Abstract
Methylobacterium chloromethanicum CM4 is an aerobic α-proteobacterium capable of growth with chloromethane as the sole carbon and energy source. Two proteins, CmuA and CmuB, were previously purified and shown to catalyze the dehalogenation of chloromethane and the vitamin B12-mediated transfer of the methyl group of chloromethane to tetrahydrofolate. Three genes located near cmuA and cmuB, designated metF, folD and purU and encoding homologs of methylene tetrahydrofolate (methylene-H4folate) reductase, methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase and formyl-H4folate hydrolase, respectively, suggested the existence of a chloromethane-specific oxidation pathway from methyl-tetrahydrofolate to formate in strain CM4. Hybridization and PCR analysis indicated that these genes were absent in Methylobacterium extorquens AM1, which is unable to grow with chloromethane. Studies with transcriptional xylE fusions demonstrated the chloromethane-dependent expression of these genes. Transcriptional start sites were mapped by primer extension and allowed to define three transcriptional units, each likely comprising several genes, that were specifically expressed during growth of strain CM4 with chloromethane. The DNA sequences of the deduced promoters display a high degree of sequence conservation but differ from the Methylobacterium promoters described thus far. As shown previously for purU, inactivation of the metF gene resulted in a CM4 mutant unable to grow with chloromethane. Methylene-H4folate reductase activity was detected in a cell extract of strain CM4 only in the presence of chloromethane but not in the metF mutant. Taken together, these data provide evidence that M. chloromethanicum CM4 requires a specific set of tetrahydrofolate-dependent enzymes for growth with chloromethane.
Aerobic methylotrophic α-proteobacteria of the genus Methylobacterium can grow with single-carbon compounds such as methanol and methylamine as the carbon and energy source (15). After oxidation to formaldehyde, a central intermediate in methylotrophic metabolism (26), carbon is either assimilated via the serine cycle or completely oxidized to carbon dioxide (15). Historically, the oxidation of formaldehyde was thought to proceed via a linear pathway involving the sequential action of formaldehyde dehydrogenase and formate dehydrogenase. However, formaldehyde dehydrogenases described for Methylobacterium species are broad-range aldehyde dehydrogenases with often low specific activities with formaldehyde. The relevance of such a pathway for growth of Methylobacterium with C1 compounds was therefore uncertain (3), and a pathway proceeding via pterin-dependent intermediates was suggested as an alternative (17). Indeed, two parallel pterin-dependent pathways were recently shown to be essential for growth with methanol in Methylobacterium extorquens AM1 (6) (Fig. 1). One of these pathways is tetrahydromethanopterin (H4MPT)-dependent and appears to be present in most methylotrophic bacteria (40). The other is tetrahydrofolate (H4folate) dependent and has so far been characterized in M. extorquens AM1 only. It is believed that the H4MPT-dependent pathway operates predominantly in the oxidative direction but that the H4folate-dependent pathway functions in either direction depending upon the cellular pools of C1 intermediates available for biosynthesis and energy generation (6, 30, 39).
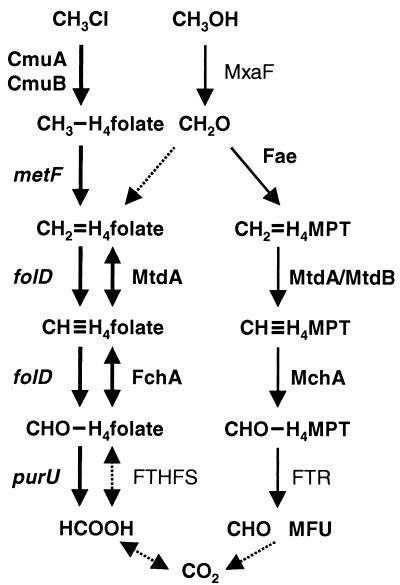
FIG. 1.
Proposed model of pterin-dependent C1 metabolism in M. chloromethanicum CM4. Transformations shown by light arrows indicate reactions involved in growth of M. extorquens AM1 with methanol (6). Transformations shown by heavy arrows are proposed to be specific to the chloromethane-degrading strain M. chloromethanicum CM4. CmuA, chloromethane-corrinoid methyltransferase (34); CmuB, methylcobalamin-H4folate methyltransferase (35); metF, methylene-H4folate reductase; folD, bifunctional methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase; purU, 10-formyl-H4folate hydrolase; MtdA, NADP-dependent methylene-H4folate-H4MPT dehydrogenase (39); FchA, methenyl-H4folate cyclohydrolase; (30); FTHFS, formyl-H4folate synthetase; Fae, formaldehyde-activating enzyme (41); MtdB, NAD(P)-dependent methylene-H4MPT dehydrogenase (10); Mch, methenyl-H4MPT cyclohydrolase; (30) FTR, formylmethanofuran-H4MPT formyltransferase (29). Enzymes indicated in boldface were detected in strain CM4 at the DNA level (reference 13 and PCR data not shown).
Methylobacterium chloromethanicum CM4 is distinct from other Methylobacterium species described thus far in its ability to grow with chloromethane as sole carbon and energy source (7). However, it is phylogenetically closely related (98% 16S ribosomal DNA sequence identity) to M. extorquens AM1 (23). Thus, central C1 metabolism is expected to be similar in both strains. Physiological and genetic studies already demonstrated that the cmuA and cmuB genes of strain CM4 are essential for growth on chloromethane and that they encode the proteins responsible for the dehalogenation of this compound (37). The two corresponding proteins were recently purified and shown to catalyze the dehalogenation of chloromethane in vitro (34, 35). Dehalogenation occurs by methyl group transfer from chloromethane onto a vitamin B12 cofactor bound to the protein CmuA (34). The methyl group is then transferred from CmuA to H4folate by CmuB (34). Importantly, the CmuA enzyme is unable to catalyze methyl transfer to H4MPT and is therefore specific for H4folate (35).
