Abstract
Cloning and sequencing of the telomere of Streptomyces griseus revealed five palindromic sequences in the terminal 116 nucleotides, all of which can make a hairpin loop structure. However, the end sequence cannot form the foldback secondary structure that is common in Streptomyces telomeres and is suggested to be necessary for terminal replication. Both inside ends of the terminal inverted repeat (TIR) were also cloned and sequenced. The results confirmed the size of the TIR to be 24 kb and identified two almost identical open reading frames that might have been involved in the formation of the TIR.
Streptomyces species are gram-positive soil bacteria with high G+C base compositions (70 to 74%) in their DNAs. They display complex morphological differentiations and produce many secondary metabolites, including medically useful antibiotics. Moreover, all of the Streptomyces species studied hitherto carry an approximately 8-Mb linear chromosome (14, 16, 18, 19, 23). Streptomyces are also known to frequently possess large linear plasmids (11). Both the linear chromosomes and plasmids contain principally similar structural features, namely, terminal proteins bound to the 5′ ends and terminal inverted repeats (TIRs) bound at both ends.
Cohen and his colleagues (1, 25) studied the replication mechanisms of Streptomyces linear plasmids pSLA2 and pSCL. They showed that the plasmid DNA replicates bidirectionally from a replication origin near the center of the molecule. Replication in the leading strand is completed at the 3′ end, but the lagging strand does not reach the 5′ end and leaves a 280-bp gap (1), which was speculated to be filled in by protein-primed DNA synthesis (24). Chen (2) proposed that a foldback secondary structure formed at the 3′ protruding end is involved in the patching of the single-stranded gap. Huang et al. (8) sequenced the telomeric regions of a total of six linear chromosomes and plasmids from Streptomyces species. All of the first 166 to 168 nucleotide (nt) sequences are similar to each other and can form a foldback structure with Y-shaped loops or a hairpin loop at the 3′ ends. Qin and Cohen (21) showed experimentally that replication at the telomere of pSLA2 was consistent with this model and was abolished by the elimination of even 4 nucleotides (nt) from the end.
We have been studying the linear chromosome of Streptomyces griseus, including its physical structure, deletion, and circularization (10, 16, 17). However, the nucleotide sequence of the telomere has not been clarified. To answer the question of whether the foldback secondary structure is always present in Streptomyces telomeres, we cloned and sequenced the telomere of Streptomyces griseus. Both inside ends of the TIR were also cloned and sequenced, which confirmed the size of the TIR and allowed us to formulate a hypothesis on its formation mechanism.
DNA preparation.
The protease-treated total DNA was prepared as follows. The mycelium from a 50-ml culture was suspended in 10 ml of TSE-sucrose medium (30 mM Tris, 50 mM NaCl, 5 mM EDTA [pH 8.0], 10.3% sucrose) to which 3 ml of lysozyme (5 mg/ml in TSE-sucrose) was added, and the mixture was incubated at 37°C for 30 min. Two milliliters of Actinase E (5 mg/ml in TSE medium; Kaken Pharmaceuticals, Tokyo, Japan) was added and the mixture was incubated at 37°C for 1 h, and then 2 ml of 10% sodium dodecyl sulfate was added and the mixture was shaken for an additional 30 min. The mixture was extracted with phenol, incubated with RNase (100 μg/ml) at 37°C for 1 h, and extracted again with phenol-chloroform. For the DNA not treated with protease, the Actinase E digestion step was omitted and phenylmethylsulfonyl fluoride (final concentration, 2 μg/ml) was added before the sodium dodecyl sulfate treatment to inhibit the strong intrinsic protease activity of S. griseus.
Cloning and analysis of the telomere.
We previously constructed a cosmid library for S. griseus strain 2247 (16) and aligned cosmids in some hundreds of kilobases at both chromosomal ends (17). In the terminal cosmid maps, cosmids 9D2 and 6E12 were located at the leftmost and rightmost positions, respectively, and the latter was found to be about 300 bp from the right end (17). The SalI end fragment was subcloned from cosmid 6E12 and named p6E12R (Fig. 1A). After subjecting the total DNA digest to probing using p6E12R, a 2.7-kb SalI fragment was deduced to be the extreme end fragment of the chromosome. Hybridization was carried out using the DIG system (Roche Diagnostics GmbH, Mannheim, Germany) overnight at 70°C in standard buffer.
