Abstract
The mitotic checkpoint ensures that chromosomes are divided equally between daughter cells and is a primary mechanism preventing the chromosome instability often seen in aneuploid human tumors. ZW10 and Rod play an essential role in this checkpoint. We show that in mitotic human cells ZW10 resides in a complex with Rod and Zwilch, whereas another ZW10 partner, Zwint-1, is part of a separate complex of structural kinetochore components including Mis12 and Ndc80–Hec1. Zwint-1 is critical for recruiting ZW10 to unattached kinetochores. Depletion from human cells or Xenopus egg extracts is used to demonstrate that the ZW10 complex is essential for stable binding of a Mad1–Mad2 complex to unattached kinetochores. Thus, ZW10 functions as a linker between the core structural elements of the outer kinetochore and components that catalyze generation of the mitotic checkpoint-derived “stop anaphase” inhibitor.
Introduction
Most chromosome imbalances during development cause embryonic lethality, and chromosome instability has been associated with tumorigenesis (Lengauer et al., 1998; Cohen, 2002). The mitotic checkpoint guards against such chromosome loss and aneuploidization by halting mitotic progression whenever as little as a single chromosome is not properly attached to a meiotic (Li and Nicklas, 1995) or mitotic spindle (Rieder et al., 1995; for review see Cleveland et al., 2003). The “stop anaphase” checkpoint signal is generated at individual unattached kinetochores, from which it diffuses into the cytoplasm to prevent activation of the anaphase promoting complex that would otherwise initiate anaphase onset and mitotic exit by destroying securin and cyclin B (Peters, 2002). The diffusible inhibitory complex is thought to consist of one or more combinations of the mitotic checkpoint proteins BubR1, Bub3, and Mad2 that sequester Cdc20, the obligatory activator of the anaphase promoting complex for recognition of mitotic substrates (Fang et al., 1998; Sudakin et al., 2001; Tang et al., 2001; Fang, 2002).
A role in generating this complex has been described for most proteins previously implicated in mitotic checkpoint control. Mad1 is required for the stable kinetochore association of a pool of Mad2 that in turn recruits and activates additional Mad2 molecules (Chen et al., 1998; Shah et al., 2004), and the kinesin-like microtubule motor centromere-associated protein (CENP)-E directly activates BubR1 kinase activity at the unattached kinetochore (Mao et al., 2003; Weaver et al., 2003). Two additional proteins, Zeste White 10 (ZW10) and Rough Deal (Rod), have been implicated in generating a sustained mitotic checkpoint; however, no molecular function in mitotic checkpoint signaling has thus far been suggested (Basto et al., 2000; Chan et al., 2000). It also remains unclear how any of these checkpoint components are connected to the underlying structural elements of the outer kinetochore.
ZW10 and Rod were originally identified in screens for mutations in Drosophila that interfere with the faithful transmission of chromosomes to daughter cells during mitosis (Smith et al., 1985; Karess and Glover, 1989), but orthologues have been recognized in C. elegans, A. thaliana, G. gallus, M. musculus, and humans (Starr et al., 1997; Chan et al., 2000; Okamura et al., 2001). The Drosophila larval neuroblasts of either mutant display identical phenotypes: anaphase figures with misaligned chromosomes and premature sister chromatid separation in cells treated with microtubule poisons (Williams et al., 1992; Scaerou et al., 1999). Several additional lines of evidence indicate that ZW10 and Rod function together. A Drosophila zw10;rod double mutant has a mitotic phenotype that is indistinguishable from the single mutants (Scaerou et al., 2001), and both proteins are found in a single complex in Drosophila embryos and human cells (Chan et al., 2000; Scaerou et al., 2001; Williams et al., 2003). In addition, ZW10 localization to kinetochores depends on Rod (Williams and Goldberg, 1994; Chan et al., 2000) and vice versa (Chan et al., 2000; Scaerou et al., 2001).
ZW10 has been implicated as a binding partner of several other proteins besides Rod. Immuno-purification of ZW10 from fly embryos isolated Zwilch, and a yeast-two-hybrid screen for human proteins that bind ZW10 identified human ZW10-interacting protein 1 (HZwint-1, hereafter referred to as Zwint-1; Starr et al., 2000; Williams et al., 2003). ZW10, Rod, and Zwilch are interdependent for kinetochore localization, can be found in a single complex in flies and human cells, and Drosophila mutants show identical mitotic phenotypes (Williams et al., 2003). No Drosophila Zwint-1 mutant has thus far been identified, but Zwint-1 is also localized to kinetochores, although it arrives there in prophase, before the arrival of ZW10 in early prometaphase (Starr et al., 2000). Another protein found to interact with ZW10 in the yeast-two-hybrid system is p50 dynamitin, a component of the cytoplasmic dynein activator dynactin (Starr et al., 1998). ZW10 can be found to interact with dynein weakly in Drosophila embryos (Williams et al., 2003), and ZW10 and Rod relocalization to Drosophila spindle microtubules during metaphase depends on functional dynein heavy chain (Wojcik et al., 2001).
ZW10 and Rod are essential for mitotic checkpoint signaling. Injection of antibodies to human ZW10 and Rod abrogates checkpoint function in human cells treated with nocodazole (Chan et al., 2000), and Drosophila mutant rod or zw10 neuroblasts are unable to elicit a checkpoint response induced by misattachment in an asp mutant strain or upon a colchicine-induced block in mitotic spindle assembly (Basto et al., 2000). Screening for mutations in alleles encoding putative chromosome instability genes in human colorectal cancers has recently identified mutations in ZW10, Rod, and Zwilch, hinting at the possibility that deregulation of ZW10 function may contribute to the chromosome instability phenotype of tumor cells (Wang et al., 2004b).
We now use immunodepletion of the ZW10–Rod complex from Xenopus egg extracts and reduction of ZW10 accumulation in mammalian cells to identify ZW10 as a bridge between the structural outer kinetochore—through its interaction with Zwint-1—and the mitotic checkpoint, where it recruits a stably bound Mad1–Mad2 complex to unattached kinetochores.
Results
ZW10 and Zwint-1 are part of distinct kinetochore subcomplexes
To purify native complexes of ZW10 or Zwint-1, cell lines were generated that stably express ZW10 or Zwint-1 fused to the localization and affinity purification (LAP) tag that contains enhanced YFP (EYFP) followed by TEV protease cleavage site and a peptide from S-protein (Cheeseman et al., 2004). Localization of the EYFP in either fusion protein in each of several independent clones verified the correct localization of the tagged fusions at the kinetochore in mitotic cells (Fig. 1, a and b). Immunoblotting revealed that ZW10LAPtag accumulated to levels comparable to that of endogenous ZW10 (Fig. 1 c). (Published antibodies to human Zwint-1 [Starr et al., 2000] failed to recognize the endogenous gene product by immunoblotting in our hands so the level of Zwint-1LAPtag relative to endogenous could not be determined.)
Figure 1.
