Abstract
We had previously isolated a facultatively anaerobic hyperthermophilic archaeon, Pyrobaculum calidifontis strain VA1. Here, we found that strain VA1, when grown under aerobic conditions, harbors high catalase activity. The catalase was purified 91-fold from crude extracts and displayed a specific activity of 23,500 U/mg at 70°C. The enzyme exhibited a Km value of 170 mM toward H2O2 and a kcat value of 2.9 × 104 s−1·subunit−1 at 25°C. Gel filtration chromatography indicated that the enzyme was a homotetramer with a subunit molecular mass of 33,450 Da. The purified catalase did not display the Soret band, which is an absorption band particular to heme enzymes. In contrast to typical heme catalases, the catalase was not strongly inhibited by sodium azide. Furthermore, with plasma emission spectroscopy, we found that the catalase did not contain iron but instead contained manganese. Our biochemical results indicated that the purified catalase was not a heme catalase but a manganese (nonheme) catalase, the first example in archaea. Intracellular catalase activity decreased when cells were grown anaerobically, while under aerobic conditions, an increase in activity was observed with the removal of thiosulfate from the medium, or addition of manganese. Based on the N-terminal amino acid sequence of the purified protein, we cloned and sequenced the catalase gene (katPc). The deduced amino acid sequence showed similarity with that of the manganese catalase from a thermophilic bacterium, Thermus sp. YS 8-13. Interestingly, in the complete archaeal genome sequences, no open reading frame has been assigned as a manganese catalase gene. Moreover, a homology search with the sequence of katPc revealed that no orthologue genes were present on the archaeal genomes, including those from the “aerobic” (hyper)thermophilic archaea Aeropyrum pernix, Sulfolobus solfataricus, and Sulfolobus tokodaii. Therefore, KatPc can be considered a rare example of a manganese catalase from archaea.
In aerobic organisms, hydrogen peroxide is produced by various enzymatic reactions, particularly the univalent reduction of molecular oxygen by oxidases and the disproportionation of superoxide by superoxide dismutases. Hydrogen peroxide is a toxic molecule as it can both oxidize and reduce organic substrates in cells. Catalases (EC 1.11.1.6) remove this toxicity by catalyzing the disproportionation of hydrogen peroxide into molecular oxygen and water. Along with superoxide dismutases, catalases play an important role in defending the cell against oxidative stress, and are distributed in almost all aerobic and facultatively anaerobic organisms.
Catalases are assigned to three phylogenetically distinct groups: two groups comprised of heme catalases and one group of nonheme catalases (29). The monofunctional (or typical) catalases, which are one type of heme catalases, have been found in many bacteria, archaea, plants, fungi, and animals. They display a broad range of subunit sizes (55 to 84 kDa) and are generally tetrameric enzymes. Monofunctional catalases usually do not harbor peroxidase activity. Representative enzymes are bovine liver catalase (31) and Escherichia coli HP II (10). The bifunctional catalase-peroxidases, the other type of heme catalases, have been found in bacteria, archaea, and fungi. Besides their difference in size (subunit size, approximately 80 kDa), they exhibit not only catalase activity but also peroxidase activity. The catalase-peroxidases are not phylogenetically related to the monofunctional catalases but rather resemble plant and fungal peroxidases. A well-known enzyme is Mycobacterium tuberculosis KatG (46). The nonheme catalases, which are sometimes referred to as pseudocatalases, constitute one minor catalase group and have a relatively small subunit size (28 to 36 kDa). They utilize manganese ions instead of ferric heme in their active site and are therefore also known as manganese catalases.
Until now, only four manganese catalases, all from bacteria, had been purified and characterized (Lactobacillus plantarum [28], Thermus thermophilus [5], Thermus sp. YS 8-13 [20], and Thermoleophilum album [1]). The three-dimensional structures of manganese catalases from T. thermophilus (3) and L. plantarum (7) have been determined at high resolution. These two enzymes have been well characterized, but the primary structure of the T. thermophilus enzyme has not been entirely published. Recently, a manganese catalase was identified as a third catalase in a pathogenic bacterium, Salmonella enterica serotype Typhimurium, and found to be regulated by RpoS (σS) (36), but its biochemical characteristics have not been analyzed.
For archaea, several heme catalases have been characterized to date. A catalase-peroxidase and a monofunctional catalase were purified from the halophilic archaeon Halobacterium halobium (11, 12), and a catalase-peroxidase was also purified from Haloarcula marismortui (14). In methanogenic archaea, monofunctional catalases were purified from Methanosarcina barkeri (41) and Methanobrevibacter arboriphilus (42). In (hyper)thermophilic archaea, a putative catalase-peroxidase gene was found in the genome of the obligately anaerobic archaeon Archaeoglobus fulgidus (26). Its gene was expressed in E. coli, and the recombinant product was characterized (25). However, an open reading frame presumed to encode a catalase was not found in the genomes of the aerobic (hyper)thermophilic archaea Aeropyrum pernix (22), Sulfolobus solfataricus (40), and S. tokodaii (21). At present, manganese catalase has not been reported as being present in archaea.
A facultatively anaerobic hyperthermophilic archaeon, Pyrobaculum calidifontis strain VA1, was previously isolated (2). P. calidifontis VA1 grows optimally at 90 to 95°C and pH 7.0 under atmospheric air. Oxygen serves as a final electron acceptor under aerobic growth conditions, while oxygen can be replaced by nitrate under anaerobic conditions. Although thiosulfate could not serve as an electron acceptor, its presence in the medium significantly enhanced the growth of strain VA1 (2). In this study, catalase activity was detected in aerobically grown VA1 cells. Therefore, we purified and characterized the enzyme and also cloned the catalase gene. Interestingly, the enzyme did not turn out to be a heme catalase but was found to be a manganese catalase. Therefore, this study reports the first characterization of a manganese catalase from archaea.
MATERIALS AND METHODS
Strains, plasmids, phages, and culture conditions.
