Abstract
The evolutionarily conserved signal recognition particle (SRP) plays an integral role in Sec-mediated cotranslational protein translocation and membrane protein insertion, as it has been shown to target nascent secretory and membrane proteins to the bacterial and eukaryotic translocation pores. However, little is known about its function in archaea, since characterization of the SRP in this domain of life has thus far been limited to in vitro reconstitution studies of heterologously expressed archaeal SRP components identified by sequence comparisons. In the present study, the genes encoding the SRP54, SRP19, and 7S RNA homologs (hv54h, hv19h, and hv7Sh, respectively) of the genetically and biochemically tractable archaeon Haloferax volcanii were cloned, providing the tools to analyze the SRP in its native host. As part of this analysis, an hv54h knockout strain was created. In vivo characterization of this strain revealed that the archaeal SRP is required for viability, suggesting that cotranslational protein translocation is an essential process in archaea. Furthermore, a method for the purification of this SRP employing nickel chromatography was developed in H. volcanii, allowing the successful copurification of (i) Hv7Sh with a histidine-tagged Hv54h, as well as (ii) Hv54h and Hv7Sh with a histidine-tagged Hv19h. These results provide the first in vivo evidence that these components interact in archaea. Such copurification studies will provide insight into the significance of the similarities and differences of the protein-targeting systems of the three domains of life, thereby increasing knowledge about the recognition of translocated proteins in general.
The transport of proteins across hydrophobic membranes is essential to the survival of all organisms. Biochemical and genetic analyses of the eukaryotic and bacterial translocation systems strongly suggest that most proteins are translocated via an evolutionarily conserved proteinaceous pore in the endoplasmic reticular (ER) and cytoplasmic membranes, respectively (17, 30, 31). However, whereas in bacteria, protein translocation occurs mainly posttranslationally and requires a cytoplasmic ATPase (SecA) to “push” the proteins through the membrane, translocation into the yeast ER occurs both co- and posttranslationally (4, 17). These processes in yeast depend on Kar2p, an ATPase in the ER that moves proteins into the lumen of this organelle (28, 36). In contrast to bacterial and yeast protein translocation, in vitro reconstitution studies have suggested that in mammalian cotranslational translocation, elongation of nascent polypeptides drives them into the ER and no exogenous energy source is required (12). Sequence analyses of all completely sequenced archaeal genomes suggest that Sec-mediated protein translocation in organisms of this domain of life involves a combination of bacterial and eukaryotic Sec component homologs and that a protein with significant sequence homology to the bacterial and eukaryotic translocation ATPases is absent (30). Thus, archaeal Sec-dependent protein secretion and membrane protein insertion may be unique, and nonhomologous components might be present in archaeal Sec systems. It is also possible that, since no homologs of known ATPases required for posttranslational protein translocation have been identified in archaea, Sec-mediated protein secretion and membrane protein insertion occur exclusively cotranslationally, alleviating the need for an additional energy source. In vivo analyses of the membrane insertion kinetics of the Halobacterium sp. strain NRC-1 polytopic membrane protein bacterioopsin suggested that the insertion of at least the first membrane-spanning segment of this protein occurs cotranslationally (9). While a second study suggested that the insertion of the Halobacterium sp. strain NRC-1 bacterioopsin fused to the Haloferax volcanii S-layer glycoprotein signal sequence into the cytoplasmic membrane occurs posttranslationally (25), this study was conducted in H. volcanii, an organism that does not naturally express bacterioopsin. Thus further characterization of archaeal cotranslational protein translocation is necessary to clarify the role of this cellular process in these organisms.
Cotranslational translocation requires the signal recognition particle (SRP), which binds the signal sequence of a nascent polypeptide chain emerging from a translating ribosome and targets this ribosome-nascent polypeptide complex to the translocon of the eukaryotic ER membrane or that of the bacterial cytoplasmic membrane. The mammalian SRP, the most well-characterized SRP of all three domains of life, is an 11S ribonucleoprotein complex consisting of a single 7S RNA plus six proteins (SRP9, -14, -19, -54, -68, and -72, corresponding to their kilodalton masses) (1, 19). While homologs of the RNA scaffold and the signal sequence binding SRP54 components are conserved in all domains of life, the bacterial SRP appears to be much simpler than that of the eukaryotes (19, 23). The eukaryotic SRP mediates the cotranslational translocation of both secreted and integral membrane proteins, while the bacterial SRP is primarily required for the insertion of proteins into the cytoplasmic membrane (10, 33b). The function of this universally conserved ribonucleoprotein complex in the archaea, however, is not well understood; thus, characterization of this targeting complex is crucial for a better understanding of its role in archaeal cotranslational protein translocation.
Analyses of archaeal genomes and reconstitution studies of heterologously expressed archaeal components, homologous to previously-identified SRP subunits, suggest that organisms of this domain contain homologs of SRP54 and 7S RNA, as well as of the eukaryotic SRP19 (3, 21, 36a). However, such approaches are unable to identify homologs of other eukaryotic and bacterial SRP subunits that share little or no sequence homology with previously identified components, or subunits that are specific to the archaeal SRP. In order to define the composition of the archaeal SRP and gain an understanding of its function, it is necessary to study this ribonucleoprotein complex in its native host. We therefore have initiated in vivo studies to purify and characterize the SRP from H. volcanii, an archaeon that is amenable to biochemical and genetic analyses (6, 8, 34). Using this system, we have been able to create an H. volcanii knockout strain and provide evidence that the archaeal SRP is essential. Furthermore, we have developed a method that not only allows us to copurify the universally conserved components from their native host, but will also provide us with the tools needed to define the composition of the archaeal SRP.
MATERIALS AND METHODS
Reagents.