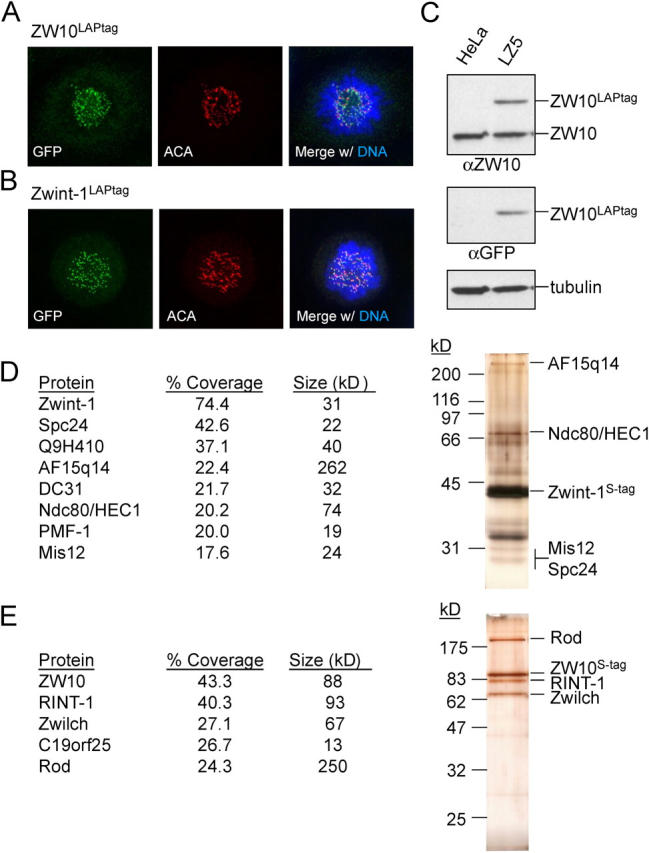
ZW10 and Zwint-1 reside in distinct kinetochore subcomplexes in HeLa cells. (A) Localization of ZW10LAPtag in a HeLa cell line stably expressing the fusion protein (clone LZ5). Cells were treated with nocodazole for 30 min before fixation, and stained for ZW10LAPtag (anti-GFP), centromeres (ACA), and DNA (DAPI). (B) Localization of Zwint-1LAPtag in clone LINT2.8. Treatment and staining was performed as in A, except cells were preextracted before fixation. (C) Immunoblot of whole cell lysates of HeLa cells and clone LZ5. Lysates were probed for ZW10, ZW10LAPtag (anti-GFP), and tubulin. (D and E) Tandem affinity purification of Zwint-1LAPtag (D) and ZW10LAPtag (E) from mitotically arrested cells. 25% of eluate was analyzed by SDS-PAGE followed by silverstain and 75% was analyzed by MudPIT mass spectrometry. Name, percent sequence coverage, and expected molecular weight of the identified proteins are indicated in the table. Suspected position of the identified proteins on the silver stained gel are indicated on the right. Unlabeled bands on silverstain likely include DC31 (∼32 kD), Q9H410 (∼40 kD), and the nonspecific proteins HSP70 (∼70 kD) and α-tubulin (∼50 kD).
Mass spectrometric analysis of eluates of the tandem affinity purification of ZW10LAPtag and Zwint-1LAPtag from mitotically arrested cells showed that the two proteins resided in distinct kinetochore complexes. Zwint-1 associated with structural kinetochore components including Mis12, Ndc80–HEC1, Spc24, and AF15q14 (the human orthologue of C. elegans kinetochore-null-1 (KNL-1), hereafter referred to as KNL-1AF15q14), along with additional recently described kinetochore proteins (Q9H410, DC31, and PMF-1; Fig. 1 d; Cheeseman et al., 2004; Obuse et al., 2004). ZW10, however, resided in a complex with known interacting partners Rod and Zwilch (Fig. 1 e; Williams et al., 2003). In addition, Rad50-interactor-1, a protein proposed to bind ZW10 in interphase (Hirose et al., 2004), was present. So too was a novel, previously hypothetical protein FLJ36666/C19ORF25. Neither Rad50-interactor-1 nor C19ORF25 bound kinetochores when fused to EYFP (unpublished data), however, supporting the likelihood that they are part of an interphase ZW10-containing protein structure.
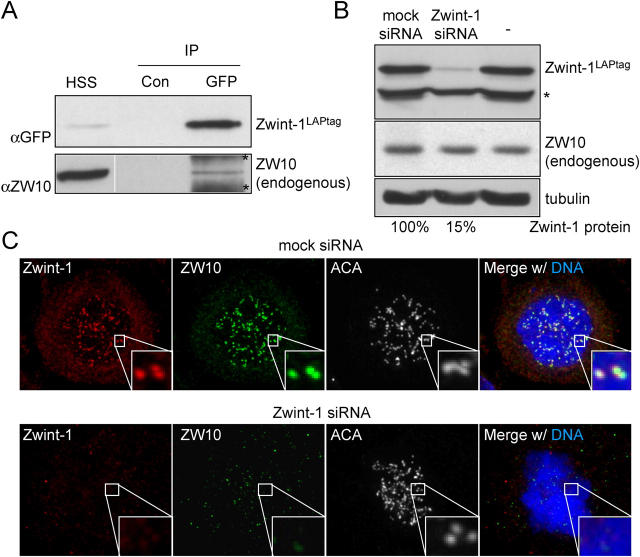
Zwint-1 was not found stably associated with our complexes containing ZW10 in the stringent LAPtag purifications. To test whether ZW10 and Zwint-1 interacted transiently or with a weak affinity, Zwint-1LAPtag was rapidly immunoprecipitated from mitotically arrested HeLa cells (clone LINT2.8; see Materials and methods) and tested for presence of endogenous ZW10. A small but significant fraction of ZW10 remained associated with Zwint-1 under these conditions (Fig. 2 a). Recombinant ZW10 also interacts with recombinant Zwint-1, and an association between the two can be found in human cultured cells that express high levels of epitope-tagged versions of both proteins (Wang et al., 2004a). That Zwint-1 is part of the structural kinetochore and binds ZW10 implies that Zwint-1 may recruit ZW10 to mitotic kinetochores. This was tested by reduction of Zwint-1 protein levels by plasmid-based small interfering RNA (siRNA) expression to ∼15% of Zwint-1 levels in cells transfected with mock siRNA (as indicated using an antibody to GFP for immunoblotting serial dilutions of LINT2.8 cell lysates expressing Zwint-1LAPtag; Fig. 2 b). Reduction of endogenous Zwint-1 yielded absence of endogenous ZW10 at kinetochores (Fig. 2 c). Because Zwint-1 apparently arrives at the kinetochore before ZW10 (Starr et al., 2000), and kinetochore localization of ZW10 depends on the presence of its Zwint-1 interaction domain (Wang et al., 2004a), these data reveal a crucial role for Zwint-1 in the recruitment of ZW10 to unattached kinetochores.
Figure 2.
Interaction between Zwint-1 and ZW10 controls ZW10 kinetochore localization. (A) Immunoblot of Zwint-1 immunoprecipitates shows weak interaction with ZW10. Cells of clone LINT2.8 were subjected to immunoprecipitation with control antibody (Con) or anti-GFP antibody to precipitate Zwint-1LAPtag and the precipitate was analyzed for the presence of endogenous ZW10. HSS, high speed supernatant before the immunoprecipitation. Bands labeled with asterisks are background due to precipitation from HSS with the anti-GFP antibody. White line indicates that intervening lanes have been spliced out. (B) Analysis of Zwint-1 knockdown efficiency by immunoblot using cells expressing Zwint-1LAPtag. Lysates of LINT2.8 cells untransfected or transfected with mock or Zwint-1 siRNA plasmid for 72 h were analyzed for Zwint-1LAPtag (anti-GFP), ZW10, and tubulin expression. Percentage of remaining protein was determined by serial dilution immunoblotting. Band labeled with asterisks is protein that cross reacts with anti-GFP in the LINT2.8 cell line. (C) Immunolocalization of ZW10 in cells depleted of endogenous Zwint-1. HeLa cells transfected as in B were treated with nocodazole for 30 min before fixation and stained for endogenous Zwint-1 and ZW10, and for centromeres (ACA) and DNA (DAPI).
The ZW10–Rod complex is essential for the mitotic checkpoint in vitro
To determine a mechanistic role for ZW10 and Rod in the mitotic checkpoint, a full-length cDNA of Xenopus ZW10 (X-ZW10) was isolated, as well as a 1836 bp fragment of Xenopus Rod encoding the carboxy-terminal 612 aa of X-Rod protein (X-RodCOOH; Fig. 3, a and b). (Despite extensive database searches [including the X. tropicalis genome] and repeated screening of multiple cDNA libraries, no additional X-Rod sequences could be identified.) Sequencing of X-ZW10 and X-RodCOOH revealed high sequence identity with their human orthologues (Fig. 3, a and b; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200411118/DC1). Both proteins were purified after expression using baculovirus-infected insect cells (Fig. 3 c) and used to generate pAbs. Affinity purified X-ZW10 and X-RodCOOH antibodies recognized ∼80- and ∼220-kD polypeptides in Xenopus oocyte extract, respectively (Fig. 3 d), as expected from the sizes of their orthologues in other species. Both were kinetochore associated in mitotic cultured Xenopus XL177 cells (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200411118/DC1) and at sister kinetochores of replicated sperm chromosomes in Xenopus oocyte extracts (Fig. 3 e). Although X-ZW10 could be only partially removed from Xenopus cytostatic factor (CSF) extracts by immunodepletion with X-ZW10 antibodies even after serial attempts, all of X-Rod was co-depleted and remained stably associated with X-ZW10 (Fig. 4 a), as was expected from our tandem affinity purification and previous studies (Chan et al., 2000; Scaerou et al., 2001). Conversely, the X-RodCOOH antibodies efficiently immunodepleted X-Rod, but again X-ZW10 was only partly co-depleted (Fig. 4 a). X-ZW10 remaining after immunodepletion with either antibody was undetectable at sperm kinetochores (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200411118/DC1). Thus, all X-Rod molecules present in the extract are bound to X-ZW10 and excess X-ZW10 not bound to X-Rod is unable to bind kinetochores.
