to the ATG start and TAA terminal codons of the inhA2 coding sequence, respectively. PCR were carried out in a reaction volume of 100 μl containing 200 μM deoxynucleoside triphosphates, 3.5 mM MgSO4, 50 pmol of each primer, 0.5 μg of B. thuringiensis strain 407 Cry− chromosomal DNA, and 0.5 U of Pwo DNA polymerase (Roche Boehringer) in a 1× reaction buffer. PCR products were purified by use of the QIA quick PCR purification Kit (Qiagen) and then eluted from agarose gel electrophoresis and sent to Genome Express (Paris, France) for sequencing.
Computer analysis of sequence data.
Preliminary sequence data for B. anthracis was obtained from the Institute for Genomic Research (TIGR) website (http://www.tigr.org). Translated open reading frames (ORFs) from TIGR were used for Blast searches of the nonredundant National Center for Biotechnology Information (NCBI) protein database (http://www.ncbi.nlm.nih.gov/). The signal PV1.1 predictor server was used to identify potential cleavage sites.
Insects and their immunization.
G. mellonella eggs were hatched at 30°C, and the larvae were reared on bees wax and pollen (Naturalim). Eggs of B. mori strain nistari were provided by INRA (Unité Nationale Séricicole, Lyon, France) and incubated at 25°C. The resulting larvae were reared on a commercially available artificial diet (Fukui and Co., Ltd., Yokohama, Japan).
Silkworm larvae on the first day of the fourth and fifth instars were immunized as follows. Cells of E. coli K12 strain TG1 was grown overnight in LB at 37°C and then washed twice and suspended in sterile water. We then injected 10 μl of the suspension, containing about 105 viable cells, through the intersegmental membrane between the fourth and the fifth abdominal legs of the larvae by using a 1-ml Terumo syringe and a microapplicator (Burckard type LV. 65.).
In vivo experimental infections.
Pathogenicity assays were carried out on B. thuringiensis vegetative cells as follows. Cells of wild-type and mutant strains of B. thuringiensis were grown in LB medium devoid of antibiotics at 30°C and with shaking. Bacterial concentrations were determined by measuring the optical density at 600 nm and verified by plating dilutions onto LB agar plates supplemented with appropriate antibiotics. We injected about 18 to 20 vegetative cells into a group of 50 silkworm larvae in the fourth instar 2 days after immunization as described above. Larvae were maintained individually in plastic containers at 25°C. They were checked daily, and mortality was recorded on the first and the second days postinfection. The killing activity of the spores of each strain was assessed on immunized B. mori larvae, as reported for vegetative cells, except that three different doses were tested on groups of 30 larvae, and the 50% lethal dose (LD50) was established. A control group was injected with sterile water.
Groups of 25 last instar G. mellonella larvae, weighing about 200 mg, were force-fed with spore-crystal suspensions in sterile water (10 μl of larva−1) by using 0.5-by-25-mm needles (Burckard Manufacturing) and a microinjector (Burckard). The larvae were kept individually in boxes containing beeswax and pollen at 25°C. Experiments were repeated three times. Mortality was recorded 1, 2, and 3 days postinfection.
In vitro assays.
In vitro experimental infections were performed in immune hemolymph pooled from fifth instar B. mori larvae 48 h after immunization. The surfaces of the larvae were cleaned with 70% ethanol and dried. Hemolymph was collected by capillary action, in a sterile, ice-cooled Eppendorf tube, by cutting off the abdominal leg. We mixed 20 μl of a dilution containing about 7 × 104 viable cells of the appropriate strains with 200 μl of immunized hemocoel and incubated the mixture at 28°C for 2 h. During this period, 20-μl portions were withdrawn at 30-min intervals and spread on LB plates for viable count assays. The experiments were repeated three times for each test. For the control experiments, the reaction mixture contained 1.1 × 105 cells of E. coli strain TG1 and 200 μl of natural or cell-free hemolymph of B. thuringiensis obtained from untreated or vaccinated insects. Cell-free hemolymph was obtained by centrifugation at 16,000 × g for 2 min.
Statistical analysis.
The mortality data following spore injections were analyzed by calculating LD50s by use of the Log-Probit program of Raymond and colleagues (45), based on work by Finney and colleagues (14).
The mortality rate caused by the injection of vegetative cells was analyzed by Fisher's exact test on 2-by-2 contingency tables as implemented by the JMP IN 3.0 software program.
The killing activities of spores following force-feeding were compared by Student's t test.
Nucleotide sequence accession number.
The nucleotide sequence of the 3,087-bp DNA region including the B. thuringiensis inhA2 gene has been submitted to the GenBank database under accession no. AF421888.
RESULTS
Identification of inhA2 metalloprotease gene in B. thuringiensis strain 407 Cry−.
Some B. thuringiensis strains possess multiple copies of the inhA gene (33). Southern blot hybridizations performed on B. thuringiensis 407 Cry− chromosomal DNA probed with an internal fragment of the inhA gene revealed two bands (data not shown).