Antibiotics were purchased from Sigma, with the exception of mevinolin (a gift from Merck Research Laboratories). The H. volcanii cosmid library (6) was generously provided by R. L. Charlebois (Department of Biology, University of Ottawa). The AlkPhos Direct kit, CDP-Star, and ECL (enhanced chemiluminescence) detection reagents were purchased from Amersham Pharmacia Biotech. The pMAL-c2 cloning vector, restriction enzymes, and amylose-agarose resin were purchased from New England Biolabs. The Ni nitrilotriacetic acid (NTA) agarose resin, anti-pentaHis antibody, and DNA purification kits were purchased from Qiagen. Trizol reagent was purchased from Life Technologies. Blotting membranes (Hybond nylon and polyvinylidene difluoride [PVDF]) were purchased from Amersham International and Micron Separations, Inc., respectively.
Strains and growth conditions.
The archaeal and bacterial strains utilized in this study are listed in Table 1. H. volcanii strains were routinely cultured at 40°C in rich medium (RM) (20) supplemented with mevinolin (20 μg/ml) and/or novobiocin (0.3 μg/ml) when required. Escherichia coli strains were routinely cultured at 37°C in Luria-Bertani medium supplemented with ampicillin (200 μg/ml), kanamycin (40 μg/ml), and/or isopropyl-β-d-thiogalactopyranoside (IPTG) (100 μM) when required.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source and/or reference |
|---|---|---|
| Strain | ||
| WFD11 | H. volcanii | 5 |
| MP12 | E. coli MC4100 | 29 |
| WAM113 | E. coli ffh conditional mutant | 27 |
| WR4 | MP12 containing pWR-4 | This study |
| WR6c | MP12 containing pWR-6c | This study |
| WAM-ffh | WAM113 containing pRB11-ffh | This study |
| WAM-hv54h | WAM113 containing pRB11-hv54h | This study |
| WRHv-NP15 | WFD11 containing pNP15 | This study |
| WRHv-6c | WFD11 containing pWR-6c | This study |
| WRHv-6c/54KO | WRHv-6c with mevR insertion in chromosomal hv54h | This study |
| WRHv-9a | WFD11 containing pWR-9a | This study |
| Plasmid/cosmid | ||
| A199chr, 347chr | Kanr; cosmid vectors (Lorist M) containing H. volcanii chromosomal MluI fragments | 6 |
| pNP15 | Ampr Nbr; H. volcanii shuttle vector | 26 |
| pMDS99 | Ampr; plasmid containing hhmevR gene | 34 |
| pBAD18 | Ampr; arabinose-inducible expression plasmid | 14 |
| pMAL-c2 | Ampr; IPTG-inducible expression plasmid encoding N-terminal maltose binding protein | 37 |
| p105-SacI | Ampr; pBAD18 containing SacI fragment of cosmid A199chr | This study |
| p64-AluI | Ampr; pBAD18 containing AluI fragment of cosmid 347chr | This study |
| pWR-2 | Ampr; pBAD18 containing hv54h ORF + 66 bp | This study |
| pWR2-hhmevR | pWR-2 with hv54h interrupted by hhmevR | This study |
| pWR-4 | Ampr; pMAL-c2 containing hv54h ORF | This study |
| pWR-6c | pNP15 containing hv54h-6xhis | This study |
| pWR-9a | pNP15 containing hv19h-6xhis | This study |
| pRB11-ffh | pRB11 containing ffh | 33 |
| pRB11-hv54h | pRB11 containing hv54h | This study |
Cloning and sequencing of the H. volcanii SRP54 (hv54h) and 7S RNA (hv7Sh) homologs.
PCR was used to generate DNA probes from H. volcanii chromosomal DNA. The sequences of the primers used were as follows: (i) hv54h probe (450 bp), degenerate forward primer 5′-CAGGGCTCSGGCAAGACSACSAC-3′ and degenerate reverse primer 5′-GAGSGCGCCGCCGCCCTTSGC-3′; and (ii) hv7Sh probe (296 bp), forward primer 5′-ACTAGGTCGGGCAGTTAGG-3′ and reverse primer 5′-GCGAGGTTGCCCGGCGTTCC-3′. These probes were used in Southern blot analyses of an MluI digest of the H. volcanii cosmid library. Positive cosmids (harboring hv54h [cosmid A199] and hv7Sh [cosmid 347]) (Table 1) were identified and confirmed to contain their respective genes by PCR. These cosmids were then digested (SacI for A199 and AluI for 347) and analyzed by Southern blotting with the probes described above. Positive bands were gel purified, and the restriction fragments containing hv54h and hv7Sh were subcloned into pBAD18 (14) (creating p105-SacI and p64-AluI, respectively) (Table 1) and sequenced.
Construction of an SRP54 phylogenetic tree.
Amino acid sequences of SRP54 from organisms representing the three domains of life (35) were obtained from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nih.gov). The ClustalW algorithm (33) (http://searchlauncher.bcm.tmc.edu) was used to align the sequences, and seven conserved regions of SRP54 (13) were used to construct the tree. The SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE programs (in this order) of the Phylogeny Inference Package (PHYLIP v.3.573c) (11) were used to construct a maximum parsimony tree, which was replicated in 100 bootstraps.
Construction of an hv54h-knockout strain.