Figure 3.
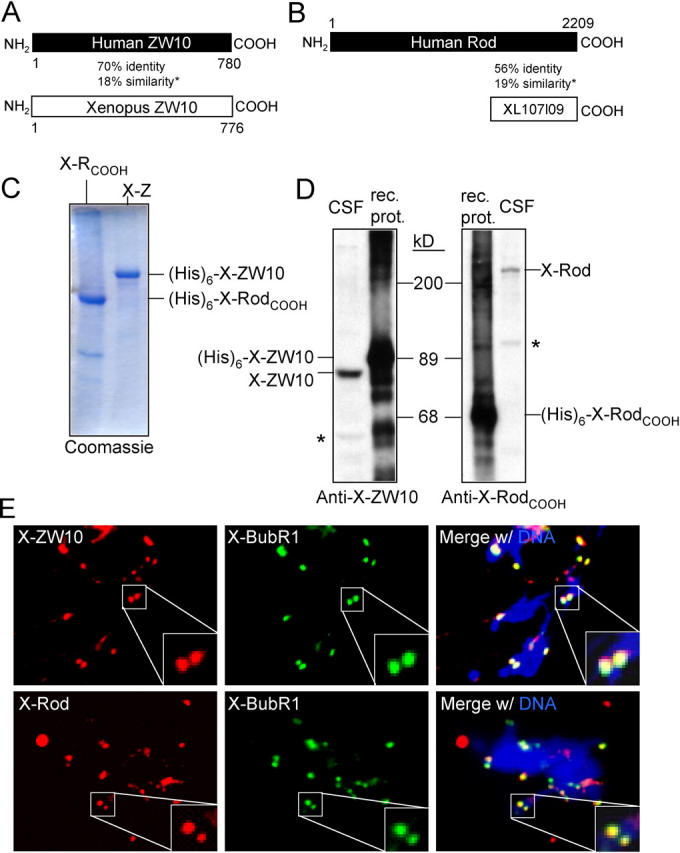
Characterization of Xenopus ZW10 and Rod. (A and B) Schematic alignment of Xenopus and human ZW10 (A) or Xenopus RodCOOH (XL107l09) and human Rod (B). Amino acid positions as well as percentage identity and additional (*) similarity on the protein level are indicated. (C) Coomassie staining of purified recombinant X-ZW10 (X-Z) and X-RodCOOH (X-RCOOH). (His)6-tagged proteins were purified from insect cells and analyzed by Coomassie blue staining. (D) Immunoblot analysis of pAbs to X-ZW10 and X-RodCOOH. 20 ng of recombinant protein (rec. prot.) and 1 μl CSF extract were analyzed by immunoblot with affinity purified anti–X-ZW10 (1348) or anti–X-RodCOOH (1351). Position of the endogenous frog proteins in the CSF extract is indicated. Cross-reacting proteins are marked by asterisks. (E) Immunolocalization of X-ZW10 and X-Rod. Sperm nuclei replicated in cycled CSF extract were immunostained for X-BubR1 and X-ZW10 or X-Rod. DNA (DAPI) is in blue. Enlarged boxes show overlap of X-BubR1 and X-ZW10–X-Rod signals on a sister kinetochore pair.
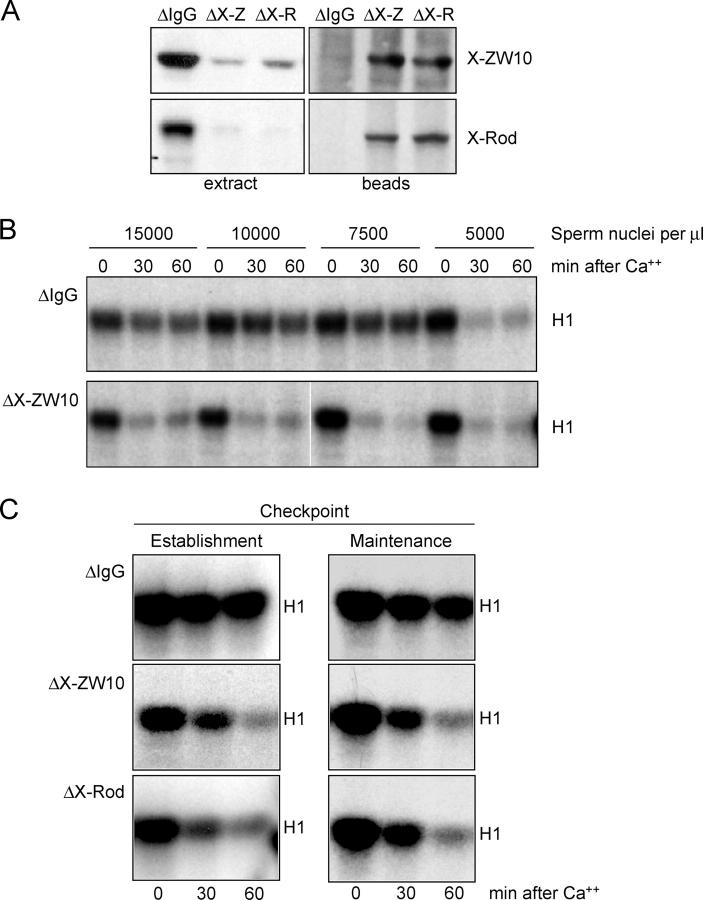
Figure 4.
The X-ZW10–X-Rod complex is essential for establishment and maintenance of the mitotic checkpoint. (A) Immunoblot of immunodepleted CSF extract. Extracts were depleted with anti–rabbit IgG (ΔIgG), anti–X-ZW10 (ΔX-Z), or anti–X-RodCOOH (ΔX-R). 1 μl of extract or the eluate of 1-μl beads from the immunodepletion were analyzed for X-ZW10–X-Rod levels. (B) Cdk1 kinase activity in Xenopus oocyte extracts. CSF extracts, mock depleted (ΔIgG) or depleted of the X-ZW10–X-Rod complex (ΔX-ZW10), were supplemented with nocodazole and the indicated amount of sperm nuclei and mitotic checkpoint activity was measured by the ability to maintain Cdk1 kinase activity toward histone H1 (H1) after inactivation of CSF by calcium for 0, 30, or 60 min. White lines indicate that intervening lanes have been spliced out. (C) CSF extracts were supplemented with sperm (15,000 per μl of extract) before (maintenance) or after (establishment) mock depletion (ΔIgG) or depletion of the X-ZW10–X-Rod complex (ΔX-ZW10 or ΔX-Rod). Mitotic checkpoint activity was measured as in B.
The X-ZW10–X-Rod complex was immunodepleted from CSF-arrested extracts and, after addition of nocodazole and various levels of sperm nuclei, the presence of an active mitotic checkpoint was measured by continued mitotic arrest with high levels of Cdk1 kinase activity upon inactivation of CSF with calcium (Chen and Murray, 1997). In contrast to mock-depleted extracts (ΔIgG), extracts depleted of the X-ZW10–X-Rod complex (Fig. 4 b, ΔX-ZW10 or Fig. 4 c, ΔX-Rod) were incapable of establishing and maintaining mitotic checkpoint signaling even in the presence of the highest concentration of unattached kinetochores (Fig. 4 b).