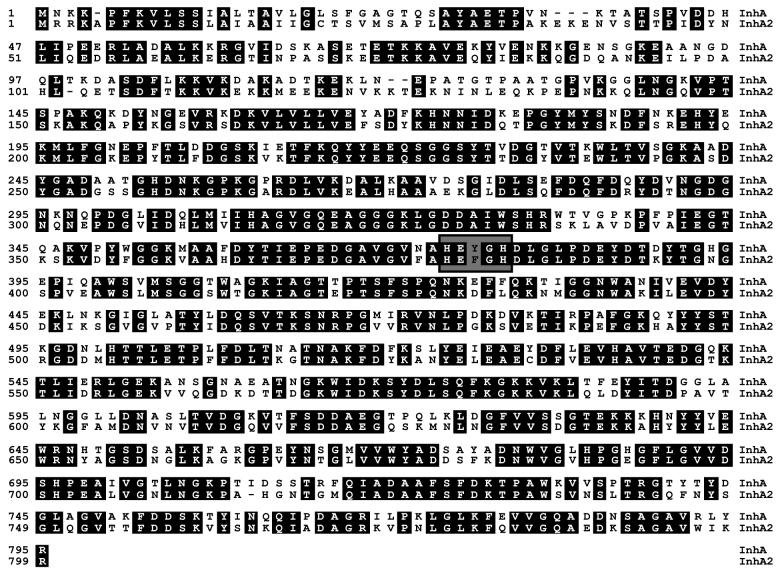
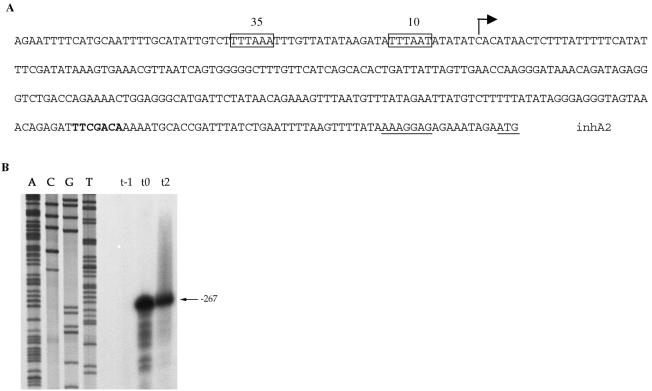
The amino acid sequence of InhA from B. thuringiensis strain 407 Cry− (17) was used to search the unfinished B. anthracis genome database obtained from the TIGR website. This revealed the presence of two highly similar peptide sequences. The most similar amino acid sequence (93% identity) was the putative B. anthracis InhA protein. The other amino acid sequence shared 67% identity with InhA. This sequence was encoded by an ORF located in B. anthracis contig number 3920. We designated this ORF the inhA2 gene. The chromosomal region of B. thuringiensis strain 407 Cry− containing the entire inhA2 gene was amplified by PCR, and both strands were sequenced. The inhA2 gene is predicted to encode a protein of 800 amino acid residues with a calculated molecular mass of 87.8 kDa. Alignment of InhA and InhA2 amino acid sequences showed a high degree of identity (66.2%) (Fig. 1). Both InhA and InhA2 amino acid sequences lack cysteine residues and contain the zinc-binding motif (HEXXH), which is characteristic of the zinc-metalloprotease family (27). A potential cleavage site was found between positions 32 and 33 (AYA-ET) of InhA2 protein by the Signal P V1.1 Predictor server (Fig. 1). The ATG codon is preceded by a typical ribosome binding sequence (AAAGGAG) at an appropriate distance (Fig. 2A). The TAA stop codon is followed by a probable rho-independent transcription terminator stem-loop sequence (TTTTCGTGTAAGAAGTATACTTCTCACAC GAAAA).
FIG. 1.
Alignment of the B. thuringiensis InhA2 amino acid sequence with that of InhA (18). Numbers indicate positions in amino acid sequence. Identical residues are shaded. The conserved zinc-binding domain (HEXXH) is boxed.
FIG. 2.
Analysis of the inhA2 promoter region. (A) Sequence of the inhA2 promoter region. The putative −10 and −35 boxes of the inhA2 promoter are boxed. The potential ribosome binding site and the start codon (ATG) are underlined. The potential reverse Spo0A box is indicated in bold. The broken arrow indicates the transcriptional start site. (B) Mapping of the inhA2 transcriptional start site by primer extension. Total RNA was isolated from B. thuringiensis strain 407 Cry− 1 h (t−1) before and 0 (t0) and 2 h (t2) after the beginning of the stationary phase. RNA was subjected to primer extension using the oligonucleotide Bext1. Lanes A, C, G, and T show the sequence of the promoter region of the inhA2 gene.
Determination of transcriptional start site of inhA2 and analysis of inhA2 gene expression in B. thuringiensis.