It appeared possible that methyl-H4folate generated from chloromethane by the conjugated action of CmuA and CmuB in strain CM4 is oxidized to formate via H4folate-linked intermediates and not oxidized via formaldehyde as other C1 growth substrates of Methylobacterium (6, 26, 42). In support of this idea, genes designated metF, folD, and purU, which encode homologs of methylene-H4folate reductase, methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase and formyl-H4folate hydrolase, respectively, were found near cmuA and cmuB in the CM4 genome (36). This suggested that the H4folate-dependent chloromethane utilization pathway in strain CM4 may be distinct from the corresponding methanol utilization pathway recently defined in M. extorquens AM1 (6), since the latter involves two proteins, methylene-H4folate dehydrogenase MtdA (39) and methenyl-H4folate cyclohydrolase FchA (30), which are unrelated to the bifunctional methylene-H4folate dehydrogenase-cyclohydrolase homolog encoded by folD found near cmuA in strain CM4.
In the present study, we report experimental evidence for the existence of a C1 oxidation pathway specific for chloromethane in M. chloromethanicum CM4, obtained by transcriptional analysis, gene inactivation studies, and enzyme measurements.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, T4 ligase and T4 polymerase were obtained from MBI Fermentas, and Pfu DNA polymerase was from Stratagene. All chemicals were from Fluka, Sigma, or Merck except where noted.
Bacterial strains and plasmids.
M. chloromethanicum CM4 wild-type (7, 23) and the cmuA, cmuB, and purU mutants (37) were described previously. Escherichia coli strains DH5α (4) and CC118(λpir) (11), pBluescript II KS(+) vector (Stratagene), the broad-host plasmid pCM62 (18), the xylE promoter probe vector pCM130 (18), and the gene inactivation plasmid pKNOCK-Km (1) were used for cloning. Conjugation of plasmids into M. chloromethanicum CM4 was performed with E. coli S17-1 (33) or S17-1(λpir) (25) and selection of transconjugants was performed as described previously (12).
Media and growth conditions.
Luria-Bertani medium (24) was used for growth of E. coli. M. chloromethanicum was grown in 500-ml rubber-stoppered serum bottles filled with 100 ml of phosphate mineral medium (37). Chloromethane gas was added with a syringe through the rubber stopper to a final concentration of 4.5 mM (2% [vol/vol]), corresponding to an initial concentration of 1.4 mM in the liquid phase, assuming a Henry constant of 0.43 (9). Methanol (sterile filtered) was added to a concentration of 20 to 40 mM. Ampicillin (100 μg/ml, final concentration) and kanamycin and tetracycline (25 μg/ml) were used where required.
DNA manipulations.
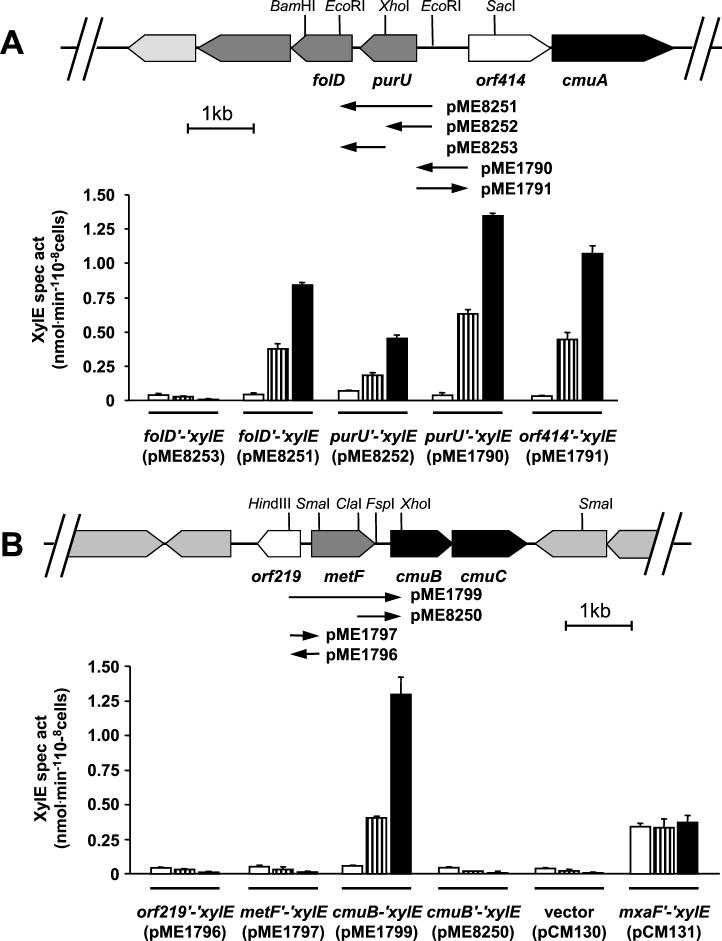
Preparation of total and plasmid DNA, recombinant DNA work, and Southern analysis were performed according to standard protocols (4). All cloning junctions and cloned PCR products were checked by sequencing by using Dye-Terminator chemistry on an ABI 310 Genetic Analyzer (Perkin-Elmer). Transcriptional xylE fusions of the folD and purU genes were obtained by the insertion of cloned DNA from strain CM4 (36) into the xylE reporter plasmid pCM130 (Fig. 2) (18). A 1.4-kb EcoRI fragment containing purU and part of the folD gene (Fig. 2A) was cloned into pBluescript (plasmid pME1798) and subcloned as a BamHI-HindIII fragment into pCM130 digested with the same restriction enzymes, yielding plasmid pME8251. A 0.7-kb HindIII/XhoI fragment of pME1798 with the XhoI site filled in was cloned into pCM130 sequentially digested with PstI, blunted, and digested with HindIII to yield plasmid pME8252. Plasmid pME8253 was constructed by digestion of plasmid pME8251 with HindIII and XhoI, filling in, and religation.
FIG. 2.
Expression of plasmid-borne xylE fusions in M. chloromethanicum CM4. Genetic organization of the gene clusters I (A) and II (B) involved in chloromethane utilization in M. chloromethanicum CM4 (36). Genes encoding methyltransferases are shown in black, genes encoding putative H4folate-dependent enzymes in C1 metabolism are shown in dark gray, vitamin B12 biosynthesis genes are shown in light gray, and genes of unknown function are shown in white. Bar diagrams show catechol dioxygenase activity in transconjugants of wild-type M. chloromethanicum CM4 with different intergenic DNA sequences fused to the promoterless xylE gene of vector pCM130 (18) grown on methanol (□), a mixture of methanol and chloromethane (hashed bars), or chloromethane (▪). Plasmid constructs (see Materials and Methods) are schematically indicated below the sequence, with the orientation of the xylE gene indicated by a black arrowhead.