ZW10–Rod recruit the Mad1–Mad2 complex and BubR1 to kinetochores in vitro
To determine why extracts depleted of the ZW10–Rod complex were mitotic checkpoint-deficient, continued recruitment of various checkpoint proteins to the unattached kinetochore was compared in the presence or absence of X-ZW10–X-Rod. X-BubR1 colocalized with X-Rod and X-ZW10 on kinetochores as expected in mock-depleted extracts, but did not bind to kinetochores depleted of the X-ZW10–X-Rod complex (Fig. 5, a–c). Similarly, X-Mad1 was absent from X-ZW10–X-Rod–depleted kinetochores (Fig. 5, a–c), as was Mad2 (Fig. 5, a–c), whose recruitment to unattached kinetochores depends on Mad1 (Chen et al., 1998). The absence of all three of these proteins at kinetochores was not due to co-depletion with X-ZW10–X-Rod, as their protein levels in depleted versus control extracts were indistinguishable (Fig. 5 d). The absence at kinetochores was selective for components of the checkpoint signaling pathway: both the inner kinetochore histone H3 variant X-CENP-A (Maddox et al., 2003) and the kinetochore microtubule depolymerase X-KCM1 (also known as MCAK; Walczak et al., 1996) were present at undiminished levels at kinetochores after X-ZW10–X-Rod depletion (Fig. 5, a–c).
Figure 5.
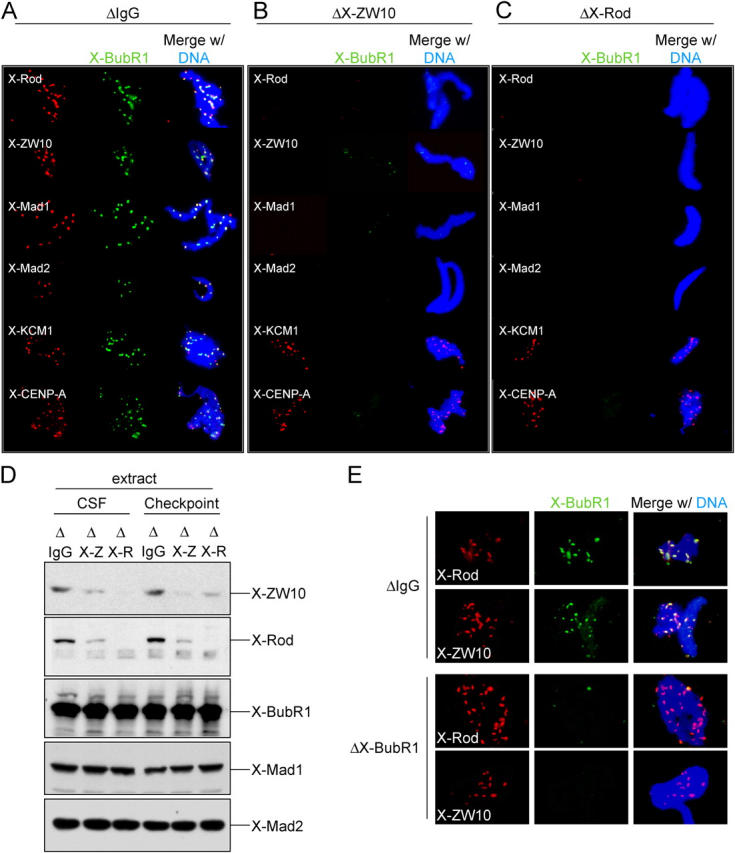
X-ZW10–X-Rod regulate kinetochore localization of X-BubR1, X-Mad1, and X-Mad2. (A–C) Immunolocalization of checkpoint proteins in depleted Xenopus extracts. Unreplicated sperm nuclei in mock (A, ΔIgG) or X-ZW10–X-Rod–depleted (B, ΔX-ZW10; or C, ΔX-Rod) extracts were immunostained with antibodies to the indicated proteins and X-BubR1. DNA (DAPI) is in blue. (D) Immunoblot of various checkpoint proteins in Xenopus extracts depleted of the X-ZW10–X-Rod complex. CSF extracts or checkpoint activated extracts were mock depleted (ΔIgG) or depleted of the X-ZW10–X-Rod complex (ΔX-Z or ΔX-R) and analyzed for presence of the indicated checkpoint proteins. (E) Immunolocalization of X-ZW10 and X-Rod in X-BubR1–depleted extracts. CSF extracts, mock depleted (ΔIgG) or depleted of X-BubR1 (ΔX-BubR1) were analyzed for kinetochore localization of X-ZW10 and X-Rod. DNA (DAPI) is in blue.
To distinguish between interdependency of checkpoint proteins for kinetochore localization and a requirement for the X-ZW10–X-Rod complex upstream of X-BubR1, X-Mad1, and X-Mad2, kinetochore localization of X-ZW10–X-Rod was determined after X-BubR1 depletion. Whereas removal of the X-ZW10–X-Rod complex mislocalized X-BubR1 (Fig. 5, b and c) and Mad2 (Mao et al., 2003), depletion of X-BubR1 had no effect on kinetochore binding of X-ZW10 and X-Rod (Fig. 5 e). Together, these data support a model in which the ZW10–Rod complex is essential for mitotic checkpoint establishment and maintenance by regulating the affinity of BubR1 and Mad1 (and thus Mad2) for unattached kinetochores.
ZW10 recruits Mad1–Mad2 to unattached human kinetochores
How ZW10 contributes to mitotic checkpoint signaling in human cells was determined by depletion of it using expression of siRNA duplexes. Knockdown was partial and heterogeneous at 2–3 d after transfection (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200411118/DC1). Significant reduction of ZW10 levels was apparent 5 d after transfection of HeLa cells, with no detectable levels of ZW10 at unattached mitotic kinetochores in ∼80% of cells (Fig. 6, a and b). Although most cells looked relatively healthy at this time point, a fraction was dying, most likely because they had sufficient knockdown at an earlier time and had undergone multiple divisions in the absence of ZW10 (see below). Phospho-histone H3 levels measured by flow cytometry in cells transfected with control siRNA constructs and treated with nocodazole for 16 h showed a 10-fold elevation in mitotic cells when compared with asynchronously growing cells (Fig. 6, c and d). In contrast, mitotic checkpoint signaling was severely compromised by reduction in ZW10, as shown previously for cells injected with anti-ZW10 antibodies (Chan et al., 2000), with the ZW10 depleted cell population yielding only a twofold increase in mitotic index (Fig. 6, c and d). Similar results were found in cells depleted of Zwint-1 (Fig. 6, c and d). Moreover, because ∼20% of the cells transfected with ZW10 siRNA had residual ZW10 at kinetochores, even this modest increase in mitotic index almost certainly reflected continued mitotic checkpoint signaling in the ZW10-containing proportion of cells and a complete absence of sustained checkpoint signaling in ZW10-depleted cells. ZW10-deficient cells underwent aberrant mitoses, which resulted in cell death after several divisions, as indicated by markedly diminished colony formation in continued presence of ZW10 siRNA (Fig. 6 e) and aberrant chromosome distribution yielding chromatin bridges and micronuclei (Fig. 6 f). This is consistent with a mitotic checkpoint defect, as the same effect was seen in HeLa cells with an inactive mitotic checkpoint after reduction in levels of BubR1 or Mad2 (Kops et al., 2004).
Figure 6.