P. calidifontis VA1 is a facultatively anaerobic hyperthermophilic archaeon, isolated from a hot spring in the Philippines (2). For the purification of catalase, cells were cultivated in a medium containing the following (per liter): 10 g of tryptone, 1 g of yeast extract, and 3 g of Na2S2O3·5H2O. Cells were grown in silicone-plugged 500-ml Sakaguchi flasks containing 200 ml of medium and incubated at 90°C in an air bath shaker (120 rpm). Anaerobic culture conditions were obtained according to a technique described in reference 4. Vials (20 ml) (Maruemu, Osaka, Japan) containing 10 ml of medium were autoclaved and then transferred into an anaerobic chamber (Tabai Espec, Osaka, Japan) filled with an anoxic gas mixture (N2-CO2-H2 [90:5:5]). In the chamber, residual oxygen in the vial was reduced by adding 0.5 mM Na2S, and then the vial was sealed with a butyl rubber stopper (Maruemu) and an aluminum cap (Maruemu). Resazurin (1 mg/liter) was used as a redox indicator. Microaerobic growth conditions were obtained by omitting the addition of Na2S. The presence of oxygen was confirmed by the color of resazurin. E. coli DH5α and the vector pUC19 were used for cloning and gene manipulation. E. coli XL1-Blue MRA (P2) (Stratagene, La Jolla, Calif.) was used as a host strain for λEMBL3 phage (Stratagene). Luria-Bertani medium was used for cultivation of E. coli, and NZYM medium was used for amplification of phage (38).
Enzyme assays.
Catalase activity was determined spectrophotometrically by a UV-1600PC spectrophotometer with a thermal control unit (Shimadzu, Kyoto, Japan). Routine assays were performed at 70°C in 50 mM potassium phosphate buffer (pH 7.0) containing 20 mM hydrogen peroxide. Decomposition of hydrogen peroxide was monitored at 240 nm (ɛ240 = 43.6 M−1 cm−1 [18]) (8). One unit of catalase activity is defined as the amount of activity required to convert 1 μmol of hydrogen peroxide to water and oxygen per min. Activity measurements of the commercially available heme catalase from bovine liver (Sigma, St. Louis, Mo.) were performed at 25°C.
Peroxidase activity was measured spectrophotometrically by a UV-1600PC spectrophotometer with a thermal control unit (Shimadzu). Assays were performed at 70°C unless stated otherwise. The reaction mixture contained 50 mM potassium phosphate buffer (pH 7.0), 3 mM hydrogen peroxide, an electron donor, and an appropriate amount of enzyme. Pyrogallol was used at a concentration of 42 mM in the reaction mixture, and the oxidation was monitored at 430 nm. One unit of peroxidase activity for pyrogallol (ɛ430 = 2.47 mM−1 cm−1) is equivalent to 1 μmol of pyrogallol oxidized per min. The oxidation of guaiacol (42 mM) was monitored at 470 nm, that of p-phenylenediamine (0.3 mM) was monitored at 485 nm, and that of o-dianisidine (0.29 mM) was monitored at 460 nm. Diaminobenzidine was used at 0.5 mM in the presence of 0.1% gelatin in the reaction mixture (17), and oxidation was monitored at 465 nm. The oxidation of NADH or NADPH (0.2 mM) was monitored at 340 nm at 30°C, while that of glutathione (2 mM) was monitored at 340 nm by coupling with glutathione reductase (1 U per ml) in the presence of 0.2 mM NADPH at 30°C.
Purification of catalase.
Purification procedures of catalase were performed at room temperature unless mentioned otherwise. Harvested cells were suspended in 50 mM potassium phosphate buffer (pH 7.0). The cells were disrupted by sonication, and the supernatant was obtained by centrifugation (12,000 × g, 30 min, 4°C). The supernatant was subjected to ammonium sulfate fractionation at 0°C. The fraction corresponding to 60 to 95% saturation was collected and adjusted to 1.5 M (35% saturation) ammonium sulfate. The fraction was applied to a hydrophobic-interaction column, Resource PHE (Amersham Pharmacia Biotech, Uppsala, Sweden), equilibrated with 50 mM potassium phosphate buffer containing 1.5 M ammonium sulfate (pH 7.0), and eluted with a decreasing linear gradient of 1.5 to 0 M ammonium sulfate in 50 mM potassium phosphate buffer. The fractions containing catalase activity were pooled and dialyzed to remove the ammonium sulfate and to convert the buffer to 50 mM Tris-HCl (pH 9.0). The desalted fractions were applied to an anion-exchange column Resource Q (Amersham Pharmacia Biotech), equilibrated with 50 mM Tris-HCl (pH 9.0), and eluted with a linear gradient of 0 to 0.5 M KCl in 50 mM Tris-HCl (pH 9.0). The fractions containing catalase activity were pooled and concentrated by using Centricon-30 (Millipore, Bedford, Mass.). The enzyme solution was applied to a gel filtration column HiLoad 16/60 Superdex 200 pg (Amersham Pharmacia Biotech) equilibrated with 50 mM potassium phosphate buffer with 0.15 M KCl (pH 7.0) and eluted with the same buffer. The fractions containing catalase activity were desalted with 50 mM potassium phosphate buffer and used as purified enzyme in following experiments. The concentration of protein was determined by the Bio-Rad (Hercules, Calif.) protein assay system with bovine serum albumin as a standard. Homogeneity of the protein was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (38). The N-terminal amino acid sequence of the purified enzyme was determined by a protein sequencer Model 270 (Perkin-Elmer Applied Biosystems, Foster City, Calif.).
Molecular properties of catalase.
Gel filtration chromatography was used to determine the native molecular mass of the purified catalase. Experiments were performed at a flow rate of 0.5 ml/min on AKTÄ explorer 10S fast-performance liquid chromatography system (Amersham Pharmacia Biotech), using a Superdex 200 HR 10/30 column (Amersham Pharmacia Biotech). The buffer used was 50 mM potassium phosphate buffer (pH 7.0) containing 0.15 M KCl. The native molecular mass of the enzyme was estimated from a calibration curve plotted by using proteins from HMW and LMW gel filtration calibration kits (Amersham Pharmacia Biotech).