FIG. 1.
Restriction (A) and Southern hybridization (B and C) analysis of the telomere of the S. griseus 2247 chromosome. (A) The restriction map of the chromosomal end. All recognition sites are shown for DraI, EcoRI, KpnI, SacI, and SalI, while only the closest sites to the end are depicted for Eco52I, ApaI, and Sau3AI. The positions of the KpnI end fragment and p6E12R are also indicated. (B) The protease-treated and nontreated total DNAs were digested with SalI plus AflII, separated on a 0.7% agarose gel, and probed by p6E12R. (C) Total DNA and the pSGE1 DNA were each digested with SacI, KpnI, ApaI, and Eco52I, separated on a 2.0% agarose gel, and probed by the KpnI end fragment. For the ApaI and Eco52I digestions, SacI and KpnI, respectively, were added to cut the multicloning site in pUC19.
To clone the extreme end of the chromosome, we first isolated an 11-kb AflII fragment, which was deduced to be the end fragment and was completely separated from bigger fragments in conventional agarose gel electrophoresis (16). Since the 11-kb AflII fragment is located inside of the 24-kb TIR region, it was impossible to distinguish the right and left end fragments. The AflII fragment was digested with SalI and then the 2.7-kb end fragment was isolated and force cloned into pUC19 which had been predigested with SalI plus SmaI. In the force cloning (7, 12, 15), the 2.7-kb SalI end fragment, which after protease treatment should still have been carrying a small peptide covalently bound to the 5′ end, was directly ligated to the vector.
To eliminate the risk that the extreme end was cut with SalI or AflII before cloning, protease-treated and nontreated total DNAs were digested with SalI plus AflII and compared on agarose gel electrophoresis. When probed by p6E12R, the nontreated sample gave a retarded hybridizing signal compared with that of the protease-treated sample (Fig. 1B). Protein-bound DNAs remain at the loading well (16, 18) or move slowly (9) in agarose gel electrophoresis. The assay result shown in Fig. 1B confirmed that the sample used for cloning contained the protein-bound extreme end. Among about 1,000 ampicillin-resistant transformants, three colonies were selected by positive hybridization to p6E12R and named pSGE1, pSGE2, and pSGE3. Nucleotide sequencing showed that pSGE1 and pSGE2 had identical sequences beginning at the SmaI cloning site, while the corresponding sequence of pSGE3 was 10 nucleotides (nt) shorter.
To prove that the extreme end was cloned in pSGE1 and pSGE2, end fragments of the chromosome and pSGE1 were compared as follows. Total strain 2247 DNA and pSGE1 were each digested with SacI, KpnI, ApaI, and Eco52I. When pSGE1 was digested with ApaI and Eco52I, SacI and KpnI were added, respectively, because ApaI and Eco52I cannot cut the multicloning site of the vector. When probed by the KpnI end fragment of pSGE1 (Fig. 1A), each pair of 2247 and pSGE1 digests gave hybridizing signals at the same position (Fig. 1C).
This result suggested two possibilities: (i) the chromosomal end is located at the left end of these fragments (Fig. 1A) or (ii) the recognition sites for all of the four restriction endonucleases are located there. There is no chance that the latter possibility is the case, because four recognition sites cannot be accommodated within the detection limit of size differences (about 10 bp in Fig. 1C). Therefore, we concluded that the chromosomal end was cloned in pSGE1 and pSGE2 and that pSGE3 lost a 10-bp end DNA during the cloning process.