The hv54h open reading frame (ORF) was PCR amplified (forward primer 5′-GGGGGAGCTCATGGTACTCGACAATCTC-3′ and reverse primer 5′-GGGGAAGCTTGCGCAGCGAGGCGAGGAAC-3′) from p105-SacI and ligated into pBAD18 (14) to generate pWR-2 (Table 1). The resistance cassette used in the disruption of hv54h was generated as follows: the Haloarcula hispanica mevinolin resistance gene (mevR) was PCR amplified (forward primer 5′-GGGGGGTACCCAAGAGCAACTTTTAAGAGT-3′ and reverse primer 5′-GGGGGCATGCCAGGAAGCACTCCTGCTC-3′) from pMDS99 (34). The 5′ and 3′ ends of this insert were digested with KpnI and SphI, respectively, and filled in with T4 DNA polymerase. pWR-2 was digested with PshA1, which produces a blunt-ended single cut at nucleotide position 762 in the hv54h ORF. The blunt-ended mevR insert was then ligated into PshA1-digested pWR-2, creating pWR2-hhmevR (Table 1). The hv54h-hhmevR insert was PCR amplified from pWR2-hhmevR (forward primer 5′-GGGGGAGCTCATGGTACTCGACAATCTC-3′ and reverse primer 5′-GGGGAAGCTTGCGCAGCGAGGCGAGGAAC-3′), purified, and used to transform spheroplasts of H. volcanii strain WRHv-6c (Table 1) as previously described (18). Transformants were selected on RM plates supplemented with mevinolin and novobiocin. Homologous recombination of the hv54h-hhmevR PCR product with the chromosomal hv54h gene was detected by PCR of transformants with a forward primer specific to the 5′ end of hhmevR (5′-GGGGGGTACCCAAGAGCAACTTTTAAGAGT-3′) and a reverse primer specific to the region downstream of hv54h (5′-GGGGAAGCTTGCGCAGCGAGGCGAGGAAC-3′), which is absent from pWR-6c.
Transfer and plating of hv54h-knockout/control cells.
Three single colonies each of WRHv-6c/54KO and WRHv-6c cells were used to inoculate three 5-ml liquid RM-mevinolin and RM samples without selection, respectively. These cultures were grown to late log phase and subsequently used to inoculate fresh liquid medium with or without mevinolin (15 such transfers were performed). At transfer numbers 3, 6, 9, 12, and 15, samples of the cultures were diluted in RM, and plated onto RM-novobiocin to test for the maintenance of pWR-6c. Each culture was plated in triplicate, and after approximately 14 days of incubation, colonies were counted with a Leica Quebec Darkfield colony counter.
Viability of WAM113 harboring ffh or hv54h.
Strain WAM113 (27), generously provided by Hongping Tian (Harvard University), was transformed with pRB11•ffh (a gift from H. Tian) or pRB11•hv54h (this study). Cells were grown at 25, 30, and 37°C in the presence and absence of IPTG (100 μM) and analyzed for cell growth. IPTG-inducible expression of Ffh and Hv54h by these strains was confirmed by Western blot analysis of whole-cell lysates with antibodies against Ffh (generously provided by Harris Bernstein, National Institutes of Health) and Hv54h (this study).
Expression of Hv54h in E. coli for generation of antiserum.
The gene containing the hv54h ORF was amplified by PCR of p105-SacI (forward primer 5′-GGGGGGAATTCATGGTACTCGACAATCTC-3′ and reverse primer 5′-GGGGAAGCTTGCGCAGCGAGGCGAGGAAC-3′) and ligated into pMAL-c2 (37), creating pWR-4 (Table 1). The Hv54h-maltose binding protein (MBP) fusion construct (Hv54h•MBP; Table 1) was purified from a 1-liter culture of strain WR4 on an amylose-agarose column as per the manufacturer's protocol. The purified protein was electrophoresed on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel, and the band corresponding to the Hv54h•MBP was excised from the gel and used to produce rabbit antiserum (anti-Hv54h) against the fusion protein.
Expression of Hv54h•6xHis in H. volcanii.
The hv54h ORF plus 66 bp upstream of the start codon was C-terminally tagged with a linker and six histidine residues (denoted as 6xHis in Hv54h•6xHis) by PCR amplification of p105 SacI (forward primer 5′-GGGGGGCCATGGCGATAGGTGTTGGCCC-3′ and reverse primer 5′-GGGGGGGGTACCTCAGTGATGGTGATGGTGATGCGGGCCGCCGAACGGCCCCATGCCGCC-3′). The resulting hv54h•6xhis insert was cloned into pNP15 (26), generating pWR-6c (Table 1), which was then used to transform a dam-negative E. coli strain. pWR-6c purified from this strain was then used to transform H. volcanii spheroplasts, creating strain WRHv-6c (Table 1). Expression of Hv54h•6xHis in WRHv-6c was monitored by immunoblotting of whole-cell lysates, by using the anti-Hv54h and anti-pentaHis antibodies.
Expression of Hv19h•6xHis in H. volcanii.
The hv19h ORF plus 50 bp upstream of the start codon (sequence reference no. RVO04094 and RVO04096) (http://wit-scranton.mbi.scranton.edu/Haloferax/genes_DNA.fasta) was C-terminally tagged with a linker and six histidine residues by PCR amplification of H. volcanii chromosomal DNA (forward primer 5′-GGGGGGCCATGGGGCTCCGTGAGACCACAACC-3′ and reverse primer 5′-GGGGGGGGTACCTCAGTGATGGTGATGGTGATGCGGGCCGCCGTCGCGGAGGATTCCGACG-3′). The resulting hv19h•6xhis insert was cloned into pNP15 (26), generating pWR-9a (Table 1), which was then used to transform a dam-negative E. coli strain. pWR-9a purified from this strain was then used to transform H. volcanii spheroplasts, creating strain WRHv-9a (Table 1). Expression of Hv19h•6xHis by WRHv-9a was monitored by immunoblotting of whole-cell lysates with the anti-pentaHis antibody.
Copurification of Hv54h•6xHis and Hv7Sh by Ni-NTA metal affinity chromatography.