The purU-orf414 intergenic region was cloned into promoter probe vector pCM130 (18) as a PCR fragment generated with primers (5′ to 3′) TTCCGCCATCTAGAGATTCC (nucleotides [nt] 4841 to 4860 in database entry AJ011316, XbaI site [in boldface] introduced by two nucleotide sequence changes [underlined]) and GGCGACATATGACGGCACC (nt 5635 to 5617 in AJ011316, introducing an NdeI site). The resulting 779-bp PCR fragment was digested with XbaI and NdeI, its ends were filled in, and it was ligated with HindIII digested and blunted pCM130, yielding plasmids pME1790 (purU′-′xylE fusion) and pME1791 (orf414′-′xylE fusion).
The cmuB′-′xylE fusions in plasmids pME1799 and pME8250 (Fig. 2B) were constructed by cloning a 1.7-kb HindIII/XhoI genomic fragment and a 0.65-kb ClaI/XhoI genomic fragment as blunt-ended fragments into CM130 as described above. The orf219-metF intergenic region was PCR-amplified with primers GCTTTCGGATCCATCAGACG, (nt 3332 to 3351 in AJ011317, with two nucleotide changes introducing a BamHI site) and CCACATATGCGGATCCGTC, (nt 3512 to 3494 in AJ011317). The obtained 159-bp PCR product was cloned as a BamHI fragment into pCM130 digested with the same enzyme, resulting in plasmid pME1796 (orf219′-′xylE fusion) and pME1797 (metF′-′xylE fusion).
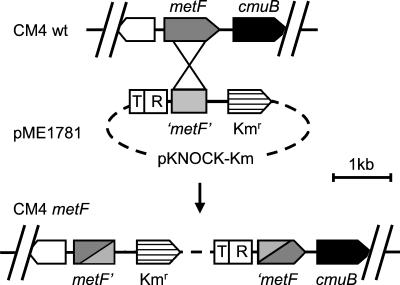
The folD gene of strain CM4 mutant was mutagenized by single-crossover recombination of plasmid pME1774, a derivative of pKNOCK-Km containing the 0.5-kb EcoRI-BamHI internal fragment of the folD gene. The metF gene of strain CM4 was mutagenized in the same way with pME1781 containing the 597-bp SmaI/ClaI internal fragment of metF in pKNOCK-Km (see Fig. 5). Derivatives of plasmid pCM62 (18) constructed for complementation studies contained the genes of interest under Plac control and the following genes (see Fig. 2B): purU (2.8-kb BamHI-SacII fragment, plasmid pME1776), metF (1.2-kb KpnI-FspI fragment, pME1789), or metF-cmuB-cmuC and the 5′ ends of orf219 and orf361 (pME1793, 4.5-kb HindIII-SmaI fragment).
FIG. 5.
Construction of the metF mutant. Abbreviations: pKNOCK-Km, 2.1-kb broad-host suicide vector (1); T, RP4 plasmid oriT region; R, R6K γ-origin of replication; Kmr, kanamycin resistance gene.
PCR detection of C1 utilization genes.
PCR was performed according to standard protocols with addition of dimethyl sulfoxide to a final concentration of 4% (vol/vol), in a volume of 50 μl with 10 ng of total DNA and 30 pmol of each primer, by using 30 cycles of 1 min in each step of denaturation (94°C), annealing (54°C), and elongation (72°C). The primer sequences used corresponded to known genes of M. extorquens AM1, i.e., TGTGGCGCTCGTCGTCAAGG and GAGCTTCAAGCCGCCGATGC for mtdA (product: 454 bp) and TGGTCTCGATGGTGTGTAACC and CGAGATGACGTTGAGATTGC (326 bp) for fchA, and to known genes of M. chloromethanicum CM4, i.e., CCCTGTGAATGCGGGTTTGC and ACCTCCGGGGCACGTATTGC for folD (504 bp), GCTCGAGCAACTTGTCCTC and CAATATCGTCGAAGCGAAGC for purU (386 bp), and CAAGAAGCTGTCTCCTACGC and GCTCGCGAAACTATATCTGC for metF (910 bp), respectively.
XylE activity measurements.
M. chloromethanicum CM4 cultures containing derivatives of xylE fusion reporter plasmid pCM130 (18) were grown at 30°C and harvested in late exponential phase (A600 of 0.6 to 0.7). Samples (1.5 ml) were centrifuged and resuspended in 0.2 ml of cold catechol-2,3-dioxygenase (XylE) assay buffer (50 mM potassium phosphate [pH 7.5], 10% acetone). A 50-μl aliquot was added to 1 ml of assay buffer, the reaction was initiated by addition of 10 μl of 0.1 M catechol-1,2(dihydroxybenzene), and the change in absorbance at 375 nm was recorded at 25°C. A molar extinction coefficient (ɛ375) of 4.4 × 104 M−1 cm−1 was used for calculating the specific activity of XylE (31), which was expressed as nmol product liberated per min per 108 cells assayed. An A600 of 1 was shown to correspond 4 × 108 cells per ml (unpublished observations).
RNA isolation.
Total RNA was isolated from M. chloromethanicum CM4 cells grown to an A600 of 0.6 to 0.7 as described previously (38). A culture aliquot (25 ml) was added to 20 ml of frozen and crushed 20 mM Tris chloride buffer [pH 7.5] containing 5 mM MgCl2 and 20 mM sodium azide. Cells were harvested by centrifugation (20,000 × g, 15 min) and resuspended in 2.5 ml of ice-cold 1 mM EDTA in 20 mM sodium acetate buffer (pH 5.5), and the aqueous phase was extracted with 2.5 ml of prewarmed (65°C) acidic phenol (pH 5.5) containing 0.5% (wt/vol) sodium dodecyl sulfate. The aqueous phase was further extracted with 2.5 ml of phenol-chloroform-isoamyl alcohol (49.5:49.5:1) and with 2.5 ml of dichloromethane. The RNA was precipitated with 2.5 volumes of ethanol and dissolved in water which had been treated with 0.1% diethyl pyrocarbonate.
Mapping of transcriptional start sites.