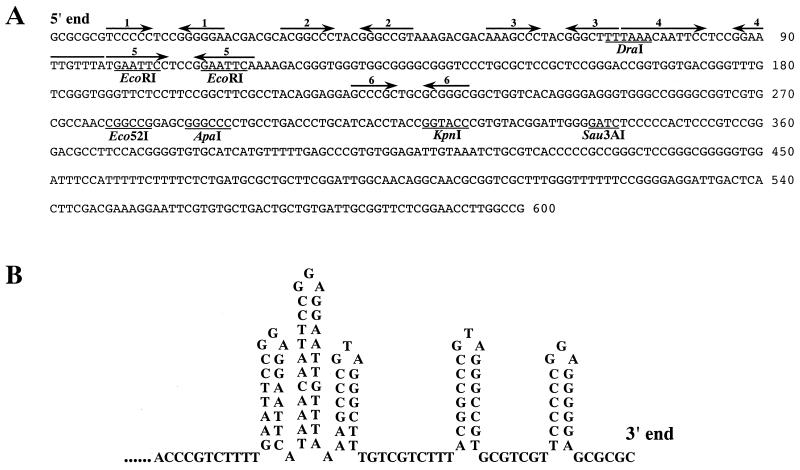
The 600-nt sequence of pSGE1 and pSGE2 is shown in Fig. 2A. Unexpectedly, a recognition site for a rare cutter DraI was found close to the end (nt 71 to 76), which added two DraI sites to the previously identified nine sites on the 2247 chromosome (16). Five palindromic sequences (I to V) were found in a 116-nt terminal sequence, all of which can form a hairpin loop structure as shown in Fig. 2B (the opposite 3′ protruding end is shown here). It is noteworthy that all six of the palindromes, including the separate palindrome VI, contain a possible sheared purine-purine pairing formed by the G and A residues and resulting in a single N-residue loop (4).
FIG. 2.
Nucleotide sequence of the 5′ end of the S. griseus 2247 chromosome (A) and five hairpin loop structures formed at the 3′ protruding end (B). (A) Palindromes I to VI (1 to 6) are indicated by paired arrows above the sequence. (B) The 3′ protruding end strand can form five hairpin loops but cannot make a foldback structure with Y-shaped loops or a hairpin loop. GGA and GTA residues are located at the centers of all of the five loops.
In the typical Streptomyces telomeres of the S. lividans chromosome and the linear plasmid SLP2, the terminal 167 nt contain seven palindromes, among which the endmost palindrome I can form a superpalindrome with the central 13 bp of palindrome IV and therefore can make a Y-shaped foldback structure together with palindromes II and III (8). This secondary structure has been suggested to play an important role in the initiation of terminal replication (8, 21). In contrast to these examples, the end sequence of the 2247 chromosome cannot form a foldback secondary structure.
We previously determined the terminal sequence of another linear plasmid, SCP1 (12), where a foldback secondary structure was likewise not possible. The end sequences of SCP1 and the 2247 chromosomes do not show any homology to the typical Streptomyces telomere sequences. We have also determined the telomere sequence of the linear plasmid pSLA2-L (7), which coexists with pSLA2-M and pSLA2-S (also known as pSLA2) in S. rochei 7434AN4 (13). It showed a similarity to the typical telomere sequences and can make a terminal foldback structure. Therefore, the foldback secondary structure is not absolutely necessary for terminal replication of Streptomyces linear genomes. The original protein-priming model reported by Salas (24) does not require it.
Cloning and analysis of the TIR.
Lezhava et al. (16) showed by restriction analysis that the S. griseus 2247 chromosome has a 24-kb TIR. In order to confirm the size of the TIR and to get a hint about its formation mechanism, cloning of both inside ends of the TIR was carried out. Previous studies revealed that the inside ends of the TIR were located on the 2.8-kb PstI fragment of the left end cosmid 9D2 and the 1.4-kb PstI fragment of the right end cosmid 6E12 (17). These two PstI fragments were subcloned to give plasmids pTL and pTR. Comparison of their restriction maps (Fig. 3A) located the left and right inside ends of the TIR on the 350-bp SacII-SmaI fragment and the 240-bp SacII fragment, respectively.
FIG. 3.
The restriction maps of pTL and pTR (A) and the nucleotide sequences (B) around the inside ends of TIR-L and TIR-R. (A) Comparison of the restriction maps of pTL and pTR revealed that the 350-bp SacII-SmaI fragment and the 240-bp SacII fragment carry the left and right inside ends of the TIR, respectively. (B) The nucleotide sequences of the 240-bp SacII fragment of pTR and the corresponding region of pTL are compared. The TIR regions are indicated by an arrow above the sequences, which extend beyond the SacII site to nt 205. Two bases are different between TIR-L and TIR-R, at nt 179 and 185. The amino acid sequence of an ORF, shown below the sequences, is directed rightward and stops at 2 nt from the inside end of the TIR.