Strains WRHv-6c and WRHv-NP15 were grown in RM supplemented with novobiocin until mid- to late-log phase. Cultures (500 ml) were centrifuged at 4,600 × g for 20 min, and cell pellets were resuspended in 2 volumes of ice-cold NSTX buffer [30 mM Mg(OAc)2, 22.5 mM imidazole, 75 mM Tris [pH 7.5], 15% glycerol, 0.015% Triton X-100, 3 mM β-mercaptoethanol, 200 U of RNAsin, 1.5 μg of pepstatin per ml, 7.5 μg of aprotinin per ml, 1.5 μg of leupeptin per ml]. Cells were lysed with a French press (4°C), and lysates were centrifuged at 15,400 × g for 10 min at 4°C. Cytoplasmic fractions were obtained by centrifugation of the cleared lysates at 265,000 × g for 1 h 8 min. Ni-NTA agarose beads (1 ml), previously equilibrated 1:1 (vol/vol) in SMTX-A buffer [1 M KCl, 20 mM Mg(OAc)2, 15 mM imidazole, 50 mM Tris (pH 7.5), 10% glycerol, 0.01% Triton X-100, 2 mM β-mercaptoethanol, 200 U of RNAsin, 1 μg of pepstatin per ml, 5 μg of aprotinin per ml, 1 μg of leupeptin per ml], was added to each cytoplasmic fraction, and the slurries were incubated with agitation for 1 h at 4°C. The slurries were applied to 5-ml columns, and the flowthrough was collected. The Ni-NTA was washed with 8 ml of SMTX-B buffer (SMTX-A with 20 mM imidazole), and the proteins were eluted with SMTX-C (SMTX-A with 200 mM imidazole).
Copurification of Hv19h•6xHis, Hv54h, and Hv7Sh by Ni-NTA metal affinity chromatography.
Strains WRHv-9a and WRHv-NP15 were grown and processed in the same manner as described above for strains WRHv-6c and WRHv-NP15.
Analysis of eluate from Ni-NTA chromatography of H. volcanii cytoplasmic fractions.
Ni-NTA elution fractions were dialyzed against two changes of sterile water at 4°C and then concentrated with a SpeedVac vacuum concentrator (Savant, Inc.). RNA and protein were purified from the fractions by using Trizol, as per the manufacturer's protocol.
Northern blot analysis.
Purified RNA was electrophoresed on a 1% denaturing agarose gel and transferred overnight to a nylon membrane by capillary blotting (32). RNA was UV cross-linked to the blot and prehybridized for 45 min in hybridization buffer at 55°C. The 296-bp Hv7Sh probe (described above) was directly labeled with alkaline phosphatase by using the Alk-phos direct system, as per the manufacturer's protocol. The labeled probe was added to the hybridization buffer, and the blot was hybridized overnight at 55°C. The blot was washed twice in primary wash buffer (2 M urea, 0.1% SDS, 50 mM sodium phosphate [pH 7.0], 150 mM NaCl, 1 mM MgCl2) at 60°C (15-min first wash, 10-min second wash) and twice in secondary wash buffer (50 mM Tris, 100 mM NaCl, 2 mM MgCl2 [pH 10.0]) at room temperature (10 min per wash). The blot was incubated in CDP-Star detection buffer and exposed to autoradiography film until a suitable image was obtained.
Western blot analysis.
Purified protein was electrophoresed on a 10% separating SDS-polyacrylamide gel, and transferred (semidry) to PVDF. After air-drying, the blots were blocked overnight in phosphate-buffered saline containing 0.1% Tween 20 (PBS-T) and 3% bovine serum albumin (BSA). The blots were then incubated in primary antibody solution (anti-Hv54 h or anti-pentaHis, 1:1,000 in PBS-T plus 3% BSA) for 1 h at room temperature. After three washes in PBS-T (5 min each), the blots were incubated in secondary antibody solution (goat anti-rabbit horseradish peroxidase [HRP] conjugate for anti-Hv54h; goat anti-mouse HRP for anti-pentaHis) for 1 h at room temperature. The blots were washed as described above, incubated in ECL detection solution, and exposed to autoradiography film until a suitable image was obtained.
Nucleotide sequence accession number.
The sequences of hv54h and hv7Sh have been submitted to the GenBank database under accession no. AF395887 and AF395888, respectively.
RESULTS
Cloning and sequence analyses of the H. volcanii SRP54 and 7S RNA homologs.
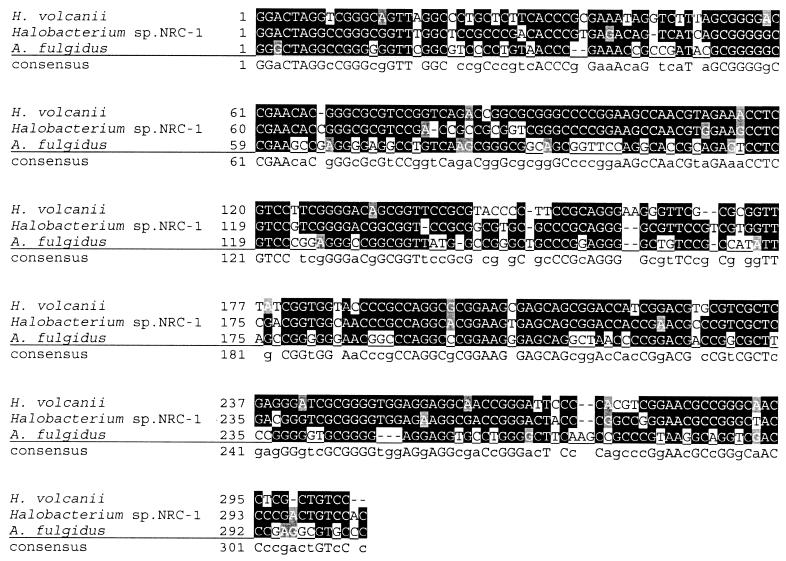
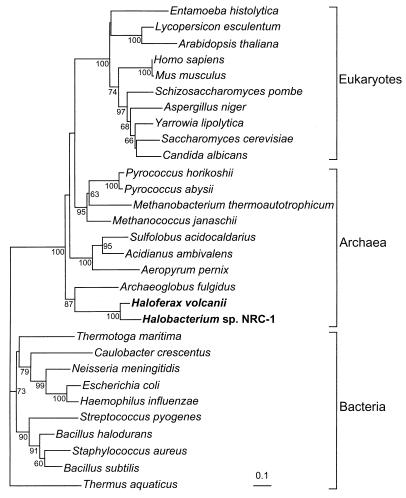
As a first step in the characterization of the archaeal SRP in its native host, it was necessary to use the H. volcanii cosmid library to clone and sequence the genes encoding the H. volcanii homologs of the universally conserved SRP components 7S RNA (hv7Sh) and SRP54 (hv54h), as the genome sequence was unavailable. Using primers designed from the extreme halophile Halobacterium sp. strain NRC-1 (24) 7S RNA homolog sequence, the hv7Sh was identified by PCR and Southern blot analysis of the H. volcanii cosmid library (6). It was found to be 83% identical to the Halobacterium sp. strain NRC-1 7S RNA homolog and 57% identical to the 7S RNA homolog of Archaeoglobus fulgidus (a thermophilic archaeon) (Fig. 1). Hv54h, which was identified by degenerate PCR of chromosomal DNA and Southern blot analysis of the cosmid library, shares 74% identity and 84% similarity with that of the recently identified SRP54 homolog of Halobacterium sp. strain NRC-1. To determine the relatedness of the haloarchaeal SRP54 homologs to those of other archaea, bacteria, and eukaryotes, we generated a phylogenetic tree by using an alignment of seven conserved domains (13) of 30 SRP54 homologs from the three domains of life. We show that Hv54h and the Halobacterium sp. strain NRC-1 SRP54 homolog branch with the archaea and are most closely related to the A. fulgidus SRP54 homolog (Fig. 2). Consistent with previous phylogenetic analyses (13), the eukaryotic and archaeal SRP54 grouped together, to the exclusion of the bacterial SRP (Fig. 2). The similarity between the archaeal SRP component homologs suggests that H. volcanii is a suitable model organism for the characterization of the archaeal SRP.