The transcriptional start site for inhA2 was mapped by primer extension analysis using a synthetic oligonucleotide Bext.1 and total RNA extracted from 407 Cry− cells growing exponentially, at the onset of (t0) and during (t2) stationary phase. At t0 and t2, a transcript was detected with its 5′ end 267 bp upstream from the presumed inhA2 start codon (Fig. 2B). The putative −10 and −35 boxes of the inhA2 promoter (Fig. 2A) resemble the σA promoter consensus (TTGACA, 16 to 18 bases, TATAAT) of Bacillus subtilis (39).
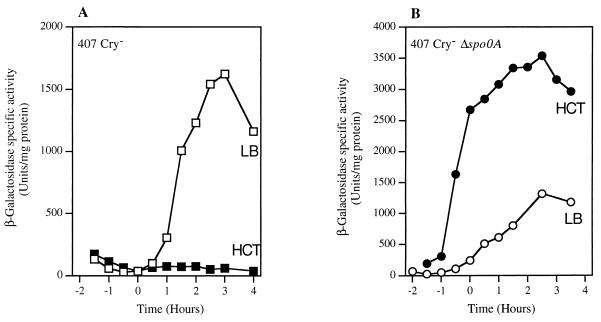
To assess the transcriptional activity of the inhA2 promoter, we constructed a transcriptional fusion between the 438-bp DNA region extending upstream from the inhA2 start codon and the lacZ gene in pHT304-18′Z. The B. thuringiensis strain 407 Cry− carrying pHT304ΩinhA2′Z was cultured in LB medium and in a sporulation-specific medium (HCT) at 30°C. β-Galactosidase production was measured at different stages of growth between t−1 (1 h before the onset of the stationary phase) and t+4 (4 h after the onset of stationary phase) (Fig. 3A).
FIG. 3.
Spo0A negatively regulates the expression of inhA2. Expression of inhA2′-lacZ in strains 407 Cry− (▪, □) (A) and 407 Cry− Δspo0A (•, ○) (B) at 30°C was determined. Open and solid symbols indicate β-galactosidase activity expressed in units/milligram of protein (Miller units) when the strains were grown in LB and HCT, respectively.
In LB medium, the level of inhA2-directed β-galactosidase synthesis was very low during exponential growth. It increased at the onset of the stationary phase and reached a maximum specific activity of 1,600 Miller units at t+3. In HCT medium, the cells did not produce β-galactosidase at any point in the growth cycle (<10 Miller units).
Effect of spo0A null mutation on inhA2 expression.
Spo0A is the key factor involved in the initiation of sporulation (22). We analyzed the expression of the inhA2′-lacZ fusion in a B. thuringiensis spo0A mutant to investigate the effect of a spo0A null mutation on inhA2 expression. The 407 Cry− Δspo0A strain carrying pHT304ΩinhA2′Z was grown in LB and HCT and β-galactosidase activity was monitored at various stages of growth (Fig. 3B). In HCT medium, the cells produced β-galactosidase from t−1 and a value exceeding 3,000 Miller units was reached at t+2. In LB medium, the pattern of inhA2′-lacZ expression was quite similar to that of the wild-type strain harboring pHT304ΩinhA2′Z (Fig. 3A and B). Analysis of the inhA2 gene promoter region revealed the presence of a reverse Spo0A box (5′-TGTCGAA-3′) (52) located 208 bp downstream from the transcriptional start site (Fig. 2A). These results suggest that the binding of Spo0A downstream from the inhA2 promoter prevents the transcription of inhA2.
Virulence of vegetative cells and spores following intrahemocoelic inoculation.
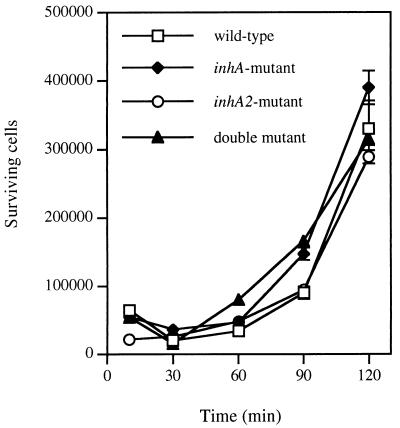
Preliminary experiments indicated that B. mori larvae were highly susceptible to B. thuringiensis 407 Cry− cells. The LD50 was less than five injected vegetative cells per larva (data not shown). Thus, this insect was used to determine the effect of inhA and inhA2 mutations on virulence. No difference was observed between the mutant strains and the wild type with respect to the killing activity of vegetative cells when normal B. mori larvae were used (data not shown). Due to the putative role of InhA in the degradation of cecropins, we performed virulence tests on immunized B. mori larvae. B. mori produces different types of antibacterial peptides, including cecropins, about 2 days after injection of E. coli K-12 cells (40, 41). About 18 to 20 exponentially growing cells (causing 90 to 95% mortality in the case of the 407 Cry− strain) of the mutant strains were injected into the hemocoel of fourth instar B. mori larvae at day 2 postvaccination. Infection of each mutant strain (ΔinhA, ΔinhA2, and ΔinhA ΔinhA2) resulted in a mortality rate of between 88 and 96% on day 2 (Table 2). No significant differences were found between the B. thuringiensis mutant and wild-type strains with respect to the killing effect of vegetative cells (Fisher's test; P value was >0.05 in all cases) (Table 2). Similarly, the inhA and the inhA2 mutations did not reduce the rate of killing: the percentage of mortality at 24 h (86 to 90%) is comparable to that observed with the wild-type cells (90 to 94%) (data not shown). The immune silkworm blood was assessed for in vitro killing activity on the various strains of B. thuringiensis. The results show that the mutant and wild-type 407 Cry− strains survive in immunized hemolymph and grow similarly (Fig. 4). In contrast, E. coli cells survived naive hemolymph but were killed a few minutes after inoculation of immunized hemolymph (data not shown).