The nucleotide sequences of these fragments were determined and are presented for comparison in Fig. 3B. The TIR regions extended 205 nt from the SacII site, which confirmed the size of the TIR to be 24 kb. Frame analysis revealed an open reading frame (ORF) in both TIR regions which started before the leftmost PstI site and stopped at 2 nt from the inside end of the TIR (Fig. 3B). Two nucleotides (nt 179 and 185) differed between these ORFs; this, however, does not affect their amino acid sequences. Homology searches of databases failed to find any proteins with significant homologies to these proteins.
Most of the linear chromosomes and plasmids so far isolated from Streptomyces have a TIR at both ends. The sizes of TIRs of linear plasmids are quite different, ranging from 44 bp for SLP2 in S. lividans (3) to 95 kb for pPZG101 in S. rimosus (6). However, Kalkus et al. (9) reported that pHG201 in Rhodococcus opacus has a TIR of only 3 bp, if a sequence of so short a length can be called a TIR. Pandza et al. (20) showed that recombination between pPZG101 and the linear chromosome of S. rimosus led to exchange of their ends. The hybrid plasmid generated seemed not to have a TIR based on its restriction map, although the possibility that an extremely short TIR was present could not be excluded. The sizes of Streptomyces linear chromosomes are also different, ranging from 24 kb for S. griseus (16) to 210 kb for S. ambofaciens (14).
The biological role and formation mechanism of TIRs have not been clarified. Fischer et al. (5) reported that recombination of two sigma factor-like genes caused chromosomal arm replacement and generated an extremely long TIR in S. ambofaciens. We also found chromosomal arm replacement in one of the deletion mutants of S. griseus (unpublished result). In the present study, two almost identical ORFs have been located at the inside ends of the TIR. These results suggest that chromosomal arm replacement frequently occurs in Streptomyces by recombination of two homologous ORFs and that this may be a general mechanism by which a TIR is produced.
This hypothesis was strongly supported by analysis of the available S. coelicolor genome sequence (Streptomyces coelicolor genome project; http://www.sanger.ac.uk/Projects/S_coelicolor/). Identical insertion sequence elements are located at the left and right inside ends of the TIR (TIR-L and TIR-R, respectively) of the S. coelicolor chromosome. The one at TIR-L is flanked by direct repeat sequences (CTTTTC) while that at TIR-R is flanked by different sequences (CTTTTC and CTTAAA). This fact indicates that TIR-R was formed by recombination of two identical insertion sequence elements which, however, carried different direct repeats, CTTTTC and CTTAAA.
Accumulated data suggest that TIR itself is not but the extreme end sequence is essential for terminal replication of Streptomyces linear genomes. The 144-nt telomeric sequence was sufficient for replication of the plasmids artificially constructed from pSLA2 (21). However, it may be possible that replication of a linear plasmid with a very short TIR is supported by another factor coded on the chromosome, as suggested for SLP2 by Redenbach et al. (22). Therefore, a final determination of the essential telomeric structure of Streptomyces linear genomes must wait for more comprehensive studies.