FIG. 1.
Nucleotide sequence alignment of the 7S RNA homologs from H. volcanii, Halobacterium sp. strain NRC-1, and A. fulgidus.
FIG. 2.
Neighbor-joining tree generated by the alignment of seven conserved domains of SRP54 for representative organisms of the three domains of life. Internal nodes are labeled with the corresponding bootstrap confidence level (BCL), based on 100 bootstrap replicates of the alignment. Bootstrap confidence levels of <60% are not shown. Scale bar represents 0.1 amino acid substitution per site.
Construction of an H. volcanii hv54h-knockout mutant.
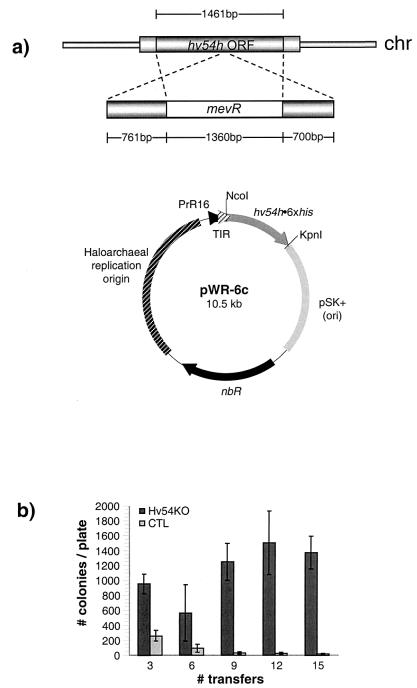
Although the SRP is essential for viability in E. coli (27), Saccharomyces cerevisiae can survive without a functional SRP (15, 22). To determine whether the H. volcanii SRP is crucial for viability, we initially attempted to obtain hv54h-knockout mutants by transforming wild-type H. volcanii with a linear form of the hv54h ORF interrupted by the mevR gene. We then selected for transformants in which chromosomal hv54h had been replaced with the mutated form by growing the cells on medium containing mevinolin. However, no viable transformants were obtained by this methodology. We therefore repeated this procedure in H. volcanii harboring a gene encoding a wild-type Hv54h tagged with six histidine residues (strain WRHv-6c; Table 1) on an expression plasmid (encoding novobiocin resistance) and selected for transformants on medium containing both novobiocin and mevinolin. Transformants that had undergone chromosomal recombination (knockouts) were identified by PCR screening (see Materials and Methods), which also demonstrated that the mevinolin resistance gene was oriented in the same direction as the hv54h gene it interrupted (data not shown). By this methodology, we were able to create the chromosomal hv54h knockout strain WRHv-6c/54KO (Table 1).
Demonstration of the essential nature of Hv54h.
We examined whether strain WRHv-6c/54KO could be cured of the plasmid expressing a six-His-tagged Hv54h (Hv54h•6xHis) in the absence of antibiotic (novobiocin) selection for the plasmid, because no regulatable promoters have been identified that would allow for the selective depletion of H. volcanii proteins in vivo. After only three transfers in medium lacking novobiocin, we observed a threefold greater number of WRHv-6c/54KO colonies when the cells were plated on medium containing novobiocin, as compared to control wild-type cells harboring the hv54h expression plasmid (WRHv-6c) (Fig. 3). By the 10th transfer, this difference had increased to greater than 2 orders of magnitude (Fig. 3). While the total number of WRHv-6c colonies decreased to almost zero over 15 transfers, the total number of WRHv-6c/54KO colonies remained relatively constant (approximately 1,000 to 1,500 per plate) (Fig. 3). The fact that WRHv-6c/54KO cells were not cured of the expression plasmid harboring hv54h•6xhis in the absence of antibiotic selection suggests that the presence of the functional Hv54h•6xHis acted as selective pressure to maintain the plasmid. Thus, not only were we able to create a chromosomal knockout of the gene in its native host and demonstrate that under the experimental conditions the Hv54h was essential, we were also able to show that the histidine-tagged form of this protein (expressed from the plasmid) was functional in vivo.
FIG. 3.