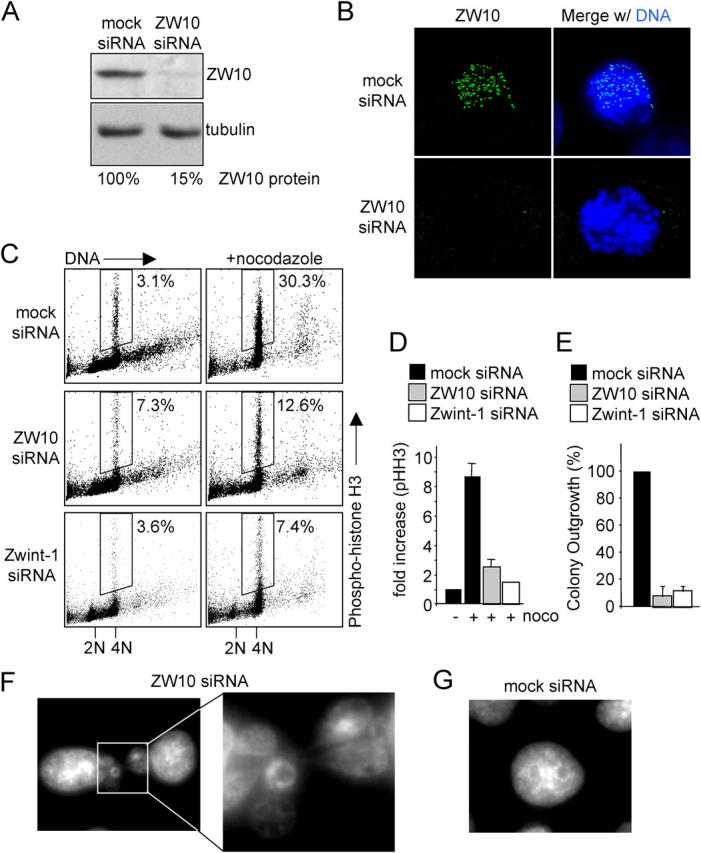
Human ZW10 is essential for mitotic checkpoint signaling. (A) Immunoblot analysis of ZW10 knockdown. HeLa cells were transfected with control or ZW10 siRNA duplexes. 5 d after transfection, total lysates were analyzed for ZW10 and tubulin protein. Percentage knockdown was determined by serial dilution immunoblotting. (B) Immunofluorescence analysis of ZW10 knockdown. Cells were transfected as in A, treated with nocodazole for 30 min before fixation and stained for ZW10 and DNA (DAPI). (C) Flow cytometric analysis of the fraction of phospho-histone H3-positive cells of mock siRNA, ZW10 siRNA and Zwint-1 siRNA cells 96 h after introduction of the siRNAs. Cells were transfected as in A, left untreated or treated with nocodazole for 16 h, and the entire population was analyzed for phospho-histone-H3 (y axis) and DNA (propidium iodide, x axis). Dot plots represent 4×104 cells for control siRNA and 104 cells for ZW10 or Zwint-1 siRNA. Percentages indicate fraction of cell population that is phospho-histone H3 positive. (D) Average fold increase of phospho-histone H3 staining after nocodazole treatment. Cells were transfected, treated, and analyzed as in C. Graph represents average of four independent experiments. (E) Graph of colony outgrowth assay. Cells were transfected as in A and retransfected for 7 d after which the colonies were stained and counted. Graph represents average of three experiments. (F and G) Aberrant mitosis in cells lacking ZW10. Cells were transfected as in A and stained with DAPI. Shown are typical interphase nuclei. (F) Cells transfected with ZW10 siRNA. (G) Cells transfected with control siRNA.
siRNA-mediated removal of ZW10 from unattached HeLa cell kinetochores resulted in mislocalization of dynein, but not core kinetochore structural components such as Mis12 or those recognized by anti-centromere auto-antibodies (unpublished data). As seen in Xenopus extracts, the Mad1–Mad2 heterodimer that stably associates with the unattached kinetochore and the dynamic Mad2 molecules that get recruited by the Mad1–Mad2 heterodimer were reduced >10-fold from unattached kinetochores in cells lacking ZW10 (Fig. 7, b, c, and e). This dependency on ZW10 was unique to Mad1–Mad2. As shown previously (Chan et al., 2000), association with unattached kinetochores of most other checkpoint components, including Bub1 (approximately twofold reduction; Fig. 7, d and e), BubR1 (Fig. 7 f), and CENP-E (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200411118/DC1) was not grossly affected by depletion of ZW10.
Figure 7.
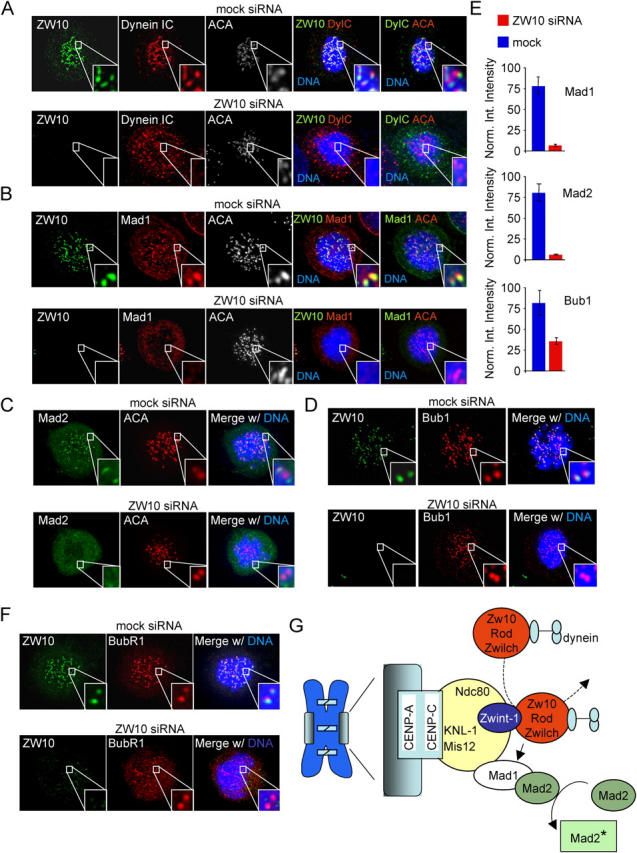
Mad1 and Mad2 are unable to bind kinetochores in absence of ZW10. (A–F) Immunolocalization of various proteins in ZW10-depleted cells. HeLa cells were transfected as in Fig. 6 A and treated with nocodazole 30 min before fixation. Cells were stained with ACA (A, B, and C), ZW10 (A, B, D, and F), and dynein intermediate chain (IC) (A), Mad1 (B), Mad2 (C), Bub1 (D), or BubR1 (F). DNA (DAPI) is in blue. Enlarged boxes show a pair of kinetochores. (E) Quantification of kinetochore fluorescence. Normalized integrated intensity (see Materials and methods) of Mad1, Mad2, and Bub1 is shown in mock siRNA and ZW10 siRNA cells. Error bars indicate the SD from measurements of three cells. (G) Model for function of the ZW10–Rod complex in mitotic checkpoint signaling. The ZW10-interactor Zwint-1 is in a structural kinetochore complex with Ndc80–HEC1 and Mis12 that is linked to the inner kinetochore by KNL-1AF15q14. The ZW10–Rod–Zwilch complex associates dynamically with the unattached kinetochore through interaction with Zwint-1, where it regulates kinetochore binding of the Mad1–Mad2 heterodimer and thus activation of Mad2.
Discussion
Our efforts here identify the ZW10–Rod complex as a bridge whose association with Zwint-1 links Mad1 and Mad2, components that are directly responsible for generating the diffusible “wait anaphase” signal, to a structural, inner kinetochore complex containing Mis12 and KNL-1AF15q14, the last of which has been proven essential for kinetochore assembly in C. elegans (Cheeseman et al., 2004). The presence of a stable complex of these components is supported by earlier affinity enrichments for Mis12 complexes which had linked it to nine other components, including Zwint-1 (Cheeseman et al., 2004; Obuse et al., 2004). Although a single affinity approach has proposed that a small proportion of two heterochromatin proteins, HP1α and HP1γ, are components of one or more Mis12 complexes, it is clear from our double affinity approach that neither of these markers for general heterochromatin is in a stable complex that includes Zwint-1. Like KNL-1AF15q14 and Mis12, Q9H410/c20orf172 and DC31/DC8 are constitutive kinetochore proteins (Cheeseman et al., 2004; Obuse et al., 2004) that are part of the inner kinetochore. Assembled onto these structures are the outer kinetochore proteins of the Ndc80–HEC1 complex and Zwint-1. Moreover, C. elegans KNL-1 has been found to associate with CENP-CHCP-4 and Mis12 colocalizes at the inner centromere with CENP-A and CENP-C (Goshima et al., 2003; Cheeseman et al., 2004; Obuse et al., 2004).
Thus, the Zwint-1–containing kinetochore subcomplex spans from the innermost components of the kinetochore across all layers of the centromere. Ultimately, mitosis-specific assembly of Zwint-1 recruits ZW10–Rod–Zwilch that serve to anchor attachment of Mad1 and Mad2 (Fig. 7 f). Although a previous study reported that ZW10–Rod inhibition after injection of Rod antibody left Mad1 and Mad2 levels still detectable on mitotic kinetochores (Chan et al., 2000), it is now clear that removal of ZW10 or Rod inactivates the mitotic checkpoint with essentially no Mad1–Mad2 recruited to unattached kinetochores in both Xenopus extracts and human cells. A requirement of ZW10–Rod for Mad1–Mad2 recruitment is further supported by genetics in Drosophila: GFP-Mad2 no longer attaches at kinetochores of colchicine-treated rod or zw10 mutant neuroblast cells (R. Karess, personal communication).
