Abstract
We report here that PBP1a can dimerize but does not interact with PBP1b to form PBP1a/PBP1b heterodimers in Escherichia coli. These findings support the idea of a relevant involvement of dimerization of both PBP1a and PBP1b during murein synthesis and suggest the existence of different peptidoglycan synthesis complexes.
In eubacteria, the final steps in peptidoglycan synthesis are mediated by a set of periplasmic membrane-bound enzymes, the penicillin-binding proteins (PBPs) (12, 19, 24). In Escherichia coli, 10 PBPs have been identified (8, 17, 21). Among them, only PBP1a and PBP1b are bifunctional enzymes organized into two catalytic domains harboring transglycosylase and transpeptidase activities (13, 19, 22). PBP1c, a new member of these bimodular enzymes, has recently been shown to display transglycosylase activity, but it does not possess demonstrated transpeptidase activity and is unable to compensate for the loss of PBP1a and PBP1b (21). PBP1a consists of 850 amino acids encoded by the ponA gene, whereas PBP1b is found as three polypeptides termed α, β, and γ, which differ in size but which are all encoded by the ponB gene (1). PBP1bα and PBP1bγ arise from two different translation initiations on the ponB mRNA, while PBP1bβ is a degradation product of PBP1bα (10, 14, 23). The precise physiological roles for the PBP1a and PBP1b proteins remain unclear. Inactivation of both PBP1a and PBP1b is lethal, but E. coli mutants lacking either one of the proteins are viable, indicating a functional complementation (15, 27). Nevertheless, several lines of evidence supported by analysis of the behavior of strains deficient either in PBP1a or in PBP1b grown in the presence of β-lactams indicate that the two proteins are not equivalent and probably play distinct roles in peptidogycan synthesis (6, 7).
Recent studies showed that PBPs could work in close association with other enzymes including hydrolases and synthetases during growth and cell division, suggesting the existence of peptidoglycan synthesis enzyme complexes (11). Little is known about the composition and function of these complexes, but it is speculated that a dimer of bifunctional PBPs (PBP1a and/or PBP1b) plays a prominent part. This hypothesis is consistent with the finding that PBP1b, the major enzyme for peptidoglycan synthesis, can indeed occur as a dimer (3, 25, 28, 29). In this study, we establish that PBP1a can also exist in a stable dimeric form, suggesting that both PBP1a and PBP1b act as dimers during cell wall synthesis. We also investigated whether PBP1a and PBP1b can interact to form a heterodimer, but we did not observe such an interaction, suggesting that PBP1a and PBP1b belong to different enzymatic complexes.
In vivo production and activity of cloned PBP1a.
PBP1a was produced in QCA2, a ponA-deleted E. coli strain (7), which was transformed with vector pPONAMyc, a moderate-copy-number plasmid producing PBP1a protein fused to the amino acid sequence EQKLISEEDL of the human oncogene product c-myc (2) at its C-terminal end. pPONAMyc was constructed from pPONB (16), which expresses PBP1b, fused to a tag peptide (YPYDVPDYA) from the hemagglutinin 1 (HA1) epitope of influenza virus (26). The ponB gene of pPONB was removed and replaced by the ponA gene and a synthetic linker encoding the c-myc peptide. Western blot analysis performed with QCA2(pPONAMyc) membrane preparations (4, 16) with the monoclonal antibody 9E10 (at a 1/1,000 dilution) (Sigma) raised against the c-myc tag peptide revealed good expression of PBP1a (Fig. 1A, lane 2).
FIG. 1.