Construction and analysis of an H. volcanii hv54h-knockout strain. The chromosomal (chr) copy of hv54h was replaced with hv54h interrupted by the H. hispanica mevinolin resistance gene (mevR) in H. volcanii strain WRHv-6c (harboring the hv54h•6xhis expression plasmid pWR6-c), creating strain WRHv-6c/54KO (a). This strain and a control strain (WRHv-6c) were then examined as to whether they could be cured of pWR-6c in the absence of antibiotic (novobiocin) selection for the plasmid (b). At each indicated transfer, cells cultured in the absence of novobiocin were grown on RM supplemented with novobiocin. Error bars represent standard deviation. Student's paired t test was used to generate P values for each indicated transfer (T): P ≪ 0.0001 for T3, T9, T12, and T15; P = 0.012 for T6. nbR, novobiocin resistance gene.
Test for complementation of an E. coli ffh-knockout strain with Hv54h.
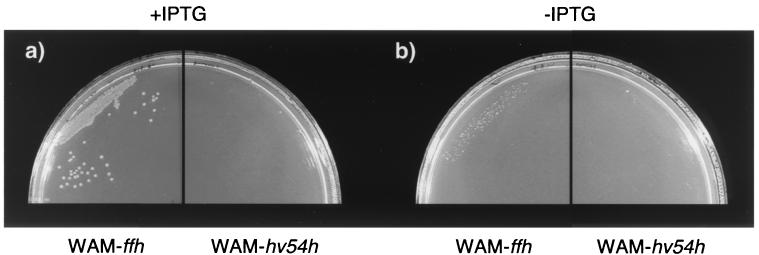
We also examined whether expression of Hv54h in an E. coli strain could complement an E. coli strain depleted of functional Ffh (WAM113 [27]) (Table 1), the E. coli SRP54 homolog. In this strain, the chromosomal copy of ffh is interrupted by a kanamycin resistance gene, and a gene encoding a functional Ffh under the control of the arabinose promoter is supplied in trans. In the absence of arabinose, Ffh is not expressed, and these cells do not grow (27). We transformed these cells with a plasmid harboring either ffh or hv54h under the control of the IPTG-inducible T7 promoter (pRB11•ffh, strain WAM-ffh; and pRB11•hv54h, strain WAM-hv54h; respectively) and grew them at 37°C in the presence and absence of IPTG (in all cases without arabinose). However, while WAM-ffh grew normally in the presence of IPTG, WAM-hv54h did not grow (Fig. 4a). Immunoblots of WAM-hv54h whole-cell lysates grown in the presence of arabinose (with and without IPTG) confirmed that hv54h was inducibly expressed (data not shown). Neither strain grew in the absence of IPTG (Fig. 4b). These phenomena were not temperature dependent, as identical results were obtained for cells grown at 25 and 30°C (data not shown). Our results demonstrate that while hv54h supplied in trans can complement a chromosomal knockout in its native host, it cannot complement a similar mutation in E. coli.
FIG. 4.
Complementation of E. coli WAM113 depleted of Ffh. WAM-ffh and WAM-hv54h (Table 1) were grown without arabinose in the presence (a) or absence (b) of IPTG.
In vivo copurification studies. (i) Copurification of Hv54h•6xHis and Hv7Sh.
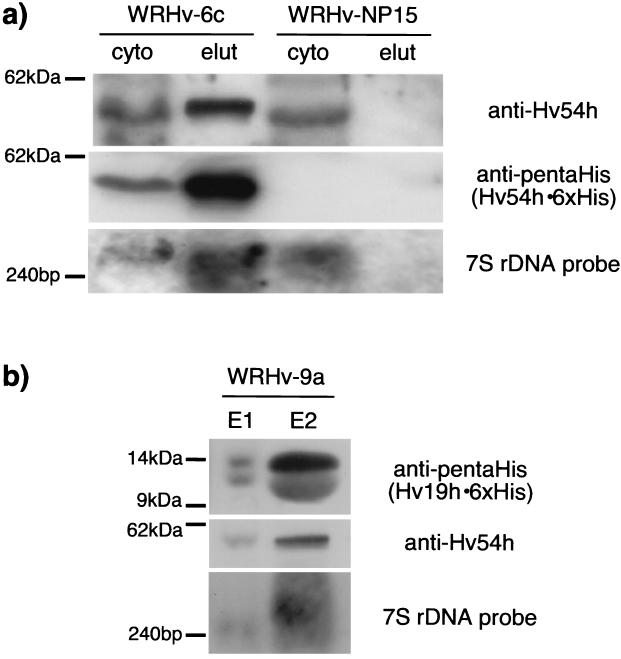
Antibodies raised against Hv54h, as well as DNA probes derived from the hv7Sh DNA sequence, allow us to monitor the copurification of these components. Furthermore, expression of the gene encoding a functional Hv54h•6xHis in H. volcanii provides a tool for the purification of the archaeal SRP. Cytoplasmic fractions of cells expressing Hv54h•6xHis (WRHv-6c) and control cells not expressing this fusion protein (WRHv-NP15) were applied to the Ni-NTA matrix, and the elution fractions were immunoblotted with the anti-Hv54h and anti-pentaHis antibodies. These analyses confirmed that Hv54h•6xHis could be purified from the cytoplasmic fraction by metal affinity chromatography (Fig. 5a, anti-Hv54h/anti-pentaHis). Northern blot analysis of RNA isolated from the elution fractions of the cells demonstrated that Hv7Sh specifically copurified with Hv54h•6xHis, indicating that a complex of at least these universally conserved components was purified from WRHv-6c (Fig. 5a, 7S ribosomal DNA probe).
FIG. 5.
In vivo copurification of the H. volcanii SRP components. (a) Copurification with Hv54h•6xHis. The cytoplasmic fractions of strains WRHv-6c (Hv54h•6xHis) and WRHv-NP15 (control) were purified with Ni-NTA, and the cytoplasmic (cyto) and elution (elut) fractions were analyzed by Western blotting with antibodies against Hv54h and 6xHis (detecting Hv54h•6xHis), as well as by Northern blotting with a DNA probe for Hv7Sh. (b) Copurification with Hv19h•6xHis. The cytoplasmic fractions of WRHv-9a (Hv19h•6xHis) and WRHv-NP15 (control, not shown) were purified with Ni-NTA, and the elution fractions (E1 and E2) were analyzed by Western blotting with antibodies against 6xHis tag (detecting Hv19 h•6xHis) and Hv54h, as well as by Northern blotting with a DNA probe for Hv7Sh. The lower band observed in the anti-pentaHis Western blot is likely a degradation product of Hv19h•6xHis.