Acknowledgments
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Chang, P.-C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 2.Chen, C. W. 1996. Complications and implications of linear bacterial chromosomes. Trends Genet. 12:192-196. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. W., T.-W. Yu, Y.-S. Lin, H. M. Kieser, and D. A. Hopwood. 1993. The conjugative plasmid SLP2 of Streptomyces lividans is a 50 kb linear molecule. Mol. Microbiol. 7:925-932. [DOI] [PubMed] [Google Scholar]
- 4.Chou, S. H., L. Zhu, and B. R. Reid. 1997. Sheared purine-purine pairing in biology. J. Mol. Biol. 267:1055-1067. [DOI] [PubMed] [Google Scholar]
- 5.Fischer, G., T. Wenner, B. Decaris, and P. Leblond. 1998. Chromosomal arm replacement generated a high level of intraspecific polymorphism in the terminal inverted repeats of the linear chromosomal DNA of Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 95:14296-14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gravius, B., D. Glocker, J. Pigac, K. Pandza, D. Hranueli, and J. Cullum. 1994. The 387 kb linear plasmid pPZG101 of Streptomyces rimosus and its interaction with the chromosome. Microbiology 140:2271-2277. [DOI] [PubMed] [Google Scholar]
- 7.Hiratsu, K., S. Mochizuki, and H. Kinashi. 2000. Cloning and analysis of the replication origin and the telomeres of the large linear plasmid pSLA2-L in Streptomyces rochei. Mol. Gen. Genet. 263:3104-3110. [DOI] [PubMed] [Google Scholar]
- 8.Huang, C.-H., Y.-S. Lin, Y.-L. Yang, S.-W. Huang, and C. W. Chen. 1998. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 28:905-916. [DOI] [PubMed] [Google Scholar]
- 9.Kalkus, J., R. Menne, M. Reh, and H. G. Schlegel. 1998. The terminal structures of linear plasmids from Rhodococcus opacus. Microbiology 144:1271-1279. [DOI] [PubMed] [Google Scholar]
- 10.Kameoka, D., A. Lezhava, H. Zenitani, K. Hiratsu, M. Kawamoto, K. Goshi, K. Inada, H. Shinkawa, and H. Kinashi. 1999. Analysis of fusion junctions of circularized chromosomes in Streptomyces griseus. J. Bacteriol. 181:5711-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinashi, H. 1994. Linear plasmids from actinomycetes. Actinomycetologica 8:87-96. [Google Scholar]
- 12.Kinashi, H., M. Shimaji-Murayama, and T. Hanafusa. 1991. Nucleotide sequence analysis of the unusually long terminal inverted repeats of a giant linear plasmid, SCP1. Plasmid 26:23-130. [DOI] [PubMed] [Google Scholar]
- 13.Kinashi, H., E. Mori, A. Hatani, and O. Nimi. 1995. Isolation and characterization of large linear plasmids from lankacidin-producing Streptomyces species. J. Antibiot. 47:1447-1455. [DOI] [PubMed] [Google Scholar]
- 14.Leblond, P., G. Fischer, F.-X. Francou, F. Berger, M. Guerineau, and B. Decaris. 1996. The unstable region of Streptomyces ambofaciens includes 210-kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol. Microbiol. 19:261-271. [DOI] [PubMed] [Google Scholar]
- 15.Levings, C. S., III, and R. R. Sederoff. 1983. Nucleotide sequence of the S-2 mitochondrial DNA from the S cytoplasm of maize. Proc. Natl. Acad. Sci. USA 80:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lezhava, A., T. Mizukami, T. Kajitani, D. Kameoka, M. Redenbach, H. Shinkawa, O. Nimi, and H. Kinashi. 1995. Physical map of the linear chromosome of Streptomyces griseus. J. Bacteriol. 177:6492-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lezhava, A., D. Kameoka, H. Sugino, K. Goshi, H. Shinkawa, O. Nimi, S. Horinouchi, T. Beppu, and H. Kinashi. 1997. Chromosomal deletions in Streptomyces griseus that remove the afsA locus. Mol. Gen. Genet. 253:478-483. [DOI] [PubMed] [Google Scholar]
- 18.Lin, Y.-S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 19.Pandza, K., G. Pfalzer, J. Cullum, and D. Hranueli. 1997. Physical mapping shows that the unstable oxytetracycline gene cluster of Streptomyces rimosus lies close to one end of the linear chromosome. Microbiology 143:1493-1501. [DOI] [PubMed] [Google Scholar]
- 20.Pandza, S., G. Biukovic, A. Paravic, A. Dadbin, J. Cullum, and D. Hranueli. 1998. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 28:1165-1176. [DOI] [PubMed] [Google Scholar]
- 21.Qin, Z., and S. N. Cohen. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 28:893-903. [DOI] [PubMed] [Google Scholar]
- 22.Redenbach, M., A. Arnold, U. Rauland, and J. Cullum. 1994. Structural instability of the Streptomyces lividans 66 chromosome and related effects. Actinomycetologica 8:97-102. [Google Scholar]
- 23.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 24.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 25.Shiffman, D., and S. N. Cohen. 1992. Reconstitution of a Streptomyces linear replicon from separately cloned DNA fragments: existence of a cryptic origin of circular replication within the linear plasmid. Proc. Natl. Acad. Sci. USA 89:6129-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]