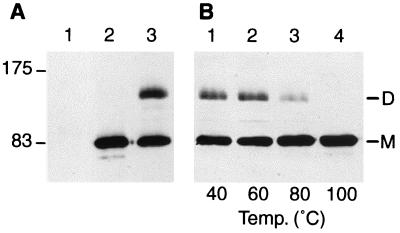
PBP1a dimer and stability. (A) Membrane fractions prepared from strain QCA2 (25 μg, lane 1) or strain QCA2(pPONAMyc) (25 μg, lanes 2 and 3) were incubated for 10 min in sample buffer at 100°C (lanes 1 and 2) or room temperature (lane 3), submitted to SDS-PAGE, and analyzed by Western blotting with the anti-c-myc 9E10 monoclonal antibody. The molecular mass markers (in kilodaltons) are displayed on the left. (B) Membrane fractions (25 μg) prepared from strain QCA2(pPONAMyc) were incubated for 10 min in sample buffer at temperatures ranging from 40 to 100°C as indicated, submitted to SDS-PAGE, and analyzed by Western blotting with the anti-c-myc 9E10 monoclonal antibody. M, monomeric form of PBP1a; D, dimeric form of PBP1a.
Since the PBP1b activity can complement PBP1a for cell growth and cannot be specifically inhibited in QCA2(pPONAMyc) cells, we constructed an E. coli strain harboring a deletion in both ponA and ponB genes to check the in vivo activity of PBP1a encoded by vector pPONAMyc. For this purpose, QCB1 cells, which lack the ponB gene (7), were first transformed with pR-PONB, a low-copy-number, temperature-sensitive derivative of pHSG415 (9), harboring ponB and chloramphenicol resistance. The chromosomal copy of the ponA gene in QCB1(pR-PONB) cells was then replaced by the inactivated ponA gene (ΔponA::Kmr) from QCA2 by P1 phage transduction (18). The resulting QC4(pR-PONB) strain harbors a chromosomal deletion of both ponA and ponB genes and thus grows at 30°C but lyses at restrictive temperature. QC4(pR-PONB) cells were then transformed with pPONAMyc and incubated overnight at 42°C to yield QC4(pPONAMyc), a pR-PONB-free strain.
QC4(pPONAMyc) bacteria grew as well as wild-type cells, indicating a fully active expressed PBP1a (data not shown). Moreover, these cells were killed with 0.3 μg of cephaloridine/ml, a specific PBP1a antibiotic inhibitor (27). This experiment showed that the cells have indeed lost the thermosensitive plasmid pR-PONB encoding PBP1b and thus rely solely on the expressed PBP1a for cell growth.
PBP1a exists in a dimeric form.
When isolated cell envelopes of the QCA2(pPONAMyc) strain were incubated in sample buffer without β-mercaptoethanol (60 mM TrisHCl [pH 6.8], 1% sodium dodecyl sulfate [SDS], 10% glycerol, and 0.01% bromophenol blue) for 10 min at room temperature for Western blot analysis, two bands were detected with the anti-c-myc antibody, a major band at a position corresponding to the monomeric form of PBP1a and an additional band at about 140 kDa (Fig. 1A, lane 3). This band, which most likely corresponds to a protein complex, was never detected with boiled samples (Fig. 1A, lane 2). Incubation of QCA2(pPONAMyc) membrane preparations with sample buffer for 10 min at several temperatures showed that this complex was stable at 60°C and partially dissociated at 80°C (Fig. 1B, lanes 1 to 4).
To determine whether this additional band was a complex between PBP1a and another protein or a dimer of PBP1a, membrane proteins prepared from QCA2(pPONAMyc) were analyzed by nondenaturing-denaturing two-dimensional gel electrophoresis. After separation of proteins by SDS-polyacrylamide gel electrophoresis (PAGE) in nondenaturing conditions as described above, a 1-cm-wide strip was excised from the gel, incubated in a denaturing buffer (125 mM TrisHCl [pH 6.8], 0.1% SDS, 0.01% bromophenol blue) at 100°C for 10 min, and put on the top of an 8% polyacrylamide gel for electrophoresis. Proteins were then transferred to a nitrocellulose sheet (Gelmansciences) for gold staining (Bio-Rad) and Western blot analysis.