Although the ZW10 complex could bind Mad1 or a Mad1–Mad2 heterodimer directly, this seems unlikely because Mad1 (as well as the Mad2 directly bound to it) is a stable component of the kinetochore before microtubule attachment (Howell et al., 2004; Shah et al., 2004), but was not found in the affinity purification of ZW10. Moreover, such a static, scaffold-like function for the ZW10 complex is hard to reconcile with the reported highly dynamic behavior of Rod in syncytial Drosophila embryos (Basto et al., 2004), albeit ZW10 and Rod dynamics have not been investigated in the context of mitotic human cells. More likely, ZW10 may affect Mad1 binding in a number of ways as only one component required for creating the Mad1-binding site(s) at the kinetochore. Ndc80–HEC1 was found in a yeast-two-hybrid screen for interactors with human Mad1, and Mad1–Mad2 kinetochore localization depends on a functional Ndc80–HEC1 complex (Martin-Lluesma et al., 2002; DeLuca et al., 2003; McCleland et al., 2003, 2004; Bharadwaj et al., 2004). No binding between Ndc80–HEC1 and Mad1 could be reconstituted in vitro with recombinant proteins (Martin-Lluesma et al., 2002). Perhaps the interaction found in the yeast-two-hybrid is too unstable to be identified by the assays we have used in vitro, which would support the idea that additional factors are required for establishing such an interaction. The ZW10 complex may fulfill that role. Tandem affinity purification of Mad1 and Ndc80–HEC1-associating factors and reconstitution using recombinant proteins or purified subcellular complexes could now directly test this.
The dependency on ZW10 of BubR1 kinetochore localization differs in frog extracts and HeLa cells. Immunodepletion of the ZW10–Rod–Zwilch complex from frog extracts may affect an activity required for proper BubR1 kinetochore localization that is unaffected by RNAi of one component (ZW10) in HeLa cells. Alternatively, the frog kinetochore in extracts may behave like an all-or-none system more so than the human kinetochore. It is newly assembled upon addition of the sperm DNA to the extract and may therefore be less mature than that of human mitotic cells. Perhaps ZW10 depletion marginally affects BubR1 localization in human cells, an effect readily detected in the less rigidly structured kinetochores that must assemble and disassembly rapidly in the very short embryonic cell cycles. One way to distinguish between these possibilities is to examine checkpoint status and checkpoint protein localization in frog extracts to which the ZW10 complex, reconstituted from recombinant proteins, was restored, a daunting task that we have not been able to achieve since even though we identified a Xenopus Zwilch homologue (GenBank/EMBL/DDBL accession no. BC070818), we have been unsuccessful in identifying full-length XRod.
Despite linkage of dynein at kinetochores to the ZW10 complex through the dynactin subunit p50 dynamitin, it is unlikely that dynein plays any role in the establishment of mitotic checkpoint signaling by ZW10. Whereas depletion of ZW10 prevents the checkpoint from being activated, inhibition of dynein function chronically activates it: overexpression of p50 dynamitin to disrupt the dynein activator dynactin in vertebrate cells or hypomorphic dynein heavy chain mutants in flies both cause checkpoint-dependent mitotic arrest that is abrogated by mutation in Drosophila Rod (Wojcik et al., 2001) or injection of antibodies to Mad2 into PtK1 cells (Howell et al., 2001). Thus, ZW10 and Rod play a role in establishing checkpoint signaling and this is independent of functional dynein.
Although mitotic arrest after inhibition of dynein has been explained by the lack of tension between sister centromeres, perhaps due to improper spindle assembly (Echeverri et al., 1996; Gaglio et al., 1997; Merdes et al., 2000), it seems most likely to us that the probable role for dynein in checkpoint signaling is a role in directly silencing it, through dynein's microtubule-dependent removal of mitotic checkpoint proteins (including the ZW10–Rod complex along with Mad1–Mad2) from kinetochores after stable microtubule capture (Howell et al., 2001; Wojcik et al., 2001). This then leads to our proposal (modeled in Fig. 7 g) that establishment and silencing of mitotic checkpoint signaling at the kinetochore is achieved by linking an activator of the checkpoint (ZW10) with a silencer (dynein) whose inhibitor activity requires microtubule capture. At unattached kinetochores, the ZW10 complex recruits checkpoint components which dynein is unable to remove. Upon bi-oriented attachment, dynein powers their removal from both sister kinetochores, breaking the relatively weak binding between the tightly kinetochore-bound Zwint-1 and Zw10 and with dynein thus pulling the ZW10–Rod–Zwilch complex poleward with it. Loss of the ZW10 complex and its contribution to tethering the stably bound Mad1–Mad2 complex serves to mediate rapid checkpoint signal silencing. Re-recruitment of the ZW10 complex during anaphase A may continue its role as a linker mediating dynein attachment at kinetochores, as poleward movement of chromosomes during anaphase is severely affected in Drosophila zw10 mutant cells (Savoian et al., 2000; Sharp et al., 2000).
Lastly, recognizing a bi-functional role for the ZW10 complex in recruitment of Mad1–Mad2 heterodimers to unattached kinetochores and their removal after attachment, combined with a role as a dynein linker for anaphase chromosome movement, it is clear that compromised function of the complex would be predicted to accelerate chromosome missegregation. Indeed, mutations in all three components of the complex are found in highly aneuploid colorectal cancers (Wang et al., 2004b), implicating disruption of one (or both) roles of this bi-functional complex as likely contributors to generation of aneuploidy in human disease.
Materials and methods
Plasmids
hZwint-1 cDNA was obtained by RT-PCR using HeLa cell cDNA and ligated into pGEMT-easy (Promega). pLAP-hZwint-1 was created by cloning a SalI–EcoRI-cut Zwint-1 PCR fragment from pGEMT-hZwint-1 into XhoI–EcoRI-cut pEYFP-LAPtag (pIC58; Cheeseman et al., 2004). pLAP-hZW10 was created by subcloning a XhoI–BamHI fragment from pBS-hZW10 into XhoI-BamHI-cut pEYFP-LAPtag. All clones were verified by automated sequencing.
Identification of XZW10 and XRod cDNA
A radiolabeled 1.2-kb fragment of an EST clone containing 798 bp of 3′ mouse ZW10 cDNA sequence was used to probe 106 plaques from a λZAP II cDNA library constructed from stage 28–30 (tailbud tadpole) head. One clone containing a 1.2-kb fragment of X-ZW10 cDNA was used to rescreen the same library and identified a clone containing an additional 700 bp of X-ZW10 cDNA. This clone was then used to rescreen the library which resulted in the identification of a clone containing full-length X-ZW10 cDNA, which was subcloned into pGEX. Two Xenopus sequences for X-Rod were identified in the EST database (clones XL107l09 [GenBank/EMBL/DDBL accession no. BJ068773] and 4202179 [GenBank/EMBL/DDBL accession no. BG233529] and were found to contain 1,716 bp or 1,836 bp of the most 3′ X-Rod coding sequence, respectively. The most 5′ 400-bp fragment of clone 4202179 was used to screen the cDNA library described above and several other libraries, but repeatedly failed to identify additional clones. All clones were verified by automated sequencing.
Antibody production
X-ZW10 and XL107l09 were subcloned into pFastBac-HT (Invitrogen) to produce (His)6-X-ZW10 and (His)6-X-RodCOOH baculovirus, respectively, that were used to infect Hi5 insect cells. 48 h after infection, cells were lysed and X-ZW10 and X-RodCOOH were purified using nickel agarose and injected into New Zealand rabbits to produce pAb 1348 and 1351, respectively. Serum from these rabbit was affinity purified by binding to recombinant protein immobilized on CNBr-activated sepharose (Sigma-Aldrich). pAb to human Mad2 was produced by injecting rabbits with (His)6-tagged full-length human Mad2 purified from bacteria. The resulting serum was affinity purified on a HiTrap-NHS column (Amersham Biosciences).
siRNA-mediated depletion of ZW10 and Zwint-1
Double-stranded 19 nt RNA duplex to ZW10 (5′-UGAUCAAUGUGCUGUUCAA-3′) and Control siRNA duplex IX were purchased from Dharmacon. Duplexes were transfected at 200 nM using Oligofectamine (Invitrogen) according to the manufacturer's protocol. pSUPERpuro-Zwint-1 was constructed as described previously (Brummelkamp et al., 2002) using the sequence: 5′-GCACGUAGAGGCCAUCAAA-3′. Plasmids were transfected using Effectene reagent (QIAGEN). Untransfected cells were removed from the cell population 24 h after transfection by treatment with puromycin (1 μg/ml) for 2 d.