(ii) Copurification of Hv19h•6xHis, Hv54h, and Hv7Sh.
We were also able to express a six-His-tagged H. volcanii SRP19 homolog (Hv19h•6xHis) in its native host. The cytoplasmic fraction of cells expressing this fusion protein (strain WRHv-9a), as well as control cells (WRHv-NP15 [data not shown]), was purified with Ni-NTA, and elution fractions were analyzed by using Western and Northern blots (Fig. 5b). Using the Hv19h•6xHis, we were able to specifically copurify both the untagged Hv54h and Hv7Sh, as demonstrated by Western and Northern blots, respectively (Fig. 5b).
These copurification studies not only demonstrate that the SRP components Hv54h, Hv19h, and Hv7Sh interact in vivo in their native host, but also provide us with a method to identify additional putative SRP components. Preliminary silver stain analysis of elution fractions from WRHv-6c, along with immunoprecipitation studies with anti-Hv54h, revealed that other proteins specifically copurified with Hv54h•6xHis (data not shown). The characterization of these components is currently being pursued.
DISCUSSION
Recent whole-genome analyses of numerous archaea have greatly expanded our understanding of organisms belonging to this domain and have provided insight into putative metabolic pathways, molecular processes, and phylogenetic relationships. These sequence analyses allowed for the identification of archaeal SRP54, 7S RNA, and SRP19 homologs. However, previous studies characterizing these archaeal SRP components have been limited to the heterologous expression of these components in nonarchaeal systems, because very few archaea are amenable to genetic and biochemical analyses. Our in vivo analyses of the H. volcanii SRP in its native host have been able to bypass these limitations. Cloning and sequencing of the universally conserved SRP subunits, as well as expression of tagged Hv54h and Hv19h in H. volcanii, provide us with the ability to characterize the complex in its native host, as this organism is amenable to both in vivo and in vitro studies and can be easily cultured under standard laboratory conditions. Phylogenetic analyses of the Hv54h amino acid sequence demonstrated that the haloarchaeal SRP54 homologs branched with those of the archaea, and grouped closely with the SRP54 homolog of the thermophilic, nonhalophilic archaeon A. fulgidus. Thus, although the SRP54 homolog of halophilic archaea is adapted to the high cytoplasmic salt concentration maintained by these organisms to balance the high salinity of the environments they inhabit (7), these results suggest that H. volcanii is a relevant model system for the study of the archaeal SRP.
We were also able to express a gene encoding a functional Hv54h•6xHis, as well as a gene encoding Hv19h•6xHis, in H. volcanii. We chose the histidine tag since its interaction with Ni-NTA matrix is stable under the high-salt conditions (1 to 3 M KCl) that are characteristic of the haloarchaeal cytoplasm (7) and may thus be necessary to maintain the integrity of the halophilic SRP. Expression of the functional Hv54h in trans allowed us to create a chromosomal knockout of the hv54h and demonstrate that these cells could not be cured of the expression plasmid. This observation represents the first evidence showing that, under the experimental conditions, the archaeal SRP is essential for survival and suggests that cotranslational translocation is an essential process in this domain.
Furthermore, we were able to express Hv54h in the ffh mutant E. coli strain. However, it was not able to functionally complement the mutation. This observation might be due to functional differences between the bacterial and archaeal SRP54 homologs, but could also be the result of misfolding of the high-salt-adapted Hv54h in a heterologous host (E. coli) in which the cytoplasmic salt concentration is markedly lower than that of H. volcanii. Previous studies have demonstrated that many halophilic proteins (e.g., halophilic β-galactosidase) require high salt concentrations for maximum functionality (16). These observations underscore the absolute need for the characterization of archaeal proteins in their native hosts.
Critical to understanding the function of the archaeal SRP is the characterization of its composition. As mentioned above, in order to identify all components of the SRP, we need to purify this complex from the archaeal cytoplasm. This may allow for the identification of not only previously identified SRP subunit homologs but also components that are archaea specific or might have low or no significant sequence homology to previously identified SRP components in bacteria and eukaryotes. Previous studies relying on in vitro reconstitution of heterologously expressed SRP components suggest an interaction of the 7SRNA, SRP54, and SRP19 homologs of thermophilic archaea, similar to that described for the eukaryotic SRP (3). Our expression of a six-His-tagged Hv54h in H. volcanii and its copurification with the Hv7Sh and our expression of a six-His-tagged Hv19h and its copurification with both Hv54h and Hv7Sh provide the first in vivo evidence that these three conserved archaeal SRP components interact. These analyses also illustrate that it is possible to successfully perform biochemical analyses under high-salt conditions, which might be required for the integrity of the haloarchaeal SRP.
With this purification procedure, we now have the ability to isolate putative SRP subunits that do not share significant sequence homology with those that are evolutionarily conserved. In addition, such analyses may allow us to identify substrates (secreted and/or membrane proteins) that are cotranslationally targeted by the SRP. Recent analyses of the Halobacterium sp. strain NRC-1 genome indicate that the majority of its secreted proteins are translocated via the twin arginine translocation (Tat) pathway (Rose et al., unpublished data), which has been shown in bacteria to translocate folded substrates (2). These analyses, as well as the findings in the present study, suggest that the haloarchaeal SRP may be primarily involved in cotranslational membrane protein insertion.
The in vivo characterization of the archaeal SRP will not only lead us to a more complete understanding of its role in the targeting of proteins to the membrane, but can also provide important information about cotranslational translocation and thus may reveal a possible energy source driving protein translocation and membrane protein insertion.