M. chloromethanicum CM4 was grown with either 40 mM methanol or 5% chloromethane, and RNA isolated as described above. Approximately 20 μg of RNA and 2 × 105 to 3 × 105 cpm of radiolabeled primer was used for primer extension experiments, which were performed as previously described (5). Extension products were purified by phenol extraction, followed by ethanol precipitation before separation on 6% denaturing polyacrylamide gels. The primers used were 5′-CGCACCTGAAACGGCAGCGACGATGC-3′ (nt 4800 to 4775 in AJ011316) for purU, 5′-CGACAGACCCGAACCTCGCCATTGG-3′ (nt 5509 to 5533 in AJ011316) for orf414, and 5′-GGGAGACCTCCAATGACAGATCGCG-3′ (nt 3627 to 3603 in AJ011317) for metF. Sequencing reactions were carried out with the same primers and a suitable plasmid as the template by using the fmol Cycle Sequencing kit (Promega).
Methylene-H4folate reductase assay.
Bacteria were grown to late exponential phase (A600 of 0.6 to 0.8) with 40 mM methanol, 5% chloromethane, or a mixture of both carbon sources or were grown with methanol and induced with 2% chloromethane for 8 h. Enzyme measurements were performed in cell extracts prepared as described previously (36). Labeled 5-[14C]-methyl-H4folate (barium salt [Amersham Biosciences], 57 mCi/mmol) was added to unlabeled (6R,S)-methyl-H4folate (Sigma) in 8 mM ascorbate buffer to a final stock concentration of 5 mM methyl-H4folate (925 dpm of 14C label/nmol) as described previously (32). Methylene-H4folate reductase activity was measured at 37°C according to previously reported methods (20, 32) with ca. 60 μg of total cell extract protein in 0.75 ml of 100 mM potassium phosphate, 0.6 mM EDTA, 0.67% bovine serum albumin buffer (pH 6.7), 0.15 ml of saturated menadione solution in 20% aqueous methanol, 0.36 ml of labeled methyl-H4folate stock solution (final concentration, 1.2 mM), and water to a total volume of 1.5 ml. Aliquots (0.25 ml) were taken at time points up to 5 min, quenched by the addition of 75 μl of 1 M sodium acetate acid containing 3 mg of dimedone/ml, and incubated at 100°C for 2 min. The formaldehyde released from methylene-H4folate and derivatized with dimedone was extracted into toluene (1 ml) by vortexing for exactly 2 min. After centrifugation (Jouan A14 centrifuge) at maximal speed (17,500 × g) for 1 min, an aliquot of the toluene phase (0.25 ml) was added to 1 ml of scintillation fluid (Microscint 40; Packard) and counted (Beckman LS1801). The reported data represent the average of three independent determinations.
RESULTS
Identification of genes encoding pterin-dependent C1 interconverting enzymes in M. chloromethanicum CM4.
Genomic DNA from M. extorquens AM1, which is unable to grow with chloromethane, was hybridized with probes for the purU, folD, and metF genes of M. chloromethanicum CM4. These genes encode homologs of methylene-H4folate reductase, methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase, and formyl-H4folate hydrolase, respectively, and were previously proposed (36) to define a specific oxidation pathway for chloromethane in strain CM4 (Fig. 1). No specifically hybridizing DNA fragments were detected (data not shown). The absence of these genes was further suggested by PCR analysis with primer pairs specific to the sequences of these genes from strain CM4 (see Materials and Methods; data not shown). That M. extorquens AM1 lacked homologs of metF, folD, and purU was also indicated by analysis of the already available sequences from the M. extorquens AM1 genome sequencing project (L. Chistoserdova and M. E. Lidstrom, unpublished data; also see http://pedant.gsf.de/cgi-bin/wwwfly.pl?Set=Mextorquens&Page=index). Conversely, however, hybridization analysis with probes corresponding to the mtdA, fae, and mchA genes of M. extorquens AM1 representing both branches of C1 utilization described in M. extorquens AM1 (6, 39, 41) (Fig. 1) showed that these genes were also present in strain CM4 and in the dichloromethane-degrading strain Methylobacterium dichloromethanicum DM4 (13). In the latter strain, sequence analysis revealed very strong sequence conservation of these genes (>96% sequence identity at the DNA level) with their homologs of strain AM1 (13). This analysis was confirmed and extended by obtaining the expected PCR products for mtdA (454 bp; see Materials and Methods) and fchA (326 bp), which define the H4folate-dependent branch of C1 characterized in M. extorquens AM1 (Fig. 1), by using total DNA from strain CM4 as a template and primers derived from the sequences of these genes that were reported for strain AM1 (data not shown). Taken together, therefore, our data indicate that strain CM4 not only possesses both the H4folate- and H4MPT-dependent pathways of C1 metabolism present in M. extorquens AM1 but also an additional set of genes encoding a third H4folate-dependent pathway for chloromethane degradation.
Analysis of transcriptional xylE fusions.
In order to gain insight into chloromethane-dependent gene regulation in M. chloromethanicum CM4, plasmids harboring transcriptional xylE fusions of the metF, folD, purU, cmuB, and orf414 genes (Fig. 2) were constructed and conjugated into wild-type strain CM4. Transconjugant strains were grown with methanol, with chloromethane, or with a mixture of both these carbon sources. Catechol dioxygenase activity was measured in exponentially growing bacteria. The controls used in these experiments were M. chloromethanicum CM4 harboring the low-background xylE promoter-probe vector pCM130 (18) or plasmid pCM131, in which the xylE gene is constitutively expressed under the control of the promoter of the large subunit methanol dehydrogenase gene (mxaF) from strain AM1 (18). The previously noted (18) high expression of the latter reporter fusion during growth on methanol was verified (Fig. 2B). Expression of XylE from both purU′-′xylE and folD′-′xylE fusions (plasmids pME8251 and pME8252) was induced in the presence of chloromethane (Fig. 2A). In contrast, the reporter folD′-′xylE construct pME8253 lacking the DNA sequence upstream of purU showed no chloromethane-induced catechol dioxygenase activity. This suggested that expression of purU and folD was induced by chloromethane from a common promoter located upstream of the purU gene (Fig. 2A). Further, a DNA fragment including the entire intergenic region beween purU and orf414 also led to high chloromethane-induced XylE activity for the purU′-′xylE fusion construct (plasmid pME1790, Fig. 2A). Construct pME1791, which contains the same insert as pME1790 but in the reverse orientation, was used as a promoter probe vector for monitoring expression of orf414 and cmuA genes as a orf414′-′xylE fusion (Fig. 2A). The catechol dioxygenase activity measured with these two last constructs were in the same range as for the other purU′-′xylE and folD′-′xylE reporter fusions.