Metal contents (manganese, iron, and calcium) of the purified catalase were analyzed by plasma emission spectroscopy (ICPS-7000; Shimadzu).
Catalytic properties of catalase.
Optimum pH conditions for the purified catalase were determined by measuring the specific activity in the following buffers: 50 mM acetic acid-sodium acetate buffer (pH 4.0 to 5.5); 50 mM potassium phosphate buffer (pH 5.5 to 8.0); 50 mM Tris-HCl buffer (pH 7.5 to 9.0); 50 mM glycine-NaOH (pH 9.0 to 9.5); and 25 mM sodium hydrogen carbonate-sodium carbonate buffer (pH 9.5 to 10.0). The assay temperature was 70°C. For each buffer, the pH was adjusted so that the pH of the solution would attain the desired value at 70°C, using published values of ΔpH/ΔT (16). The results are shown as percentage ratios relative to the specific activity of the catalase in 50 mM glycine-NaOH buffer (pH 9.5).
In order to determine the optimum temperature for the purified catalase activity in 50 mM potassium phosphate buffer (pH 7.0), the reaction mixture was preincubated at the desired temperature for 3 min prior to measurements. Reaction temperatures were maintained during measurements with a thermal control unit (described above).
To determine thermostability, enzyme samples (0.1 mg/ml) were incubated at various temperatures in 50 mM potassium phosphate buffer (pH 7.0). At various intervals, aliquots of the enzyme were taken and assayed for residual activity in 50 mM potassium phosphate buffer (pH 7.0) at 70°C.
Isolation of P. calidifontis VA1 catalase-encoding gene (katPc gene).
A genomic library of P. calidifontis VA1 was prepared by ligating genomic DNA partially digested with Sau3AI into BamHI-digested arms of λEMBL3 (Stratagene). Two oligonucleotide primers were designed. One primer (5′-ATGTAYCTNAGRATHGAYCGNCTNCARAT-3′, where R is A or G, Y is C or T, H is A, C, or T, and N is A, T, G, or C) was derived from the determined N-terminal amino acid sequence of the purified enzyme, and the other primer (5′-YTCDATRTGNCCNAGYTCYTCNGT-3′, where R is A or G, Y is C or T, D is A, G, or T, and N is A, T, G, or C) was derived from a consensus sequence obtained from an alignment among manganese catalase genes and some orthologous genes. A DNA fragment containing a part of the katPc gene was obtained by PCR using these primers and P. calidifontis VA1 genomic DNA as a template. A phage clone, which carried the complete katPc gene, was screened from the genomic library by plaque hybridization using the DNA fragment as a probe.
DNA manipulation and sequencing.
Routine DNA manipulations were performed by standard methods (38). For isolation of plasmid and phage DNA, Plasmid Mini-, Midi- and Lambda Kits (Qiagen, Hilden, Germany) were used. Restriction enzymes and modifying enzymes were purchased from Toyobo (Osaka, Japan), Takara Shuzo (Kyoto, Japan), and Boehringer Mannheim (Mannheim, Germany). DNA sequencing on both strands of DNA was carried out using the ABI PRISM kit and Model 310 capillary DNA sequencer (Perkin-Elmer Applied Biosystems). Sequence data were analyzed and compared using GENETYX-WIN Version 4 software package (Software development, Tokyo, Japan). The multiple alignment of amino acid sequences was performed by the program ALIGN contained within the CLUSTAL W program (44) provided by DNA Data Bank of Japan (DDBJ).
Nucleotide sequence accession number.
The katPc gene sequence is available under accession no. AB076020 in the GenBank, EMBL, and DDBJ databases.
RESULTS
Catalase activity in P. calidifontis VA1 cells grown under various culture conditions.
P. calidifontis VA1 was cultivated under various culture conditions, and catalase activities in the respective cell extracts were measured. Anaerobically or microaerobically grown cells showed low levels of catalase activity (≤10 U/mg). Cells grown under atmospheric air, but without shaking, also displayed similar levels of catalase activity. However, when cells were grown under atmospheric air and vigorous shaking, the cell extracts exhibited 230 to 270 U of catalase activities per mg. Therefore, P. calidifontis VA1 harbors a catalase that is induced when grown under aerobic conditions.
Purification of the catalase.
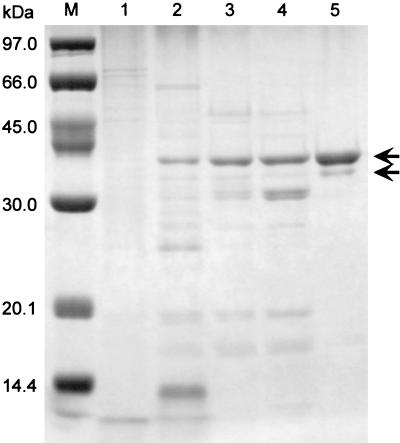
P. calidifontis VA1 cells grown under atmospheric air with vigorous shaking were used to purify the catalase. The catalase activity in the cell extract displayed a specific activity of 259 U/mg. Catalase was purified from the cell extract 91-fold by ammonium sulfate fractionation, followed by hydrophobic-interaction, anion-exchange, and gel filtration column chromatography (Table 1 and Fig. 1). The N-terminal amino acid sequence of the purified enzyme was MYLRIDRLQIQLPA(P/D)KE(P/Y)D. It showed significant homology (79% identity) to the N-terminal amino acid sequence of the manganese catalase from Thermus sp. YS 8-13 (accession no. AB008786). One minor band was observed below the major band in Fig. 1, lane 5. The N-terminal amino acid sequence of the minor band (MYLRI) was identical to that of the major band. Furthermore, the relative densities of the two bands differed under various denaturing conditions, indicating that the bands represented different denaturation states of the same polypeptide. We have previously encountered many cases in which thermostable proteins showed aberrant migration rates on SDS-PAGE gels, due to incomplete denaturation of the protein in SDS gel-loading buffer (32). The purified catalase (KatPc) exhibited a specific activity of 23,500 U/mg at 70°C.