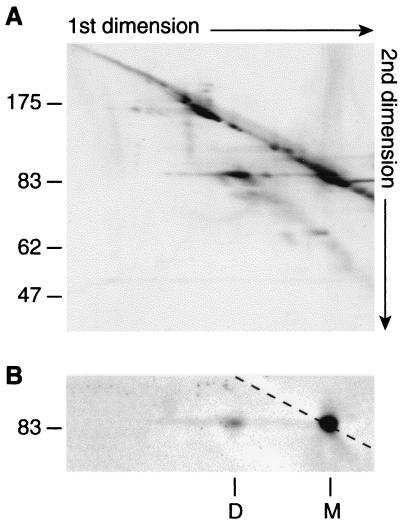
Gold staining revealed that the 140-kDa complex observed in the first dimension dissociated in a single spot which migrated at a position corresponding to the mass of the monomeric PBP1a (Fig. 2A). Western blot analysis performed with the anti-c-myc antibody confirmed that it corresponds to the PBP1a protein (Fig. 2B). These data indicate that the 140-kDa complex consists of two PBP1a monomers. We cannot totally rule out the possibility that it corresponds to a complex between PBP1a and another protein that we were not able to detect by gold staining. However, the spot of PBP1a resulting from the dissociation of the 140-kDa band was strongly labeled, and it is unlikely that the presence of another protein in a similar amount would have escaped detection. Moreover, the apparent molecular mass of the complex fits with the hypothesis of a PBP1a dimer, since it migrates at a position corresponding to a molecular mass of about 140 kDa, close to the 150 kDa observed with PBP1b dimers.
FIG. 2.
Analysis of the PBP1a complex by nondenaturing-denaturing two-dimensional electrophoresis. (A) Proteins of membrane fractions prepared from strain QCA2(pPONAMyc) (20 μg) were separated by nondenaturing-denaturing two-dimensional gel electrophoresis. Proteins were subsequently transferred onto a nitrocellulose sheet which was stained with colloidal gold. The molecular mass markers (in kilodaltons) of the second dimension are displayed on the left. (B) The nitrocellulose sheet shown in panel A was immunoblotted with the anti-c-myc 9E10 monoclonal antibody. M, monomeric form of PBP1a; D, dimeric form of PBP1a.
This dimeric form of PBP1a is fairly stable, since it requires a 10-min incubation at 80°C to be dissociated. In addition, the use of a sample buffer lacking β-mercaptoethanol indicates that the dimerization does not rely on the formation of disulfide bonds. Interestingly, these characteristics resemble those described for PBP1b dimers, suggesting that for both PBP1a and PBP1b, dimerization occurs in a similar way. Zijderveld et al. (28) showed that the transglycosylation domain of PBP1b is probably involved in complex formation. Unfortunately, our attempts to localize the interaction domain in PBP1a dimer, by using deletion or fusion mutants, failed due to protein degradation in the cells.
PBP1a and PBP1b do not form heterodimers.
According to previous studies and the results described above, both PBP1a and PBP1b proteins are able to dimerize. We then investigated whether these two proteins, which complement for cell growth, can interact and act as a heterodimer within the multienzyme complexes involved in the synthesis of peptidoglycan. We constructed the QC4(pY1a/pM46L) strain, which expresses PBP1a fused to the c-myc tag peptide, and the large form of PBP1b (PBP1bα), fused to the HA1 tag peptide. This strain was obtained by transforming QC4(pR-PONB) cells with pM46L vector, a pPONB derivative where mutation M46L in PBP1b abolished the production of PBP1bγ without affecting that of PBP1bα, the large component of PBP1b (4). Transformants were incubated at 42°C to select for pR-PONB-free cells, and the resulting QC4(pM46L) bacteria were subsequently transformed with plasmid pY1a, a pACYC184 (5) derivative carrying the ponA gene fused to the c-myc tag-encoding sequence.
QC4 harboring vector pY1a alone grew normally, indicating that PBP1a produced by the pY1a vector was fully active in vivo (data not shown). In addition, treatment of the QC4(pY1a/pM46L) strain with cephaloridine (0.3 μg/ml) had no effect on cell growth, confirming that PBP1bα produced by pM46L was fully active, as previously described (4).