TABLE 2.
Bombyx mori mortality after injection of vegetative cells of B. thuringiensis mutant strainsa
| Time after injection (h) | 407 Cry− ΔinhA
|
407 Cry− ΔinhA2
|
407 Cry− ΔinhA ΔinhA2
|
|||
|---|---|---|---|---|---|---|
| % Mortalityb | Pc | % Mortality | P | % Mortality | P | |
| 24 | 86 (43/50) | 0.317 | 90 (45/50) | 0.715 | 88 (44/50) | 1.0000 |
| 48 | 90 (45/50) | 0.204 | 96 (48/50) | 1.0000 | 88 (44/50) | 0.48 |
All experiments were carried out on immunized B. mori larvae. Each larva was vaccinated with 105 viable cells of E. coli strain TG1. Two days after immunization, the larvae were challenged with 18 to 20 cells of B. thuringiensis mutant strains. The same dose of wild-type cells caused 90 to 95% mortality. The control was injected with sterile water that caused 0% killing in all assays. Fisher's exact test was used to evaluate the significance of changes in mortality with respect to that caused by the wild-type strain.
Number of dead larvae/number of infected larvae.
P value was calculated by Fisher's exact test.
FIG. 4.
Surviving B. thuringiensis cells after incubation with hemolymph from immunized B. mori larvae. The reaction mixture contained 200 μl of hemolymph and 20 μl of a suspension containing about 7 × 104 viable cells. Aliquots were withdrawn at the time points indicated and the surviving bacteria were counted. Experiments were repeated three times. Vertical bars indicate the standard error of the mean.
We also examined the effect of injecting mutant and wild-type spores on the mortality of immunized B. mori larvae (Table 3). In all the cases, the injection of spores resulted in a high mortality rate; 24 h after injection, the LD50 ranged from 9 to 15 spores injected per larva. The LD50s were not significantly different, as the confidence intervals overlapped each other.
TABLE 3.
Pathogenicity of spores from B. thuringiensis wild-type and mutant strains injected into hemocoel of immunized B. mori larvae
| Strains | LD50 (no. of spores/ injected larva)a
|
|
|---|---|---|
| Mean | 95% CIb | |
| 407 Cry− | 11.94 | 7.47-15.39 |
| 407 Cry− ΔinhA | 9.49 | 4.85-12.85 |
| 407 Cry− ΔinhA2 | 14.56 | 5.61-20.81 |
| 407 Cry− ΔinhA ΔinhA2 | 9.22 | 7.34-10.89 |
Values are LD50s calculated by log-probit analysis at 24 h after infection.
95% confidence intervals (14).
Virulence of spores following force-feeding.
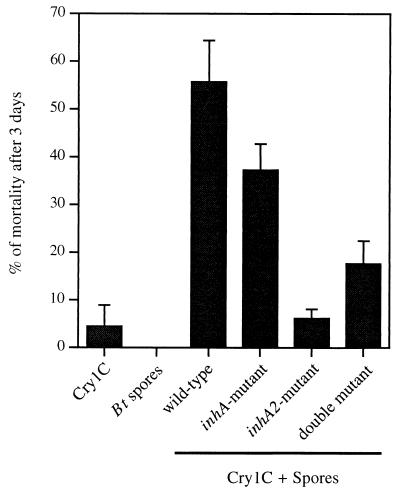
It has been previously reported that B. thuringiensis 407 Cry− spores have a significant synergistic effect on the toxicity of sublethal doses of the Cry1C toxin in G. mellonella larvae following infection via the oral route (46). Based on these results, the synergism of the various ΔinhA and ΔinhA2 mutant spores was assessed and compared to the synergistic effect of the wild-type spores (Fig. 5). Very low mortality (<10%) was obtained following the ingestion of the crystals (3.2 μg larva−1) or spores (106 spores larva−1) alone. A clear pattern of synergism was obtained when the Cry1C protein was mixed with the parental 407 Cry− strain spores. The level of synergism with inhA-deficient mutant spores was not significantly different from that obtained with the wild-type (Student's t test; P > 0.01). Synergism was, however, significantly attenuated with the spores of 407 Cry− ΔinhA2 and 407 Cry− ΔinhA ΔinhA2 (Student's t test; P < 0.01) with a larger decrease in mortality for the inhA2-deficient mutant.