Regarding the genes found in cluster II, the data obtained with the two transcriptional fusions metFcmuB′-′xylE and cmuB′-′xylE (plasmids pME1799 and pME8250, Fig. 2B) indicated that expression of cmuB is controlled by a promoter located upstream of the metF gene rather than in the noncoding region between metF and cmuB. Interestingly, however, the short intergenic sequence between the start codons of the divergent orf219 and metF genes was by itself insufficient to induce XylE expression in either direction (Fig. 2B, plasmids pME1796 and pME1797).
Determination of chloromethane induced transcription initiation sites in M. chloromethanicum CM4.
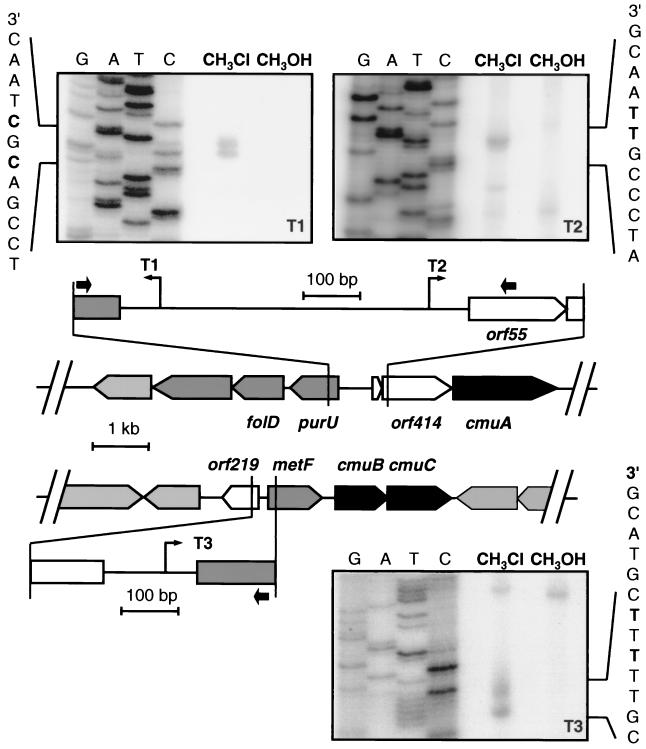
Expression studies with transcriptional fusions suggested the presence of at least three chloromethane-inducible promoters located upstream of purU, metF, and orf414, respectively (Fig. 2). Transcriptional start sites were mapped by primer extension with RNA isolated from wild-type M. chloromethanicum CM4 grown with either chloromethane or methanol. Specific elongation products were obtained with RNA from chloromethane-grown cells that were absent in RNA from cultures grown with methanol, and their 3′ ends mapped upstream of genes purU, metF, and orf414 (Fig. 3). In contrast, no extension products were obtained with primers designed to detect transcriptional start sites upstream of cmuA, folD, and orf219 (data not shown). The mapping of a transcriptional start site 340 nt upstream of the translational start of orf414 revealed a previously overlooked short open reading frame of 165 nt, orf55, which could be transcriptionally coupled with orf414 since the stop codon of orf55 overlaps with the predicted start codon of orf414 (Fig. 3). The putative promoter sequences upstream of the chloromethane-induced transcripts exhibited striking sequence conservation (Fig. 4). Their −35 region was identical to the minus −35 region of the dichloromethane dehalogenase promoter of M. dichloromethanicum DM4 (14) and that of the E. coli consensus promoter. Conservation was less extensive in the −10 region, and the deduced consensus had no similarity to the −10 promoter regions previously identified for Methylobacterium genes (Fig. 4).
FIG. 3.
Transcriptional start sites of chloromethane induced genes in M. chloromethanicum CM4. RNA was isolated from M. chloromethanicum CM4 grown with chloromethane or methanol and reverse transcribed with the primers indicated by black arrows on the schematic view of the two gene clusters (see Fig. 2). Lanes A, T, C, and G show the sequencing ladders obtained with the same primers. The identified transcriptional start sites are marked in boldface (hooked arrows; T1, AJ011316, nt 4918 and 4920; T2, AJ011316, nt 5383 and 5384; and T3, AJ011317, nt 3483 and 3485).
FIG. 4.
Alignment of chloromethane specific promoter regions in M. chloromethanicum CM4. The putative −35 and −10 regions are indicated in boldface, and experimentally determined transcription initiation sites are underlined. A likely consensus promoter is shown. Promoter regions of previously identified Methylobacterium genes involved in one-carbon metabolism are also shown for comparison. PdcmA, dichloromethane dehalogenase promoter of M. dichloromethanicum (14); PmxaF(AM1), methanol dehydrogenase promoter of M. extorquens AM1 (2); PmxaF(XX), methanol dehydrogenase promoter of Methylobacterium organophilum XX (16). The E. coli σ70 consensus promoter sequence (43) is also given for comparison.
Analysis of gene disruptions in purU, folD, and metF.
The previously obtained minitransposon purU mutant (37), although unable to grow with chloromethane, exhibited wild-type chloromethane dehalogenase activity (36). In comparison, cmuA mutants (36) were unable to dehalogenate chloromethane but showed the same growth phenotype as the purU mutant. The growth yield of a cmuA mutant with a mixture of methanol and chloromethane was only two-thirds that of the wild-type CM4 (Table 1), since the cmuA mutant metabolized the methanol but not the chloromethane present in the medium (Table 1). In contrast, the growth yield of the purU mutant was in the same range as that of the wild type during growth on a mixture of chloromethane and methanol (Table 1). This indicated that the purU mutant was able to assimilate carbon derived from the dehalogenation of chloromethane in the presence of methanol even if it did not grow with chloromethane as the sole carbon source. Providing the purU gene in trans by ways of plasmid pME1776 was sufficient to restore growth of the purU mutant with chloromethane. Southern blot analysis confirmed that plasmid pME1776 was stably maintained in strain CM4 and did not recombine into the chromosome (data not shown).
TABLE 1.