TABLE 1.
Purification of P. calidifontis VA1 catalase
| Method | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 174,000 | 670 | 259 | 100 | 1 |
| Ammonium sulfate (60-95%) | 101,000 | 132 | 762 | 58 | 3 |
| Resource PHE | 76,500 | 24 | 3,150 | 44 | 12 |
| Resource Q | 64,100 | 11 | 5,960 | 37 | 23 |
| Superdex 200 | 30,500 | 1.3 | 23,500 | 18 | 91 |
FIG. 1.
SDS-PAGE of samples during the purification process of the P. calidifontis VA1 catalase. Lane M, molecular mass markers; lane 1, crude extract (5 μg); lane 2, ammonium sulfate fraction (5 μg); lane 3, peak fraction after hydrophobic-interaction chromatography (5 μg); lane 4, flowthrough fraction after anion-exchange chromatography (5 μg); lane 5, purified enzyme obtained from gel filtration chromatography (5 μg). The arrows indicate the major and minor bands described in the text.
Subunit assembly.
The subunit composition of the purified enzyme was determined by gel filtration chromatography. The native enzyme had a molecular mass of 154 kDa (data not shown). The subunit molecular mass was estimated to be approximately 38 kDa by SDS-PAGE (Fig. 1) and 33,450 Da from the deduced amino acid sequence of its gene (see below). These results suggested that P. calidifontis VA1 catalase was a homotetrameric enzyme.
Metal content.
Typical heme catalases have a specific absorbance at 407 nm (Soret band), which is due to the heme prosthetic group. The UV and visible light spectrum of P. calidifontis VA1 catalase (250 to 700 nm; not shown) displayed only a single peak at 280 nm, without a Soret band. We further examined the metal content of the catalase by plasma emission spectroscopy. We could not detect any iron in the catalase. However, manganese was present at 1.32 ± 0.03 atoms per subunit (a subunit molecular mass of 33,450 Da was used to calculate the metal content). These results suggested that P. calidifontis VA1 catalase was not a heme catalase but a manganese catalase.
Comparison with heme catalase.
Most catalases contain a heme group at their active site. As most heme enzymes are sensitive to low concentrations of azide or cyanide, an easy method to distinguish heme and manganese catalases is to examine the inhibitory effects of azide or cyanide on the catalase activity. We investigated azide sensitivity of P. calidifontis VA1 catalase, compared with the heme catalase from bovine liver (Sigma). When aliquots of the heme catalase from bovine liver were assayed in the presence of sodium azide at concentrations ranging from 0 to 2 μM, a drastic decrease in activity was observed (e.g., 40% decrease in the presence of 1 μM sodium azide). In contrast, the P. calidifontis VA1 catalase was hardly inhibited in the same concentration range. Ninety percent of activity could still be detected in the presence of 1,000 μM sodium azide. Although a manganese catalase has not been reported to be present in archaea, our biochemical results clearly indicate that the catalase from P. calidifontis VA1 is a manganese catalase.
Enzymatic properties.
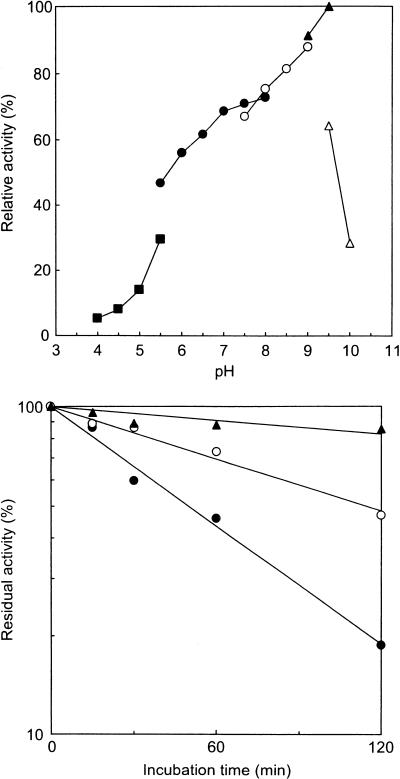
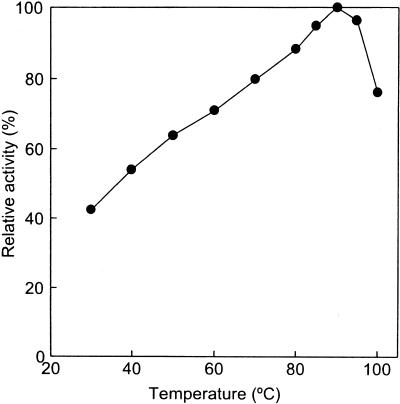
Measurements of catalase activity were performed at various pHs and temperatures. Catalase activities were detected from pH 4.0 to 10.0 (Fig. 2A). The highest activity was detected in glycine-NaOH buffer, pH 9.5. The catalase displayed significant activity from 30 to 100°C, with an optimal temperature of 90°C (Fig. 2B). In order to determine the thermostability of the catalase, aliquots of the enzyme (0.1 mg/ml) were incubated at various temperatures. The residual activities after heat treatment were assayed at 70°C (Fig. 2C). The half-lives of the catalase were 50, 114, and 432 min at 100, 95, and 90°C, respectively.
FIG. 2.