FIG. 5.
Results from three pooled independent experiments. Last instar G. mellonella larvae were force-fed with spores alone, crystals alone (Cry1C), or spore/crystal mixtures. In all experiments, the doses of spores and crystals were 106 and 3.2 μg larva−1, respectively. For all the strains, spores alone caused no mortality. Vertical bars indicate the standard error of the mean.
DISCUSSION
We describe the identification and the transcriptional analysis of inhA2, a gene encoding a protein that is highly similar to the B. thuringiensis InhA metalloprotease. The inhA and inhA2 genes coexist in B. thuringiensis strain 407 Cry−. Both InhA and InhA2 amino acid sequences contained the canonical zinc-binding consensus motif (HEXXH), which is highly conserved in the zinc-containing metalloproteases (27). In this motif, the two histidine residues function as the first and the second zinc ligands. According to the recent classification system proposed for the zinc-requiring metalloproteases, InhA and InhA2 belong to the zincin superfamily. This superfamily has been divided into at least 10 families on the basis of the location of the third (E) and the fourth (Y) ligands in the sequence around the HEXXH domain (38). However, examination of the InhA and InhA2 amino acid sequences failed to find any relationship with the previously reported metalloprotease families. This suggests that the B. thuringiensis InhA-like metalloproteases may form a new family. Examination of the amino acid sequence of InhA2 revealed a putative signal peptide cleavage site between positions 32 and 33, suggesting that InhA2 is exported. Several other bacterial zinc metalloproteases have been shown to be extracellular proteins that need signal and leader sequences to aid transport across the bacterial cell membrane (38). Alignment of InhA and InhA2 proteins showed that the putative cleavage sites are located in the same positions.
The identification of the 5′ end of the inhA2 transcript and the determination of the putative promoter region suggest that inhA2 is transcribed by an RNA polymerase containing the major sigma factor, sigma A. This is similar to the situation found for the inhA gene (18). However, in sharp contrast with inhA, which is activated by Spo0A via AbrB (18), the transcription of inhA2 is repressed by Spo0A. The presence of a reverse Spo0A box downstream from the transcription start suggests that Spo0A acts directly on inhA2 transcription (52). Thus, the transcription of the two inhA metalloprotease genes in B. thuringiensis strain 407 Cry− is controlled in two opposite manners. This suggests that inhA is preferentially expressed when the cells are grown and sporulate in a relatively poor medium (i.e., HCT medium), whereas the inhA2 gene is preferentially expressed when the cells are grown and sporulate in a rich medium (i.e., LB medium). The functional significance of the duplication of the inhA gene and their complementary regulation systems in the B. thuringiensis 407 Cry− might reflect a physiological regulatory mechanism, which enables the bacteria to cope with adverse environmental conditions, especially biotic variations.
The zinc-requiring metalloprotease InhA specifically degrades attacins and cecropins (8). This suggests that InhA is implicated in the invasive mechanisms of B. thuringiensis by allowing bacteria to interfere with the immune system of the host. To establish the contribution of this biochemical role in virulence, we tested the pathogenicity of single and double inhA and inhA2 mutants in their insect hosts. Spores and vegetative cells of the single and double mutant strains were found to be equally as virulent as those of the parental strain when injected into immunized B. mori larvae: the time of appearance of mortality, about 18 h postinfection (results not shown), and the mortality levels were similar for all of the strains. Moreover, exponentially growing cells of all mutants displayed the same in vitro colonization potential for the vaccinated hemolymph. These results suggest that the inactivation of B. thuringiensis inhA and inhA2 genes does not affect the ability of the bacteria to kill insects and it did not prevent it from resisting and multiplying in immune hemolymph in vitro. We can thus conclude that InhA-like metalloproteases are not essential for the virulence of B. thuringiensis when injected into B. mori larva via the intrahemocoelic route. These results prompted us to investigate whether InhA is especially targeted for cecropin degradation and, if so, whether this biochemical function is responsible for cecropin resistance in B. thuringiensis. Inhibition zone assay, consisting of plating B. thuringiensis cells onto LB medium and adding synthetic cecropin A, showed that cecropin did not prevent the growth of the parental strain or that of the mutant strains (data not shown). This suggests that InhA and InhA2 are not essential for cecropin resistance in B. thuringiensis. Culture filtrates from the mutant strains collected 2 h after entering stationary phase protected E. coli from cecropins and immune hemolymph (data not shown). This indicates that protection against cecropins is not exclusively achieved by an InhA-like specific proteolytic activity, which is in contrast with previous reports (8). However, it is consistent with the fact that cecropins are high susceptible to diverse proteases (P. Bulet, personal communication).