Growth yields of M. chloromethanicum CM4 wild-type and mutantsa
| CM4 strain | Growth yield (g [dry weight] mol of C−1)
|
||
|---|---|---|---|
| MeOH | MeOH-CH3Cl | CH3Cl | |
| Wild type | 10.0 ± 0.2 | 9.5 ± 0.2 | 10.6 ± 0.7 |
| cmuA | 9.7 ± 0.3 | 6.2 ± 0.2 | NG |
| purU | 9.9 ± 0.3 | 9.6 ± 0.2 | NG |
| folD | 8.9 ± 0.6 | 9.1 ± 0.2 | 11.3 ± 1.1 |
The average of three independent cultures grown with either 20 mM methanol (MeOH), 20 mM methanol and 10 mM chloromethane (MeOH-CH3Cl), or 10 mM chloromethane (CH3Cl) is given. NG, no growth.
A mutant in metF, encoding a methylene-H4folate reductase homolog suspected to represent the next enzyme of the chloromethane oxidation pathway after dehalogenation of chloromethane to methyltetrahydrofolate (34, 36), was also constructed by insertional mutagenesis with a derivative of the suicide broad-host-range vector pKNOCK-Km (1) containing an internal fragment of the metF gene. Southern blot analysis demonstrated that the gene knockout plasmid had inserted into the CM4 genome as expected, causing a disruption of metF (Fig. 5 and data not shown). The growth rate of the metF mutant on methanol was in the same range as that of the wild type, whereas no growth was observed on chloromethane or, interestingly, on a mixture of chloromethane and methanol (data not shown). This suggested a toxic effect of chloromethane dehalogenation in this mutant. Growth on chloromethane was restored by mating the metF mutant with derivatives of plasmid pCM62 (18) containing the metF gene. Plasmid pME1793 containing metF, together with the downstream cmuB and cmuC genes, was stably maintained in the metF mutant growing with chloromethane. However, plasmid pME1789 containing only the metF gene recombined into the CM4 genome upon cultivation of the transconjugant on chloromethane (data not shown). Taken together, these observations confirmed not only that the observed growth phenotype of the metF mutant was caused by disruption of the metF gene but also that the genes cmuB and cmuC essential for growth of strain CM4 with chloromethane (36) were cotranscribed with metF.
The folD gene, encoding a homolog of the bifunctional enzyme methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase and suggested from experiments with reporter gene fusions to be cotranscribed with purU, was disrupted by the same strategy as for the metF mutant. The resulting folD mutant strain was shown by hybridization analysis to lack an intact folD gene as a consequence of the recombination of the plasmid into the chromosome (data not shown). This mutant, however, grew with chloromethane as well as the wild type (Table 1).
Measurements of methylene-H4folate reductase activity.
Cell extracts of wild-type CM4, its metF mutant, and the metF mutant complemented by expression of metF, cmuB, and cmuC genes from plasmid pME1793 were assayed for methylene-H4folate reductase activity by measuring the conversion of [14C]methyl-H4folate to methylene-H4folate by a previously described method (19, 32). This provided unequivocal evidence for chloromethane-induced methylene-H4folate reductase activity of MetF (Table 2). Cell extracts of wild-type CM4 grown with chloromethane showed significant methylene-H4folate reductase activity, unlike those of the strain grown with methanol. Strain CM4 grown on a mixture of both growth substrates showed an intermediate level of activity. Importantly, low but significant activity was also detected in an extract of strain CM4 grown with methanol and then induced with chloromethane for 8 h. In contrast, the corresponding extract of the metF mutant showed no detectable activity under the same conditions (Table 2). Finally, the very high and specific induction of enzyme activity in the complemented mutant grown in the presence of chloromethane (Table 2) confirmed that the metF gene from strain CM4 encodes a methylene tetrahydrofolate reductase whose expression is specifically regulated by chloromethane.
TABLE 2.
Methylene-H4folate reductase activity in cell extracts of bacteria grown with different C1 sources
| CM4 strain | Sp act (nmol min−1 mg−1)
|
||
|---|---|---|---|
| MeOH | MeOH-CH3Cl | CH3Cl | |
| Wild type | <0.7 | 158 ± 20 | 245 ± 44 |
| 6.5 ± 0.6a | <0.5b | ||
| metF | <0.4 | <0.5a | NGc |
| metF(pME1793) | 8.5 ± 0.9 | 931 ± 52 | 1,378 ± 77 |
Grown with 20 mM methanol and induced with 2% CH3Cl for 8 h.
Extract was boiled for 5 min before measurement.
NG, no growth.
DISCUSSION
The data obtained with xylE reporter fusions of metF, purU, and folD (Fig. 2) and the chloromethane-regulated transcriptional units that were identified (Fig. 3 and Fig. 4) provide evidence for the existence of a chloromethane-induced H4folate-dependent pathway for the oxidation of chloromethane that involves metF, folD, and purU (Fig. 1). This pathway runs alongside to that defined by methylene-H4folate dehydrogenase MtdA (39) and methenyl-H4folate cyclohydrolase FchA (30), which are both essential for growth of strain AM1 with methanol and, at the genetic level at least, are also present in strain CM4.
A database search using the promoter consensus proposed in Fig. 4 as a pattern failed to detect any other such sequence motifs in available sequences from M. chloromethanicum CM4, in the gene upstream sequences available for the chloromethane degraders Aminobacter sp. strain IMB-1 (metF, cmuB, and cmuA homologs [44]) and Hyphomicrobium chloromethanicum CM2 (folD homolog [22]) or indeed in any other Methylobacterium sequences deposited in databases. Thus, the identified promoter consensus is probably neither utilized for the regulation of chloromethane genes in methylotrophic bacteria nor for driving gene expression in Methylobacterium species in general. However, the existence of a consensus promoter sequence for chloromethane-regulated genes in strain CM4 suggests that the expression of these genes could be driven by a common regulator. Experiments aiming at the identification of such a regulator protein are in progress.