(A) Optimum pH of the P. calidifontis VA1 catalase. The buffers used for each pH range were 50 mM acetic acid-sodium acetate buffer (pH 4.0 to 5.5, closed squares); 50 mM potassium phosphate buffer (pH 5.5 to 8.0, closed circles); 50 mM Tris-HCl buffer (pH 7.5 to 9.0, open circles); 50 mM glycine-NaOH (pH 9.0 to 9.5, closed triangles); and 25 mM sodium hydrogen carbonate-sodium carbonate buffer (pH 9.5 to 10.0, open triangles). The assay temperature was 70°C. The results are shown as percentage ratios relative to the specific activity of the catalase in 50 mM glycine-NaOH buffer (pH 9.5). (B) Optimum temperature of the P. calidifontis VA1 catalase. (C) Thermostability of the P. calidifontis VA1 catalase at various temperatures. Closed circles, incubation at 100°C; open circles, incubation at 95°C; closed triangles, incubation at 90°C.
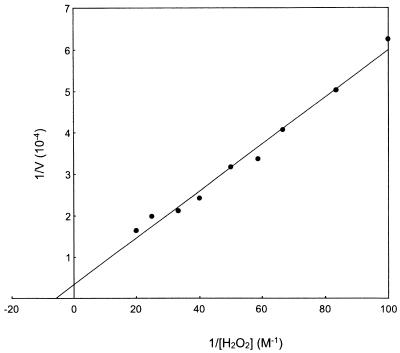
In order to determine the kinetic parameters of the purified catalase, we investigated enzyme activity at various concentrations of H2O2. An assay was performed at 25°C in order to accurately compare enzymatic properties with those of previously reported manganese and heme catalases. The enzyme followed Michaelis-Menten kinetics, and from the Lineweaver-Burk plot (Fig. 3), we deduced a Km value of 170 mM and a kcat value of 2.9 × 104 s−1·subunit−1.
FIG. 3.
Lineweaver-Burk plot of the P. calidifontis VA1 catalase with the substrate hydrogen peroxide. Activity measurements were performed at 25°C for comparison with other manganese and heme catalases.
Peroxidase activities were tested for various substrates including pyrogallol, guaiacol, p-phenylenediamine, o-dianisidine, diaminobenzidine, NADH, NADPH, and glutathione. No peroxidase activity was detected when guaiacol, p-phenylenediamine, o-dianisidine, diaminobenzidine, NADH, NADPH, or glutathione was used as a substrate under our assay conditions. However, the P. calidifontis VA1 catalase displayed peroxidase activity toward pyrogallol with a specific activity of 604 U/mg at 70°C.
Effects of manganese and hydrogen peroxide on intracellular catalase activity.
As described above, the catalase activity in the cell extracts of strain VA1 was induced under aerobic cultivation with vigorous shaking. In order to gain further insight on the factors that affect the intracellular levels of catalase, we measured the activity in the cells grown with additional compounds such as hydrogen peroxide and manganese. It was previously reported that the removal of thiosulfate, a possible antioxidant, from the medium led to a drastic decrease in cell growth (2). Here, we measured the catalase activity in these cells and found a 3.4-fold-greater induction in intracellular activity in cells grown without thiosulfate (869 U/mg) compared to that in cells grown with thiosulfate (253 U/mg). We then added hydrogen peroxide, paraquat (methyl viologen, a well-known reductor of oxygen to superoxide anion), or manganese ions in the medium without thiosulfate. Growth was not observed with the addition of 100 μM hydrogen peroxide, 1 μM paraquat, or 10 μM manganese chloride. We were able to measure catalase activity in cells grown in the presence of 10 μM hydrogen peroxide, 0.1 μM paraquat, or 1 μM manganese chloride. No influence on catalase activity was observed with 10 μM hydrogen peroxide (850 U/mg) or 0.1 μM paraquat (876 U/mg). However, addition of 1 μM manganese led to a further increase in intracellular catalase activity (1,460 U/mg).
Catalase-encoding (katPc) gene.
In order to obtain a homologous probe for katPc gene encoding the catalase, two PCR primers were designed. One primer was designed from the determined N-terminal amino acid sequence of the purified P. calidifontis VA1 catalase. The other primer was designed from a consensus sequence (68TEELGHIE75 in Thermus sp. YS 8-13 manganese catalase) obtained from an alignment with manganese catalase genes and orthologous genes. A partial fragment (approximately 0.2 kbp) of katPc gene was amplified by PCR using these two primers and P. calidifontis VA1 genomic DNA as a template. After confirming the sequence, this amplified fragment was used as a probe in plaque hybridization.
A genomic library was constructed from P. calidifontis VA1 by using the λEMBL3 phage vector system. A phage clone, which carried the complete katPc gene, was identified from the genomic library by plaque hybridization using the DNA fragment as a probe. This phage DNA was digested with SalI and subcloned into pUC19. One of these plasmids containing a 3.5-kbp inserted fragment was found to include the entire katPc gene and was sequenced.
The katPc gene was composed of 897 bp and corresponded to a protein of 298 amino acids with a deduced molecular mass of 33,450 Da. This value was comparable with the apparent molecular mass of 38 kDa determined by SDS-PAGE for the purified catalase subunit. Furthermore, the N-terminal amino acid sequence of the purified catalase determined by Edman degradation was identical to the deduced sequence from the katPc gene.
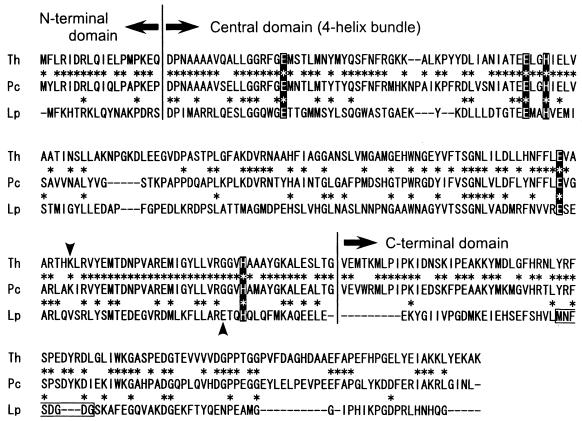
We compared the deduced amino acid sequence from the katPc gene with other previously reported manganese catalases (Fig. 4). It showed high similarity with a manganese catalase from a thermophilic bacterium, Thermus sp. YS 8-13 (61% identity). In contrast, it showed lower similarities with manganese catalases from mesophilic bacteria (L. plantarum, 24% identity; Salmonella enterica serotype Typhimurium, 19% identity). Although the complete primary structure has not been published, the three-dimensional structure of T. thermophilus catalase has been determined. The structure has revealed that Glu36, Glu70, His73, Glu155 and His188 were coordinated to manganese ions in the active site (6). These residues were conserved in the P. calidifontis VA1 catalase (Fig. 4).