The B. thuringiensis InhA and InhA2 metalloproteases are not primary virulence factors in intrahemocoelic infections. However, our results strongly suggested that InhA2 is essential for providing a synergistic effect to B. thuringiensis spores on the toxicity of the Cry1C protein against G. mellonella following infection via the oral route. The inactivation of inhA has not shown a significant effect on synergism. One hypothesis explaining the difference between InhA and InhA2 in providing synergism would be that only inhA2 is expressed in vivo. Indeed, the insect host is a rich nutritional medium for B. thuringiensis, and we demonstrated that inhA2 is expressed preferentially to inhA in rich media, such as LB. Similar results, with respect to synergism, were reported by Salamitou et al. (46) for a plcR deletion mutant. Spores of a ΔplcR mutant were virulent following intrahemocoelic infection but were unable to provide a synergistic effect against G. mellonella larvae. PlcR is a pleotropic regulator that positively regulates the transcription of various genes encoding extracellular virulence factors including phospholipases C (PlcA and PlcB), enterotoxins (Hbl and Nhe), and proteases (1, 30, 44). The degradative enzymes encoded by PlcR-regulated genes might be required so that the bacterium can cross the intestinal defense barrier in the insect. A similar mode of action might also exist for the B. thuringiensis InhA2 metalloprotease. Dalhammar and Steiner (8) reported that InhA can hydrolyze the collagen-containing substrate Hide Powder. Collagen is a biologically important substance in eukaryotic organisms. It is the predominant constituent of many mammalian tissues, and its degradation probably leads to the loss of tissue integrity, which has significant implications for health (19). Collagen has been found in the basement membranes and other connective tissues in insects (4). Many extracellular zinc-containing proteases from pathogenic organisms have been shown to cause necrotic or hemorrhagic tissue damage in the host by digesting structural components of the substances, such as collagen. This is especially the case for the zinc metalloproteases belonging to the thermolysin family. For example, Vibrio vulnificus VvpE specifically degrades type IV collagen (37), thereby disrupting the backbone structure of the basal layer of capillary vessels. Pseudomonas aeruginosa elastase and Aeromonas hydrophila AhyB, two zinc metalloproteases sharing high sequence similarity, degrade several important biological substances of protective tissues, such as elastin and collagen (7, 9, 56).
It has been postulated that the massive tissue disintegration caused by these metalloproteases, especially those possessing a collagenase function, may help bacteria cross the host barrier and gain access to deeper tissues (20, 34, 38). If InhA2 protease was to have a collagenase activity, like that of InhA, the passage of bacteria across the basement membrane underlining the midgut epithelium of the insect would be facilitated, thus explaining the synergy with the crystals following infection by the oral route. The role of the InhA2 metalloprotease in the infectious process following natural inoculation is reminiscent of that of the Vibrio anguillarum extracellular zinc metalloprotease, which is highly homologous to the elastase of P. aeruginosa. The virulence of mutants deficient in this metalloprotease was lower in immersion infection experiments than in intraperitoneal-inoculation experiments (35). This illustrates that these zinc metalloproteases are primarily involved in the early steps of infection when they interact with the host barrier. Our results show that B. thuringiensis InhA and InhA2 metalloproteases are not essential in septicemia caused by intrahemocoelic infection. However, this did not exclude the possibility that these proteases contribute to virulence following this mode of infection. Indeed, InhA had a lethal effect following intrahemocoelic injection (33, 51). It has been shown that toxicity is a physiological consequence of a highly specific proteolytic action. Although the precise cause of the lethality was not elucidated, it is likely that the InhA metalloprotease interacts with a particularly important component of the host, leading to death. As illustrated by many examples (34, 38), various zinc metalloproteases from pathogens actively contribute to pathology. Indeed, in addition to damaging the host tissues during infection, these proteases act as toxic factors that degrade many blood components and enhance vascular permeability, cytotoxicity, etc. However, in most cases, attempts to evaluate the role of metalloprotease genes in virulence have failed to obtain conclusive results with respect to a major role in virulence for these metalloproteases (13, 25, 43, 50).
Acknowledgments
We thank Myriam Gominet and Leyla Slamti for their help with primer extension. We are grateful to Angela Jackson for carrying out the force-feeding assays, to Denis Bourguet and Michel Gohar for their help with the statistical analysis, and to Alex Edelman for English correction.