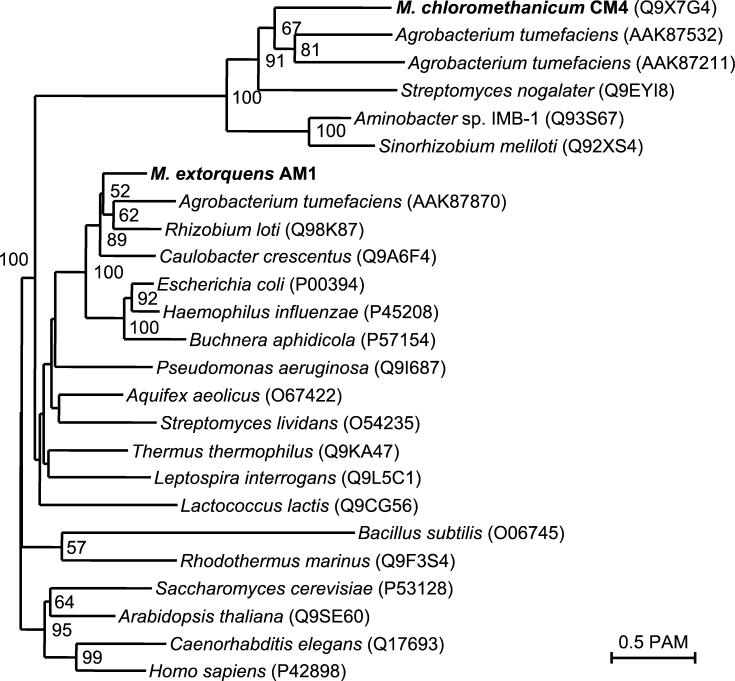
Evidence for the specific involvement of metF and purU in a chloromethane oxidation pathway to formate in strain CM4 stems from the phenotypic properties of the corresponding mutants and the absence of these genes in strain AM1. The observation that the metF mutant of strain CM4 was not only unable to grow with chloromethane but was sensitive to its presence during growth with methanol is worth emphasizing. We speculate that in the metF mutant, folate accumulates as methyl-H4folate upon dehalogenation of chloromethane and that this is detrimental for the function of biosynthetic pathways that rely on C1 moieties provided by H4folate. Such a “methyl trap” model was already proposed for the clinical picture observed in human patients with pernicious anemia, in which the presence of a dysfunctional methionine synthase leads to the accumulation of methyl-H4folate (19) and to poor availability of the H4folate cofactor for other important physiological processes. Strikingly, MetF of strain CM4 displays only low sequence similarity to the majority of known methylene-H4folate reductases (see Fig. 6). The MetF homolog of the halomethane-utilizer Aminobacter sp. strain IMB-1 (44) is more closely related to the CM4 MetF rather than that encoded in the M. extorquens AM1 genome sequence (26% sequence identity [Chistoserdova and Lidstrom, unpublished, and Fig. 6]). This might reflect the fact that in chloromethane degradation, the role of MetF is to enable the efficient oxidation of methyl-H4folate to methylene-H4folate (Fig. 1, Table 2). The main function of MetF is usually in the opposite direction, affording one-carbon precursors for the synthesis of methionine from homocysteine, as in E. coli, for example (21).
FIG. 6.
Phylogenetic analysis of MetF proteins from bacteria. A multiple alignment of representative MetF sequences produced with T-Coffee (27) was used to generate a phylogenetic tree (325 aligned positions) with FITCH from PHYLIP (8). The tree was drawn with NJPlot (28). Nodes confirmed by bootstrapping analysis are indicated by the percent conservation, and the accession numbers of protein sequences are given in parentheses.
Like the metF mutant, the purU mutant is unable to grow with chloromethane alone. However, its growth yield on a mixture of chloromethane and methanol is undistinguishable from that of the wild-type (Table 1), indicating that the tetrahydrofolate-bound carbon derived from chloromethane can be metabolized in a productive fashion despite the lack of this gene. No purU gene could be detected in strain AM1 (our DNA hybridization and PCR data; Chistoserdova and Lidstrom, unpublished), and it appears that the functionally homologous enzyme involved in the interconversion of formyl-H4folate and formate in strain AM1 is not related to PurU (J. Vorholt, unpublished data). We therefore assume at present that in CM4 purU encodes a specific chloromethane-induced formyl-H4folate hydrolase and that in the absence of purU formyl-H4folate can be transformed to formate by another enzyme common to strain AM1 and CM4 which is expressed during growth in the presence of methanol but not during growth with chloromethane alone.
The evidence for a role of folD in the chloromethane-degrading pathway of folD, suggested by its sequence to be a methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase, is more indirect. Indeed, the retained ability of the folD mutant to grow with chloromethane, as well as the complementation of the growth phenotype of the purU mutant by an intact copy of purU in trans without the need for the downstream folD gene, actually shows that the folD gene is dispensable for chloromethane metabolism. However, its coinduction with purU in strain CM4 (Fig. 2) and its location near the chloromethane dehalogenase gene cmuA in H. chloromethanicum CM2 (22) support the idea that folD is involved in C1 oxidation during chloromethane utilization. Moreover, no folD homolog was yet detected in strain AM1 (unpublished data and Chistoserdova and Lidstrom, unpublished). At present, therefore, we assume that in the CM4 folD mutant growing with chloromethane, the two reactions from methylene-H4folate dehydrogenase to formyl-H4folate are performed by MtdA (39) and FchA (30) since the corresponding genes are also present in strain CM4 and that this explains the lack of phenotype of the folD mutant. The construction of mtdA and fchA mutants in both the background of strain CM4 and that of its folD mutant should help to shed light on this question.
In conclusion, the evidence presented here for an additional C1 utilization pathway specific for chloromethane in strain CM4 adds to the emerging complexity of C1 metabolism in Methylobacterium species. On the other hand, the existence of such an additional pathway in strain CM4 may allow us to obtain and investigate, in the background of strain CM4, mutants in C1 utilization genes previously found to be essential in M. extorquens AM1 if these can be complemented by homologous genes of the chloromethane-specific pathway.
Acknowledgments
A.S. and C.M. contributed equally to this work.
We thank Ludmila Chistoserdova and Mary Lidstrom for access to unpublished sequence information from the M. extorquens AM1 genome, Julia Vorholt for discussions, and Chris Marx and Mikhail Alexeyev for generous gifts of plasmids.
This work was supported by ETH grant 0-20436-97 to T.L.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-828. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. J., C. J. Morris, D. N. Nunn, C. Anthony, and M. E. Lidstrom. 1990. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene 90:173-176. [DOI] [PubMed] [Google Scholar]
- 3.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, England.
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology, vol. 1. Greene Publishing Associates/John Wiley & Sons, Inc., New York, N.Y.