FIG. 4.
Amino acid sequence alignment of KatPc (Pc) with manganese catalases from Thermus sp. YS 8-13 (Th, accession no. AB008786) and L. plantarum (Lp, accession no. D87070). Alignment was performed with CLUSTAL W. Conserved residues are indicated with asterisks. Domain boundaries were determined by using the structure of the T. thermophilus catalase (3). Residues highlighted with a solid background are residues coordinated with manganese ions. The lysine (Pc and Th) or glutamate (Lp) residues indicated by arrowheads are considered to contribute to catalytic proton transfer. The calcium binding site of the catalase from L. plantarum is boxed.
DISCUSSION
In this report, we describe the first characterization of an archaeal manganese catalase, KatPc, from a hyperthermophilic archaeon, Pyrobaculum calidifontis VA1. This was supported by (i) the absence of a Soret band; (ii) the lack of inhibition by sodium azide; (iii) the presence of manganese, but not iron, indicated by plasma emission spectroscopy; and (iv) the primary structure of the protein. During the purification procedure, we could not detect any additional protein fractions with catalase activity. Furthermore, no decrease in catalase activity in the cell extracts was observed after addition of sodium azide (data not shown). These observations strongly indicate that the manganese catalase purified in this study is the only catalase in P. calidifontis VA1.
The subunit molecular mass of the catalase was estimated to be approximately 38 kDa by SDS-PAGE (Fig. 1) and calculated to be 33,450 Da from the deduced amino acid sequence of the katPc gene. The native molecular mass was estimated to be 154 kDa by gel filtration chromatography. Therefore, KatPc was considered to be a homotetramer. This is also the case for the enzyme from T. album, which is a homotetrameric enzyme composed of 34-kDa subunits (1). In contrast, manganese catalases from L. plantarum (28.3 kDa) (28), T. thermophilus (35 kDa) (5), and Thermus sp. (36 kDa) (20) are hexameric enzymes.
KatPc exhibited a specific activity of 23,500 U/mg at 70°C. This value was comparable to the specific activities of other manganese catalases (L. plantarum, 7,800 U/mg at 25°C [9]; Thermus sp. YS 8-13, 8,000 U/mg at 65°C [20]; T. album, 17,745 U/mg at 25°C [1]). Kinetic analysis of KatPc revealed that the enzyme had a kcat value of 2.9 × 104 s−1·subunit−1 and a Km value of 170 mM toward H2O2. These parameters were also comparable to those of previously reported manganese catalases (Table 2). In general, heme catalases can be regarded as superior catalysts, with a kcat value of approximately 107 s−1·subunit−1, and a kcat/Km value of approximately 4 × 108 (45).
TABLE 2.
Kinetic parameters of manganese catalases
Previously reported crystal structures of manganese catalases indicated the presence of two manganese atoms per subunit in the catalytic centers (3, 6, 7). In this study, plasma emission spectroscopy detected only 1.32 ± 0.03 atoms per subunit in the P. calidifontis VA1 catalase. This low value may be due to a partial loss of manganese ions during purification or systematic errors of plasma emission spectroscopy and/or protein concentration assay. Another factor may be the presence of catalase apoenzymes in the cells without the proper coordination of manganese ions. Although cell growth was low, we observed a 1.7-fold increase in intracellular catalase activity when 1 μM Mn2+ was added to the medium in the absence of thiosulfate. This may reflect an increase of the amount of holoenzyme in the cells, as well as an induction in gene expression. This aberrantly low value has also been observed in previous studies using atomic absorption spectroscopy. In these cases, although crystal structures reveal the presence of 2 manganese atoms/subunit, manganese contents of manganese catalases were 1.12 ± 0.37 atoms/subunit (mean ± standard deviation) (L. plantarum [28]), 1.4 ± 0.4 atoms/subunit (T. album [1]), and 1.2 atoms/subunit (Thermus sp. [20]). It is reasonable to assume that the catalase from P. calidifontis VA1 also contains two manganese atoms per subunit.
Previously reported three-dimensional structures of manganese catalases reveal that the enzymes are composed of three distinct structural units: an N-terminal polypeptide, a central four-helix bundle that serves as the scaffolding for the catalytic active site, and a C-terminal tail (7). The structure of the central domain, including the amino acid residues coordinated to the manganese atoms, is relatively well conserved (Fig. 4). However, while a lysine residue (Lys162) was indicated to contribute to catalytic proton transfer in the T. thermophilus enzyme (3), this residue was found to be functionally substituted by a glutamate residue (Glu178) in the L. plantarum enzyme (7). From sequence alignment (Fig. 4), we have found that the lysine residue, and not the glutamate residue, was conserved in the P. calidifontis VA1 enzyme. Concerning the C-terminal domain, the primary structure of VA1 catalase resembled that of the T. thermophilus enzyme and was distinct from that of the L. plantarum enzyme. The L. plantarum catalase has been reported to contain one calcium atom per subunit in this domain (7), while calcium was not found in the T. thermophilus enzyme (3). As plasma emission spectroscopy revealed the absence of calcium in the VA1 enzyme, the P. calidifontis VA1 catalase may harbor a structure similar to that of the T. thermophilus catalase in this domain.