This work was supported by a grant from the Département de la Santé des Plantes et de l'Environnement (INRA). Sinda Fedhila was supported by a grant from the Tunisian government and by a grant from INRA.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Økstad, A. B. Kolstø, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse, H., and D. Lereclus. 1994. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J. Bacteriol. 176:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agaisse, H., and D. Lereclus. 1996. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20:633-643. [DOI] [PubMed] [Google Scholar]
- 4.Ashhurst, D. E. 1985. Connective tissues, p. 249-284. In G. A. Kerkut and L. I. Gilbert (ed.), Comprehensive insect physiology biochemistry and pharmacology, vol. 3. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 5.Boman, H. G., and D. Hultmark. 1987. Cell-free immunity. in insects Annu. Rev. Microbiol. 41:103-126. [DOI] [PubMed] [Google Scholar]
- 6.Bravo, A., H. Agaisse, S. Salamitou, and D. Lereclus. 1996. Analysis of cryIAa expression in sigE and sigK mutants of Bacillus thuringiensis. Mol. Gen. Genet. 250:734-741. [DOI] [PubMed] [Google Scholar]
- 7.Cascon, A., J. Yugueros, A. Temprano, M. Sanchez, C. Hernanz, J. M. Luengo, and G. Naharro. 2000. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 68:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalhammar, G., and H. Steiner. 1984. Characterization of inhibitor A, a protease from Bacillus thuringiensis which degrades attacins and cecropins, two classes of antibacterial proteins in insects. Eur. J. Biochem. 139:247-252. [DOI] [PubMed] [Google Scholar]
- 9.de Bentzmann, S., M. Polette, J. M. Zahm, J. Hinnrasky, C. Kileztky, O. Bajolet, J. M. Klossek, A. Filloux, A. Lazdunski, and E. Puchelle. 2000. Pseudomonas aeruginosa virulence factors delay airway epithelial wound repair by altering the actin cytoskeleton and inducing overactivation of epithelial matrix metalloproteinase-2. Lab. Investig. 80:209-219. [DOI] [PubMed] [Google Scholar]
- 10.Donovan, W. P., J. C. Donovan, and J. T. Engleman. 2001. Gene knockout demonstrates that vip3A contributes to the pathogenesis of Bacillus thuringiensis toward Agrotis ipsilon and Spodoptera exigua. J. Invertebr. Pathol. 78:45-51. [DOI] [PubMed] [Google Scholar]
- 11.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edlund, T., I. Siden, and H. G. Boman. 1976. Evidence for two immune inhibitors from Bacillus thuringiensis interfering with the humoral defense system of saturniid pupae. Infect. Immun. 14:934-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein, R. A., M. Boesman-Finkelstein, Y. Chang, and C. C. Hase. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, United Kingdom.
- 15.Gibson, T. J. 1984. Ph.D thesis. University of Cambridge, Cambridge, United Kingdom.
- 16.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 17.Grandvalet, C., V. de Crecy-Lagard, and P. Mazodier. 1999. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol. Microbiol. 31:521-532. [DOI] [PubMed] [Google Scholar]
- 18.Grandvalet, C., M. Gominet, and D. Lereclus. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805-1813. [DOI] [PubMed] [Google Scholar]
- 19.Harrington, D. J. 1996. Bacterial collagenase and collagen-degrading enzymes and their potential role in human disease. Infect. Immun. 64:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hase, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heierson, A., I. Sidén, A. Kivaisi, and H. G. Boman. 1986. Bacteriophage-resistant mutants of Bacillus thuringiensis with decreased virulence in pupae of Hyalophora cecropia. J. Bacteriol. 167:18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoch, J. A. 1993. Regulation of the onset of the stationary phase and sporulation in Bacillus subtilis. Adv. Microb. Physiol. 35:111-133. [DOI] [PubMed] [Google Scholar]
- 23.Hultmark, D., A. Engstrom, H. Bennich, R. Kapur, and H. G. Boman. 1982. Insect immunity: isolation and structure of cecropin D and four minor antibacterial components from Cecropia pupae. Eur. J. Biochem. 127:207-217. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki, S., M. Miyasono, M. Yamamoto, K. Ohba, T. Ishiguro, R. Takeda, and Y. Hayashi. 1992. Induction of antibacterial activity against Bacillus thuringiensis in the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Appl. Entomol. Zool. 27:565-570. [Google Scholar]
- 25.Jeong, K. C., H. S. Jeong, J. H. Rhee, S. E. Lee, S. S. Chung, A. M. Starks, G. M. Escudero, P. A. Gulig, and S. H. Choi. 2000. Construction and phenotypic evaluation of a Vibrio vulnificus vvpE mutant for elastolytic protease. Infect. Immun. 68:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. E., and W. H. McGaughey. 1996. Contribution of Bacillus thuringiensis spores to toxicity of purified Cry proteins towards Indianmeal moth larvae. Curr. Microbiol. 33:54-59. [DOI] [PubMed] [Google Scholar]
- 27.Jongeneel, C. V., J. Bouvier, and A. Bairoch. 1989. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 242:211-214. [DOI] [PubMed] [Google Scholar]
- 28.Lecadet, M. M., M. O. Blondel, and J. Ribier. 1980. Generalized transduction in Bacillus thuringiensis var. berliner 1715, using bacteriophage CP54 Ber. J. Gen. Microbiol. 121:203-212. [DOI] [PubMed] [Google Scholar]