- 5.Babst, M., H. Hennecke, and H. M. Fischer. 1996. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol. Microbiol. 19:827-839. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C-1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 7.Doronina, N. V., A. P. Sokolov, and Y. A. Trotsenko. 1996. Isolation and initial characterization of aerobic chloromethane-utilizing bacteria. FEMS Microbiol. Lett. 142:179-183. [Google Scholar]
- 8.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 9.Gossett, J. M. 1987. Measurement of Henry's law constants for C1 and C2 chlorinated hydrocarbons. Environ. Sci. Technol. 21:202-208. [Google Scholar]
- 10.Hagemeier, C., L. Chistoserdova, M. Lidstrom, R. K. Thauer, and J. Vorholt. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 267:3762-3769. [DOI] [PubMed] [Google Scholar]
- 11.Herrero, M., V. De Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kayser, M. F., M. T. Stumpp, and S. Vuilleumier. 2000. DNA polymerase I is essential for growth of Methylobacterium dichloromethanicum DM4 with dichloromethane. J. Bacteriol. 182:5433-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kayser, M. F., Z. Ucurum, and S. Vuilleumier. 2002. Dichloromethane metabolism and C1 utilization genes in Methylobacterium strains. Microbiology 148:1915-1922. [DOI] [PubMed] [Google Scholar]
- 14.La Roche, S. D., and T. Leisinger. 1990. Sequence analysis and expression of the bacterial dichloromethane dehalogenase structural gene, a member of the glutathione S-transferase supergene family. J. Bacteriol. 172:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lidstrom, M. E. 1992. The aerobic methylotrophic bacteria, p. 431-445. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, vol. 3. Springer-Verlag, New York, N.Y.
- 16.Machlin, S. M., and R. S. Hanson. 1988. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J. Bacteriol. 170:4739-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marison, I. W., and M. M. Attwood. 1982. A possible alternative mechanism for the oxidation of formaldehyde to formate. J. Gen. Microbiol. 128:1441-1446. [Google Scholar]
- 18.Marx, C. J., and L. Chistoserdova. 2001. Development of versatile broad-host-range vectors for use in methylotrophs and other gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 19.Matthews, R. G. 1984. Methionine biosynthesis, p. 497-553. In R. L. Blakely and S. J. Benkovic (ed.), Folates and pterins: chemistry and biochemistry of folates, vol. 1. Wiley, New York, N.Y.
- 20.Matthews, R. G. 1986. Methylenetetrahydrofolate reductase from pig liver. Methods Enzymol. 122:372-381. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, R. G. 1996. One-carbon metabolism, p. 600-611. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 22.McAnulla, C., C. A. Woodall, I. R. McDonald, A. Studer, S. Vuilleumier, T. Leisinger, and J. M. Murrell. 2001. Chloromethane utilization gene cluster from Hyphomicrobium chloromethanicum strain CM2T and development of functional gene probes to detect halomethane-degrading bacteria. Appl. Environ. Microbiol. 67:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald, I. R., N. V. Doronina, Y. A. Trotsenko, C. McAnulla, and J. C. Murrell. 2001. Hyphomicrobium chloromethanicum sp. nov., Methylobacterium chloromethanicum sp. nov., chloromethane-utilizing bacteria isolated from a polluted environment. Int. J. Syst. Evol. Microbiol. 51:119-122. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae require toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell, J. C., and I. R. McDonald. 2000. Methylotrophy, p. 245-255. In J. Lederberg (ed.), Encyclopedia of microbiology, vol. 3. Academic Press, Inc., New York, N.Y.
- 27.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 305:202-217. [DOI] [PubMed] [Google Scholar]
- 28.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 29.Pomper, B. K., and J. A. Vorholt. 2001. Characterization of the formyltransferase from Methylobacterium extorquens AM1. Eur. J. Biochem. 268:4769-4775. [DOI] [PubMed] [Google Scholar]
- 30.Pomper, B. K., J. A. Vorholt, L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1999. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur. J. Biochem. 261:475-480. [DOI] [PubMed] [Google Scholar]
- 31.Rothmel, R. K., A. M. Chakrabarty, A. Berry, and A. Darzins. 1991. Genetic systems in Pseudomonas. Methods Enzymol. 204:485-514. [DOI] [PubMed] [Google Scholar]
- 32.Sheppard, C. A., E. E. Trimmer, and R. G. Matthews. 1999. Purification and properties of NADH-dependent 5,10-methylenetetrahydrofolate reductase (MetF) from Escherichia coli. J. Bacteriol. 181:718-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. BioTechniques 1:784-790. [Google Scholar]
- 34.Studer, A., E. Stupperich, S. Vuilleumier, and T. Leisinger. 2001. Chloromethane:tetrahydrofolate methyl transfer by two proteins from Methylobacterium chloromethanicum strain CM4. Eur. J. Biochem. 268:2931-2938. [DOI] [PubMed] [Google Scholar]
- 35.Studer, A., S. Vuilleumier, and T. Leisinger. 1999. Properties of the methylcobalamin:H4folate methyltransferase involved in chloromethane utilization by Methylobacterium sp. strain CM4. Eur. J. Biochem. 264:242-249. [DOI] [PubMed] [Google Scholar]
- 36.Vannelli, T., M. Messmer, A. Studer, S. Vuilleumier, and T. Leisinger. 1999. A corrinoid-dependent catabolic pathway for growth of a Methylobacterium strain with chloromethane. Proc. Natl. Acad. Sci. USA 96:4615-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vannelli, T., A. Studer, M. Kertesz, and T. Leisinger. 1998. Chloromethane metabolism by Methylobacterium sp. strain CM4. Appl. Environ. Microbiol. 64:1933-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Völker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Völker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 39.Vorholt, J. A., L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1998. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 180:5351-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vorholt, J. A., L. Chistoserdova, S. M. Stolyar, R. K. Thauer, and M. E. Lidstrom. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuilleumier, S. 2002. Coping with a halogenated one-carbon diet: aerobic dichloromethane-mineralizing bacteria, p. 105-131. In W. Reineke and S. Agathos (ed.), Bio/technology for the environment. Focus on Biotechnology Series, vol. 3A. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 43.Wagner, R. 2000. Transcription in prokaryotes. Oxford University Press, New York, N.Y.
- 44.Woodall, C. A., K. L. Warner, R. S. Oremland, J. C. Murrell, and I. R. McDonald. 2001. Identification of methyl halide-utilizing genes in the methyl bromide-utilizing bacterial strain IMB-1 suggests a high degree of conservation of methyl halide-specific genes in gram-negative bacteria. Appl. Environ. Microbiol. 67:1959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]