Some catalases are known to harbor peroxidase activity in addition to their catalase activity (29). We found that the P. calidifontis catalase exhibited peroxidase activity toward pyrogallol. Among previously reported manganese catalases, the T. album enzyme displayed peroxidase activity specific to p-phenylenediamine (1), while no peroxidase activity was detected in the L. plantarum enzyme (27). The presence and absence of peroxidase activity among manganese catalases have been proposed to reflect structural differences in the vicinity of the active center. In particular, Leu154 and Gly185 in the Thermus catalase were conserved as Leu151 and Gly182 in the P. calidifontis VA1 catalase. These residues are supposed to generate an additional short, wide solvent channel to the protein surface in the Thermus enzyme that is not present in the L. plantarum catalase (7).
In archaea, several heme-containing catalases have been characterized. (11, 12, 14, 41, 42), but there have been no previous reports of a manganese catalase. Recently, an abundant number of complete genome sequences from various archaeal strains have been determined. Therefore, we searched the database of each archaeal genome sequenced to date for both heme-containing and manganese catalases. To our surprise, we found very few archaeal genomes with catalase orthologues. One catalase-peroxidase gene was found in the entire genome of the anaerobic hyperthermophilic archaeon A. fulgidus (26), and characterization of the recombinant protein has been reported (25). Another catalase-peroxidase gene was found in the genome of the mesophilic halophilic archaeon Halobacterium sp. NRC-1 (33). However, no catalase orthologue gene was found in all other genomes, including those of the (hyper)thermophilic archaea Methanobacterium thermoautotrophicum (43), Methanococcus jannaschii (13), Pyrococcus horikoshii (23), Pyrococcus abyssi (http://www.genoscope.cns.fr/Pab/), Thermoplasma acidophilum (37), and Thermoplasma volcanium (24). Moreover, the complete genomes of the “aerobic” (hyper)thermophilic archaea A. pernix (22), S. solfataricus (40), and S. tokodaii (21) also did not harbor putative catalase genes in spite of having superoxide dismutases which produced hydrogen peroxides. Jenny et al. found superoxide reductase activity in the anaerobic hyperthermophilic archaeon Pyrococcus furiosus, which reduces superoxide to hydrogen peroxide without forming molecular oxygen (19). The enzyme was considered a key component of a novel oxidative stress protection system. In this system, rubredoxin oxidoreductase or neelaredoxin was considered to detoxify superoxide as a superoxide reductase (30). Superoxide dismutase or superoxide reductase orthologues were found in all of the archaeal complete genome sequences. However, putative catalase genes or rubrerythrin genes, which are considered to detoxify hydrogen peroxide (15, 30), were not found in the genomes of aerobic hyperthermophile A. pernix (22) and facultatively anaerobic thermophiles T. acidophilum (37) and T. volcanium (24), although these organisms have superoxide dismutase genes. They might withstand hydrogen peroxide stress using other peroxiredoxin systems (e.g., alkyl hydroperoxide reductase) (35). As at present, the hyperthermophilic archaeon P. calidifontis VA1 is the only archaeal strain that seems to harbor a manganese catalase, the strain can be considered a valuable tool for investigating the physiological roles and evolution of (manganese) catalases. The presence of the gene in P. calidifontis VA1 may also represent an interesting example of lateral gene transfer. Studies on the physiological roles and biochemical characteristics of the catalase are under way.
REFERENCES
- 1.Allgood, G. S., and J. J. Perry. 1986. Characterization of a manganese-containing catalase from the obligate thermophile Thermoleophilum album. J. Bacteriol. 168:563-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amo, T., M. L. F. Paje, A. Inagaki, S. Ezaki, H. Atomi, and T. Imanaka. Pyrobaculum calidifontis sp. nov., a novel hyperthermophilic archaeon which grows under atmospheric air. Archaea, in press. [Online.] http://www.archaea.ws/. [DOI] [PMC free article] [PubMed]
- 3.Antonyuk, S. V., V. R. Melik-Adamyan, A. N. Popov, V. S. Lamzin, P. D. Hempstead, P. M. Harrison, P. J. Artymyuk, and V. V. Barynin. 2000. Three-dimensional structure of the enzyme dimanganese catalase from Thermus thermophilus at 1 Å resolution. Crystallography Rep. 45:105-116. [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barynin, V. V., and A. I. Grebenko. 1986. T-catalase is nonheme catalase of extremely thermophilic bacterium Thermus thermophilus HB-8. Dolk. Akad. Nauk. USSR 286:461-464. (In Russian.) [Google Scholar]
- 6.Barynin, V. V., P. D. Hempstead, A. A. Vagin, S. V. Antonyuk, W. R. Melik-Adamyan, V. S. Lamzin, P. M. Harrison, and P. J. Artymiuk. 1997. The three-dimensional structure of the di-Mn catalase and the environment of the di-Mn sites in different redox states. J. Inorg. Biochem. 67:196. [Google Scholar]
- 7.Barynin, V. V., M. M. Whittaker, S. V. Antonyuk, V. S. Lamzin, P. M. Harrison, P. J. Artymiuk, and J. W. Whittaker. 2001. Crystal structure of manganese catalase from Lactobacillus plantarum. Structure 9:725-738. [DOI] [PubMed] [Google Scholar]
- 8.Beers, R. F., Jr., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. [PubMed] [Google Scholar]
- 9.Beyer, W. F., Jr., and I. Fridovich. 1985. Pseudocatalase from Lactobacillus plantarum: evidence for a homopentameric structure containing two atoms of manganese per subunit. Biochemistry 24:6460-6467. [DOI] [PubMed] [Google Scholar]
- 10.Bravo, J., M. J. Mate, T. Schneider, J. Switala, K. Wilson, P. C. Loewen, and I. Fita. 1999. Structure of catalase HPII from Escherichia coli at 1.9 Å resolution. Proteins 34:155-166. [DOI] [PubMed] [Google Scholar]
- 11.Brown-Peterson, N. J., and M. L. Salin. 1993. Purification of a catalase-peroxidase from Halobacterium halobium: characterization of some unique properties of the halophilic enzyme. J. Bacteriol. 175:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown-Peterson, N. J., and M. L. Salin. 1995. Purification and characterization of a mesohalic catalase from the halophilic bacterium Halobacterium halobium. J. Bacteriol. 177:378-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H. P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 14.Cendrin, F., H. M. Jouve, J. Gaillard, P. Thibault, and G. Zaccai. 1994. Purification and properties of a halophilic catalase-peroxidase from Haloarcula marismortui. Biochim. Biophys. Acta 1209:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Coulter, E. D., N. V. Shenvi, and D. M. Kurtz, Jr. 1999. NADH peroxidase activity of rubrerythrin. Biochem. Biophys. Res. Commun. 255:317-323. [DOI] [PubMed] [Google Scholar]
- 16.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for Biochemical Research, 3rd ed. Clarendon Press, Oxford, United Kingdom.