- 29.Lereclus, D., H. Agaisse, M. Gominet, and J. Chaufaux. 1995. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Bio/Technology 13:67-71. [DOI] [PubMed] [Google Scholar]
- 30.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2749-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lereclus, D., O. Arantes, J. Chaufaux, and M.-M. Lecadet. 1989. Transformation and expression of a cloned [pdiff]-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 60:211-218. [DOI] [PubMed] [Google Scholar]
- 32.Li, R. S., P. Jarrett, and H. D. Burges. 1987. Importance of spores, crystals, and [pdiff]-endotoxins in the pathogenicity of different varieties of Bacillus thuringiensis in Galleria mellonela and Pieris brassicae. J. Invertebr. Pathol. 50:277-284. [Google Scholar]
- 33.Lövgren, A., M. Zhang, A. Engström, G. Dalhammar, and R. Landén. 1990. Molecular characterization of immune inhibitor A, a secreted virulence protease from Bacillus thuringiensis. Mol. Microbiol. 4:2137-2146. [DOI] [PubMed] [Google Scholar]
- 34.Maeda, H. 1996. Role of microbial proteases in pathogenesis. Microbial. Immunol. 40:685-699. [DOI] [PubMed] [Google Scholar]
- 35.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyasono, M., S. Inagaki, M. Yamamoto, K. Ohba, T. Ishiguro, R. Takeda, and Y. Hayashi. 1994. Enhancement of δ-endotoxin activity by toxin-free spore of Bacillus thuringiensis against the diamonback moth, Plutella xylostella. J. Invertebr. Pathol. 63:111-112. [Google Scholar]
- 37.Miyoshi, S., H. Nakazawa, K. Kawata, K. Tomochika, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 66:4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 39.Moran, C. P. 1993. RNA polymerase and transcription factors, p. 653-667. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 40.Morishima, I., S. Suginaka, T. Bougaki, M. Inoue, and T. Ueno. 1988. Induction and partial characterization of antibacterial proteins in the hemolymph of the silkworm Bombyx mori. Agric. Biol. Chem. 52:929-934. [Google Scholar]
- 41.Morishima, I., S. Suginaka, T. Ueno, and H. Hirano. 1990. Isolation and structure of cecropins, inducible antibacterial peptides, from the silkworm Bombyx mori. Comp. Biochem. Physiol. 95:551-554. [DOI] [PubMed] [Google Scholar]
- 42.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172:824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogierman, M. A., A. Fallarino, T. Riess, S. G. Williams, S. R. Attridge, and P. A. Manning. 1997. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J. Bacteriol. 179:7072-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Økstad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A.-B. Kolstø. 1999. Sequence analysis of three Bacillus cereus loci under PlcR virulence gene regulator control. Microbiology 145:3129-3138. [DOI] [PubMed] [Google Scholar]
- 45.Raymond, M., G. Prato, and D. Ratsira. 1993. PROBIT analysis of mortality assays displaying quantal response. Licence L93019. Avenix, St George d'Orques, France.
- 46.Salamitou, S., F. Ramisse, M. Brehelin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 47.Sanchis, V., H. Agaisse, J. Chaufaux, and D. Lereclus. 1996. Construction of new insecticidal Bacillus thuringiensis recombinant strains by using the sporulation non-dependent expression system of cryIIIA and a site specific recombination vector. J. Biotechnol. 48:81-96. [DOI] [PubMed] [Google Scholar]
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao, C.-P., and L.-I. Hor. 2000. Metalloprotease is not essential for Vibrio vulnificus virulence in mice. Infect. Immun. 68:3569-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siden, I., G. Dalhammar, B. Telander, H. G. Boman, and H. Somerville. 1979. Virulence factors in Bacillus thuringiensis: purification and properties of a protein inhibitor of immunity in insects. J. Gen. Microbiol. 114:45-52. [DOI] [PubMed] [Google Scholar]
- 52.Spiegelman, G. B., T. H. Bird, and V. Voon. 1995. Transcription regulation by the Bacillus subtilis response regulator Spo0A, p. 159-179. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 53.Steiner, H. 1985. Role of the exoprotease InA in the pathogenicity of Bacillus thuringiensis in pupae of Hyalophora cecropia. J. Invertebr. Pathol. 46:346-347. [Google Scholar]
- 54.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5′-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 55.Villafane, R., D. H. Bechhofer, C. S. Narayanan, and D. Dubnau. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wretlind, B., and O. R. Pavlovskis. 1983. Pseudomonas aeruginosa elastase and its role in pseudomonas infections. Rev Infect. Dis. 5(Suppl. 5):S998-S1004. [DOI] [PubMed] [Google Scholar]
- 57.Yusuke, K., T. Kiyoko, H. Hirohiko, and Y. Minoru. 1993. Expression and characterization of cDNAs for cecropin B, an antibacterial peptide from the silkworm. Bombyx mori. Insect Biochem. Mol. Biol. 23:285-290. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, M.-Y., A. Lövgren, M. G. Low, and R. Landén. 1993. Characterization of an avirulent pleitropic mutant of the insect pathogen Bacillus thuringiensis: reduced expression of flagellin and phospholipases. Infect. Immun. 61:4947-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]