Acknowledgments
We thank Harris Bernstein, Fevzi Daldal, and Marjan van der Woude for valuable comments on the manuscript.
Support was provided to R.W.R. by a predoctoral fellowship from the American Heart Association (reference no. 0110093U) and to M.P. by a National Science Foundation grant (MCB-9816411).
REFERENCES
- 1.Althoff, S., D. Selinger, and J. Wise. 1994. Molecular evolution of SRP cycle components: functional implications. Nucleic Acids Res. 22:1933-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 3.Bhuiyan, S. H., K. Gowda, H. Hotokezaka, and C. Zwieb. 2000. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 28:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabelli, R. J., L. Chen, P. C. Tai, and D. B. Oliver. 1988. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell 55:683-692. [DOI] [PubMed] [Google Scholar]
- 5.Charlebois, R. L., W. L. Lam, S. W. Cline, and W. F. Doolittle. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. USA 84:8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlebois, R. L., L. C. Schalkwyk, J. D. Hofman, and W. F. Doolittle. 1991. Detailed physical map and set of overlapping clones covering the genome of the archaebacterium Haloferax volcanii DS2. J. Mol. Biol. 222:509-524. [DOI] [PubMed] [Google Scholar]
- 7.Christian, J. H. B., and J. A. Waltho. 1962. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta 65:506-508. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, A., W. L. Lam, R. L. Charlebois, and W. F. Doolittle. 1991. Localizing genes on the map of the genome of Haloferax volcanii, one of the archaea. Proc. Natl. Acad. Sci. USA 89:1602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dale, H., and M. P. Krebs. 1999. Membrane insertion kinetics of a protein domain in vivo. The bacterioopsin N-terminus inserts co-translationally. J. Biol. Chem. 274:22693-22698. [DOI] [PubMed] [Google Scholar]
- 10.DeGier, J.-W. L., Q. A. Valent, G. Von Heijne, and J. Luirink. 1997. The E. coli SRP: preferences of a targeting factor. FEBS Lett. 408:1-4. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle.
- 12.Gorlich, D., and T. A. Rapoport. 1993. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75:615-630. [DOI] [PubMed] [Google Scholar]
- 13.Gribaldo, S., and P. Cammarano. 1998. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targeting machinery. J. Mol. Evol 47:508-516. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hann, B. C., and P. Walter. 1991. The signal recognition particle in S. cerevisiae. Cell 67:131-144. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, M. L., R. K. Scopes, R. L. Moritz, R. J. Simpson, C. Englert, F. Pfeifer, and M. L. Dyall-Smith. 1997. Purification and analysis of an extremely halophilic beta-galactosidase from Haloferax alicantei. Biochim. Biophys. Acta 1337:276-286. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, A. E., and M. A. van Waes. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15:799-842. [DOI] [PubMed] [Google Scholar]
- 18.Jolley, K. A., E. Rapaport, D. W. Hough, M. J. Danson, W. G. Woods, and M. L. Dyall-Smith. 1996. Dihydrolipoamide dehydrogenase from the halophilic archaeon Haloferax volcanii: homologous overexpression of the cloned gene. J. Bacteriol. 178:3044-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lütcke, H. 1995. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem 228:531-550. [DOI] [PubMed] [Google Scholar]
- 20.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162:461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moll, R., S. Schmidtke, and G. Schäfer. 1999. Domain structure, GTP-hydrolyzing activity and 7S RNA binding of Acidianus ambivalens Ffh-homologous protein suggest an SRP-like complex in archaea. Eur. J. Biochem. 259:441-448. [DOI] [PubMed] [Google Scholar]
- 22.Mutka, S. C., and P. Walter. 2001. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell 12:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura, K., S. Yahagi, T. Yamazaki, and K. Yamane. 1999. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem. 274:13569-13576. [DOI] [PubMed] [Google Scholar]
- 24.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortenberg, R., and M. Mevarech. 2000. Evidence for post-translational membrane insertion of the integral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii. J. Biol. Chem. 275:22839-22846. [DOI] [PubMed] [Google Scholar]
- 26.Patenge, N., and J. Soppa. 1999. Extensive proteolysis inhibits high-level production of eukaryal G protein-coupled receptors in the archaeon Haloferax volcanii. FEMS Microbiol. Lett. 171:27-35. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744-746. [DOI] [PubMed] [Google Scholar]
- 28.Plath, K., W. Mothes, B. M. Wilkinson, C. J. Stirling, and T. A. Rapoport. 1998. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94:795-807. [DOI] [PubMed] [Google Scholar]
- 29.Pohlschroder, M., C. Murphy, and J. Beckwith. 1996. In vivo analyses of interactions between SecE and SecY, core components of the Escherichia coli protein translocation machinery. J. Biol. Chem. 271:19908-19914. [DOI] [PubMed] [Google Scholar]
- 30.Pohlschröder, M., W. A. Prinz, E. Hartmann, and J. Beckwith. 1997. Protein translocation in the three domains of life: variations on a theme. Cell 91:563-566. [DOI] [PubMed] [Google Scholar]
- 31.Rapoport, T. A., B. Jungnickel, and U. Kutay. 1996. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65:271-303. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Tian, H., and J. Beckwith. 2002. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition pathway. J. Bacteriol. 184:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33b.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 34.Wendoloski, D., C. Ferrer, and M. L. Dyall-Smith. 2001. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147:959-964. [DOI] [PubMed] [Google Scholar]
- 35.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Toward a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, B. P., R. A. Craven, P. J. Reid, M. Willer, and C. J. Stirling. 2001. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 20:262-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Zweib, C., and J. Eichler. 2002. Getting on target: the archaeal signal recognition particle. Archaea 1:27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwick, M. B., L. L. Bonnycastle, K. A. Noren, S. Venturini, E. Leong, C. F. Barbas III, C. J. Noren, and J. K. Scott. 1998. The maltose-binding protein as a scaffold for monovalent display of peptides derived from phage libraries. Anal. Biochem. 264:87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]