Affinity purification of ZW10 and Zwint-1 complexes
HeLa cells stably expressing ZW10LAPtag or Zwint-1LAPtag were established as described previously (Shah et al., 2004). In short, ZW10LAPtag and Zwint-1LAPtag were subcloned to pBabe-blasticidin, and retrovirus was produced from 293-GP cells and used to infect HeLa cells. After 2 wk of selection with blasticidin (2 μg/ml), single cells clones were selected by flow cytometry and analyzed for EYFP localization. Clones LZ5, LZ7, LZ2.8 (ZW10LAPtag), LINT2.8, and LINT 2.14 (Zwint-1LAPtag) were grown in 20 15-cm dishes each and blocked in 200 ng/ml nocodazole for 20 h. Cells were collected and mixed for further purification of native complexes as described previously (Cheeseman et al., 2004). Mass spectrometry was conducted essentially as described previously (Cheeseman et al., 2002). The samples were analyzed using a ThermoFinnigan LTQ mass spectrometer using Mudpit with the following gradients of 500 mM ammonium acetate: 10, 25, 35, 50, 65, 80, and 100%. Results were searched by Sequest against an ncbi human-mouse-rat database and filtered with DTA Select.
Cell culture
HeLa cells were grown in DME supplemented with 5 μg/ml pen/strep (Invitrogen) and 10% FBS. Nocodazole was used at 200 ng/ml.
Xenopus extracts
CSF-arrested extracts were prepared from unfertilized Xenopus eggs as described previously (Murray, 1991). Checkpoint extracts were prepared from these by a 30-min incubation with ∼12,000 demembranated sperm nuclei/μl and 10 μg/ml nocodazole (Chen and Murray, 1997). Exit from CSF arrest was induced by addition of 0.5 mM CaCl2. For immunodepletion, 100 μg (anti–X-ZW10) or 50 μg (anti–X-RodCOOH) of affinity-purified antibody or nonimmune rabbit IgG were bound to 100 μl Dynabeads protein A (Dynal). 100 μl CSF egg extract was added for 1 h at 4°C. Immunodepletion of X-BubR1 was done as described previously (Mao et al., 2003). Checkpoint establishment experiments were done by depleting CSF extract for 1 h before adding sperm and nocodazole, whereas checkpoint maintenance experiments were done by depleting extracts for 1 h after sperm and nocodazole were added. Immunofluorescence with Xenopus egg extracts was performed as described previously (Abrieu et al., 2000), using absence of X-BubR1 signal at kinetochores as a marker for the successful depletion of X-ZW10 or X-Rod. Immunoprecipitation was performed as described previously (Mao et al., 2003). Antibodies to X-CENPE (1:200), X-BubR1 (1:1,000), X-Mps1 (1:200), and X-Mad2 (1:200) have been described previously (Abrieu et al., 2000, 2001; Mao et al., 2003). Anti–X-CENPA (1:100) was a gift of A. Straight (Stanford University, Stanford, CA), anti–X-KCM1 (1:100) was a gift of A. Desai (Ludwig Institute for Cancer Research, La Jolla, CA), and anti–X-Mad1 (1:200) was a gift of R.-H. Chen (Cornell University, Ithaca, NY).
Immunolocalization
Images were acquired on a DeltaVision deconvolution microscope (Applied Precision) equipped with a CoolSnap CCD camera (Roper Scientific) at 20°C. z-sections were acquired at 2-μm steps using a 100×, 1.3 NA Olympus U-Planapo objective with 1 × 1 binning and 640 × 640 pixel area. All images are z-stack projections. HeLa cells, grown on poly-l-lysine–coated coverslips, were washed once in MTSB buffer (100 mM Pipes, 1 mM EGTA, 1 mM MgSO4, 30% glycerol) and preextracted in MTSB/0.5% Triton X-100 at 37°C for 5 min, unless stated otherwise. Cells were fixed with ice-cold methanol for 10 min and all coverslips were blocked in PBS/3% BSA for 1 h. Primary antibody incubations were in PBS/3% BSA for 1 h at RT, followed by four washes of PBS/0.1% Triton X-100. Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories and diluted 1:200 in PBS/3% BSA. Coverslips were washed, submerged in PBS containing DAPI, washed once with PBS, and mounted using ProLong antifade reagent (Molecular Probes). Anti-Bub1 (1:500) was purchased from Abgent. Anti-GFP was used at 1:1,000. Anti-centromere antiserum (ACA; 1:100) was purchased from Antibodies, Inc. Anti-hZw10 (1:50) was described previously (Hirose et al., 2004), anti-hZwint1 (1:100) was a gift of M. Goldberg (Cornell University), anti-BubR1 (5F9; 1:500) was a gift of S. Taylor (University of Manchester, Manchester, UK), and mouse anti-Mad1 (9B10; 1:100) was a gift of T. Yen (Fox Chase Cancer Center, Philadelphia, PA).
Quantitation of immunofluorescence
Images acquired on the DeltaVision microscope (see previous section) were quantitatively deconvolved, converted to maximum projections and saved as 16-bit TIFF files without scaling, after which ACA staining was used to draw regions containing all kinetochores using MetaMorph software. Regions were then transferred onto images of Mad1, Mad2, or Bub1 to be quantitated. Area, average pixel intensity, and integrated intensity of each kinetochore-containing region (KCR) was acquired and exported to Microsoft Excel. Values for a 400-pixel square background region from each cell were also obtained and several background values for each cell were averaged. Integrated intensity was calculated by multiplying the area of the KCR times the average intensity of the background square and subtracting this value from the total integrated intensity of the KCR. Resulting integrated intensities were normalized against the highest value obtained and then averaged and graphed.
Immunoprecipitation and immunoblotting
Immunoprecipitation of Zwint-1LAPtag was done as described for the affinity purification, using rabbit anti-GFP antibody coupled and cross-linked to Affi-Prep protein A support (Bio-Rad Laboratories). The matrix was washed and resuspended in 2× Laemmli sample buffer. SDS-PAGE and Western blotting were standard. Antibodies and dilutions used were anti-GFP (1:1,000), anti-tubulin (DM1a; 1:2,000), and anti-hZW10 (1:500). All antibodies to Xenopus proteins were at 1:1,000.
Flow cytometry
Cells were washed once with PBS and fixed in 70% ice-cold ethanol for 16 h. Cells were washed with PBS/0.1% Triton X-100 (PBST), incubated with anti–phospho-Ser10-Histone H3 (Upstate Biotechnology) in PBST (final concentration of 3 μg/ml) for 1 h on ice and washed again with PBST. Incubation with FITC-conjugated donkey-anti–rabbit secondary antibody (Jackson ImmunoResearch Laboratories) was for one hour on ice and in the dark. After a final wash with PBST, DNA was stained with propidium iodide and measured on a FACS-Vantage flow cytometer (Becton Dickinson).
Colony outgrowth assay
To measure general toxicity of ZW10 and Zwint-1 depletion, cells were transfected with siRNA duplexes and 4 d after transfection cells were split to different confluencies. 16 h after that, cells were retransfected with the oligos. Surviving colonies were stained with crystal violet and counted 7 d after retransfection.
Online supplemental material
Fig. S1 shows the alignment of Xenopus ZW10 and Rod-COOH with human orthologues. Fig S2 shows the characterization of subcellular location of X-Rod in mitotic Xenopus XL177 cells. Fig. S3 shows the immunolabeling of the X-ZW10–X-Rod complex on Xenopus sperm DNA in X-Rod–depleted Xenopus egg extracts. Fig. S4 shows the time course of siRNA-mediated depletion of ZW10 in HeLa cells. Fig. S5 shows the CENP-E kinetochore localization in ZW10-depleted HeLa cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200411118/DC1.
Acknowledgments
The authors thank Aaron Straight, Arshad Desai, Rey-Huei Chen, Michael Goldberg, Tim Yen, and Stephen Taylor for providing reagents; Jagesh Shah and Dennis Young for help with generating stable cell lines; Iain Cheeseman for assistance with the tandem affinity purifications; Lars Jansen for cross-Atlantic technical assistance; Jagesh Shah and Iain Cheeseman for critical reading of the manuscript; and the Cleveland laboratory for helpful discussions.