- 17.Herzog, V., and H. D. Fahimi. 1973. A new sensitive colorimetric assay for peroxidase using 3,3′-diaminobenzidine as hydrogen donor. Anal. Biochem. 55:554-562. [DOI] [PubMed] [Google Scholar]
- 18.Hildebrandt, A. G., and I. Roots. 1975. Reduced nicotinamide adenine dinucleotide phosphate (NADPH)-dependent formation and breakdown of hydrogen peroxide during mixed function oxidation reactions in liver microsomes. Arch. Biochem. Biophys. 171:385-397. [DOI] [PubMed] [Google Scholar]
- 19.Jenney, F. E., Jr., M. F. J. M. Verhagen, X. Cui, and M. W. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 20.Kagawa, M., N. Murakoshi, Y. Nishikawa, G. Matsumoto, Y. Kurata, T. Mizobata, Y. Kawata, and J. Nagai. 1999. Purification and cloning of a thermostable manganese catalase from a thermophilic bacterium. Arch. Biochem. Biophys. 362:346-355. [DOI] [PubMed] [Google Scholar]
- 21.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, K. Kubota, Y. Nakamura, N. Nomura, Y. Sako, and H. Kikuchi. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101, 145-152. [DOI] [PubMed] [Google Scholar]
- 23.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, T. Yoshizawa, Y. Nakamura, F. T. Robb, K. Horikoshi, Y. Masuchi, H. Shizuya, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 24.Kawashima, T., N. Amano, H. Koike, S. Makino, S. Higuchi, Y. Kawashima-Ohya, K. Watanabe, M. Yamazaki, K. Kanehori, T. Kawamoto, T. Nunoshiba, Y. Yamamoto, H. Aramaki, K. Makino, and M. Suzuki. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. USA 97:14257-14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kengen, S. W. M., F. J. Bikker, W. R. Hagen, W. M. de Vos, and J. van der Oost. 2001. Characterization of a catalase-peroxidase from the hyperthermophilic archaeon Archaeoglobus fulgidus. Extremophiles 5:323-332. [DOI] [PubMed] [Google Scholar]
- 26.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 27.Kono, Y., and I. Fridovich. 1983. Inhibition and reactivation of Mn-catalase. J. Biol. Chem. 258:13646-13648. [PubMed] [Google Scholar]
- 28.Kono, Y., and I. Fridovich. 1983. Isolation and characterization of the pseudocatalase of Lactobacillus plantarum. J. Biol. Chem. 258:6015-6019. [PubMed] [Google Scholar]
- 29.Loewen, P. C., M. G. Klotz, and D. J. Hassett. 2000. Catalase--an “old” enzyme that continues to surprise us. ASM News 66:76-82. [Google Scholar]
- 30.Lumppio, H. L., N. V. Shenvi, A. O. Summers, G. Voordouw, and D. M. Kurtz, Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melik-Adamyan, W. R., V. V. Barynin, A. A. Vagin, V. V. Borisov, B. K. Vainshtein, I. Fita, M. R. Murthy, and M. G. Rossmann. 1986. Comparison of beef liver and Penicillium vitale catalases. J. Mol. Biol. 188:63-72. [DOI] [PubMed] [Google Scholar]
- 32.Nakatani, M., S. Ezaki, H. Atomi, and T. Imanaka. 2000. A DNA ligase from a hyperthermophilic archaeon with unique cofactor specificity. J. Bacteriol. 182:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. H. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penner-Hahn, J. E. 1992. Structural properties of the manganese site in the manganese catalases, p. 29-45. In V. L. Pecoraro (ed.), Manganese redox enzymes. VCH, New York, N.Y.
- 35.Poole, L. B., C. M. Reynolds, Z. A. Wood, P. A. Karplus, H. R. Ellis, and M. Li Calzi. 2000. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267:6126-6133. [DOI] [PubMed] [Google Scholar]
- 36.Robbe-Saule, V., C. Coynault, M. Ibanez-Ruiz, D. Hermant, and F. Norel. 2001. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σS). Mol. Microbiol. 39:1533-1545. [DOI] [PubMed] [Google Scholar]
- 37.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Shank, M., V. Barynin, and G. C. Dismukes. 1994. Protein coordination to manganese determines the high catalytic rate of dimanganese catalases. Comparison to functional catalase mimics. Biochemistry 33:15433-15436. [DOI] [PubMed] [Google Scholar]
- 40.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Y. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shima, S., A. Netrusov, M. Sordel, M. Wicke, G. C. Hartmann, and R. K. Thauer. 1999. Purification, characterization, and primary structure of a monofunctional catalase from Methanosarcina barkeri. Arch. Microbiol. 171:317-323. [DOI] [PubMed] [Google Scholar]
- 42.Shima, S., M. Sordel-Klippert, A. Brioukhanov, A. Netrusov, D. Linder, and R. K. Thauer. 2001. Characterization of a heme-dependent catalase from Methanobrevibacter arboriphilus. Appl. Environ. Microbiol. 67:3041-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J. I. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoder, D. W., J. Hwang, and J. E. Penner-Hahn. 2000. Manganese catalases. Met. Ions Biol. Syst. 37:527-557. [PubMed] [Google Scholar]
- 46.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]