This work has been supported by grants from the National Institutes of Health to D.W. Cleveland (GM 29513) and to J.R. Yates III (RR11823-09). G.J.P.L. Kops is supported by a fellowship from the Dutch Cancer Society (KWF Kankerbestrijding). Salary support for D.W. Cleveland is provided by the Ludwig Institute for Cancer Research.
G.J.P.L. Kops's present address is Dept. of Experimental Oncology, UMC Utrecht, 3584 CG Utrecht, Netherlands.
Abbreviations used in this paper: ACA, anti-centromere antiserum; CENP, centromere-associated protein; CSF, cytostatic factor; EYFP, enhanced YFP; KCR, kinetochore-containing region; LAP, localization and affinity purification; siRNA, small interfering RNA; Zwint-1, ZW10-interacting protein 1.
References
- Abrieu, A., J.A. Kahana, K.W. Wood, and D.W. Cleveland. 2000. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 102:817–826. [DOI] [PubMed] [Google Scholar]
- Abrieu, A., L. Magnaghi-Jaulin, J.A. Kahana, M. Peter, A. Castro, S. Vigneron, T. Lorca, D.W. Cleveland, and J.-C. Labbe. 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 106:83–93. [DOI] [PubMed] [Google Scholar]
- Basto, R., R. Gomes, and R.E. Karess. 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol. 2:939–943. [DOI] [PubMed] [Google Scholar]
- Basto, R., F. Scaerou, S. Mische, E. Wojcik, C. Lefebvre, R. Gomes, T. Hays, and R. Karess. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14:56–61. [DOI] [PubMed] [Google Scholar]
- Bharadwaj, R., W. Qi, and H. Yu. 2004. Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 279:13076–13085. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., S.A. Jablonski, D.A. Starr, M.L. Goldberg, and T.J. Yen. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates III, C.S. Chan, D.G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., and A. Murray. 1997. Characterization of spindle assembly checkpoint in Xenopus egg extracts. Methods Enzymol. 283:572–584. [DOI] [PubMed] [Google Scholar]
- Chen, R.H., A. Shevchenko, M. Mann, and A.W. Murray. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores. From epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- Cohen, J. 2002. Sorting out chromosome errors. Science. 296:2164–2166. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., B.J. Howell, J.C. Canman, J.M. Hickey, G. Fang, and E.D. Salmon. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103–2109. [DOI] [PubMed] [Google Scholar]
- Echeverri, C.J., B.M. Paschal, K.T. Vaughan, and R.B. Vallee. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132:617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 13:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., H. Yu, and M.W. Kirschner. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio, T., M.A. Dionne, and D.A. Compton. 1997. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 138:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, H., K. Arasaki, N. Dohmae, K. Takio, K. Hatsuzawa, M. Nagahama, K. Tani, A. Yamamoto, M. Tohyama, and M. Tagaya. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23:1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., B.F. McEwen, J.C. Canman, D.B. Hoffman, E.M. Farrar, C.L. Rieder, and E.D. Salmon. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., B. Moree, E.M. Farrar, S. Stewart, G. Fang, and E.D. Salmon. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964. [DOI] [PubMed] [Google Scholar]
- Karess, R.E., and D.M. Glover. 1989. Rough deal: a gene required for proper mitotic segregation in Drosophila. J. Cell Biol. 109:2951–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops, G.J., D.R. Foltz, and D.W. Cleveland. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 101:8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer, C., K.W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature. 396:643–649. [DOI] [PubMed] [Google Scholar]
- Li, X., and R.B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632. [DOI] [PubMed] [Google Scholar]
- Maddox, P., A. Straight, P. Coughlin, T.J. Mitchison, and E.D. Salmon. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y., A. Abrieu, and D.W. Cleveland. 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 114:87–98. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., V.M. Stucke, and E.A. Nigg. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1–Mad2. Science. 297:2267–2270. [DOI] [PubMed] [Google Scholar]
- McCleland, M.L., R.D. Gardner, M.J. Kallio, J.R. Daum, G.J. Gorbsky, D.J. Burke, and P.T. Stukenberg. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland, M.L., M.J. Kallio, G.A. Barrett-Wilt, C.A. Kestner, J. Shabanowitz, D.F. Hunt, G.J. Gorbsky, and P.T. Stukenberg. 2004. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14:131–137. [DOI] [PubMed] [Google Scholar]
- Merdes, A., R. Heald, K. Samejima, W.C. Earnshaw, and D.W. Cleveland. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. [PubMed] [Google Scholar]
- Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda, and M. Yanagida. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141. [DOI] [PubMed] [Google Scholar]
- Okamura, A., C. Pendon, M.M. Valdivia, T. Ikemura, and T. Fukagawa. 2001. Gene structure, chromosomal localization and immunolocalization of chicken centromere proteins CENP-C and ZW10. Gene. 262:283–290. [DOI] [PubMed] [Google Scholar]
- Peters, J.M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell. 9:931–943. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., R.W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian, M.S., M.L. Goldberg, and C.L. Rieder. 2000. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat. Cell Biol. 2:948–952. [DOI] [PubMed] [Google Scholar]
- Scaerou, F., I. Aguilera, R. Saunders, N. Kane, L. Blottiere, and R. Karess. 1999. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J. Cell Sci. 112:3757–3768. [DOI] [PubMed] [Google Scholar]
- Scaerou, F., D.A. Starr, F. Piano, O. Papoulas, R.E. Karess, and M.L. Goldberg. 2001. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J. Cell Sci. 114:3103–3114. [DOI] [PubMed] [Google Scholar]
- Shah, J.V., E. Botvinick, Z.Q. Bonday, F. Furnari, M.W. Berns, and D.W. Cleveland. 2004. Dynamics of centromere and kinetochore proteins: implications for checkpoint activation and silencing. Curr. Biol. 14:942–952. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., G.C. Rogers, and J.M. Scholey. 2000. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2:922–930. [DOI] [PubMed] [Google Scholar]
- Smith, D.A., B.S. Baker, and M. Gatti. 1985. Mutations in genes encoding essential mitotic functions in Drosophila melanogaster. Genetics. 110:647–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., B.C. Williams, Z. Li, B. Etemad-Moghadam, R.K. Dawe, and M.L. Goldberg. 1997. Conservation of the centromere/kinetochore protein ZW10. J. Cell Biol. 138:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., B.C. Williams, T.S. Hays, and M.L. Goldberg. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., R. Saffery, Z. Li, A.E. Simpson, K.H. Choo, T.J. Yen, and M.L. Goldberg. 2000. HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 113:1939–1950. [DOI] [PubMed] [Google Scholar]
- Sudakin, V., G.K. Chan, and T.J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., R. Bharadwaj, B. Li, and H. Yu. 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1:227–237. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., T.J. Mitchison, and A. Desai. 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 84:37–47. [DOI] [PubMed] [Google Scholar]
- Wang, H., Z. Dou, X. Hu, X. Ding, A.W. Shaw, M.L. Goldberg, D.W. Cleveland, and X. Yao. 2004a. Human Zwint-1 specifies localization of ZW10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279:54590–54598 10.1074/jbc.M407588200. [DOI] [PubMed]
- Wang, Z., J.M. Cummins, D. Shen, D.P. Cahill, P.V. Jallepalli, T.L. Wang, D.W. Parsons, G. Traverso, M. Awad, N. Silliman, et al. 2004. b. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 64:2998–3001. [DOI] [PubMed] [Google Scholar]
- Weaver, B.A., Z.Q. Bonday, F.R. Putkey, G.J. Kops, A.D. Silk, and D.W. Cleveland. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., and M.L. Goldberg. 1994. Determinants of Drosophila zw10 protein localization and function. J. Cell Sci. 107:785–798. [DOI] [PubMed] [Google Scholar]
- Williams, B.C., T.L. Karr, J.M. Montgomery, and M.L. Goldberg. 1992. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 118:759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., Z. Li, S. Liu, E.V. Williams, G. Leung, T.J. Yen, and M.L. Goldberg. 2003. Zwilch, a new component of the ZW10–ROD complex required for kinetochore functions. Mol. Biol. Cell. 14:1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik, E., R. Basto, M. Serr, F. Scaerou, R. Karess, and T. Hays. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007. [DOI] [PubMed] [Google Scholar]