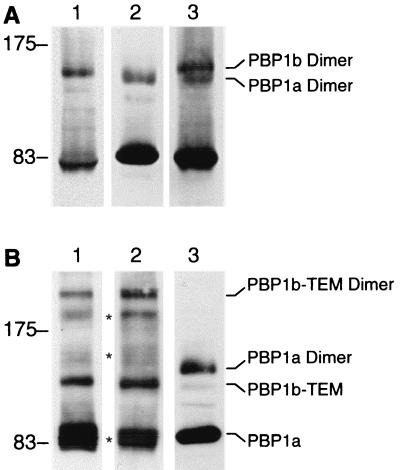
Membrane proteins of QC4(pY1a/pM46L) cells were separated by SDS-PAGE and analyzed by Western blotting performed with either the 12CA5 anti-HA1 antibody (1/50,000) (a generous gift from J. Grassi, CEA, Saclay, France), the 9E10 anti-c-myc antibody (1/1,000), or a combination of both. The use of the anti-HA1 antibody alone revealed two bands corresponding to the monomeric and dimeric forms of PBP1bα (4) (Fig. 3A, lane 1). A similar pattern was observed when the anti-c-myc antibody was used in the experiment to detect the PBP1a protein (Fig. 3A, lane 2). Surprisingly, although PBP1a has a higher molecular mass than the large form of PBP1b, the dimeric form of PBP1a was detected at a position corresponding to a slightly lower apparent molecular mass than the dimeric form of PBP1bα (Fig. 3A, lanes 1 and 2), suggesting perhaps that PBP1a dimers are more compact. Western blotting analysis with both anti-HA1 and anti-c-myc antibodies confirmed the presence of two different bands corresponding to the PBP1a and PBP1b dimers (Fig. 3A, lane 3). No additional band responding against the two antibodies could be detected, suggesting that PBP1a and PBP1b are not able to form heterodimers.
FIG. 3.
Western blot analysis of PBP1a and PBP1b complexes in QC4(pY1a/pM46L) and QC4(pY1a/pM46LBla) cells. (A) Membrane fractions prepared from strain QC4(pY1a/pM46L) (25 μg) were incubated for 10 min in sample buffer at room temperature, submitted to SDS-PAGE, and transferred onto a nitrocellulose sheet which was subsequently cut in bands corresponding to the lanes. Bands of nitrocellulose were analyzed by Western blotting with the 12CA5 antibody against the HA1 tag peptide of PBP1b (lane 1), the 9E10 antibody against the c-myc tag peptide of PBP1a (lane 2), or with both antibodies (lane 3). The molecular mass markers (in kilodaltons) are displayed on the left. (B) Membrane fractions prepared from strain QC4(pM46LBla) (25 μg, lane 1) or strain QC4(pY1a/pM46LBla) (25 μg, lanes 2 and 3) were analyzed by Western blotting with the 12CA5 anti-HA1 antibody (lanes 1 and 2) or the 9E10 anti-c-myc antibody (lane 3), as described above. The molecular mass markers (in kilodaltons) are displayed on the left. ∗, degraded forms of PBP1b-TEM and PBP1b-TEM dimers.
To rule out the possibility that the detection of a PBP1a/PBP1b heterodimer was hampered by the diffuse bands of the PBP1a and PBP1b homodimers, we constructed plasmid pM46Lbla by inserting part of the bla gene which encodes for the mature β-lactamase TEM-1 (amino acids 24 to 293) downstream from the ponB gene of pM46L. The resulting plasmid expresses a PBP1bα-HA1-TEM fusion protein that can be detected with an anti-HA1 antibody. QC4(pM46Lbla) cells grew normally and remained viable in the presence of ampicillin (100 μg/ml), indicating that the fusion protein was produced with a proper folding of the PBP1b and TEM domains.
Western blotting analysis performed with QC4(pY1a/pM46Lbla) membrane proteins with the anti-HA1 antibody showed that the PBP1b-TEM protein migrates at a position corresponding to an apparent molecular mass of 120 kDa and is able to dimerize (Fig. 3B, lane 2). Additional bands located between the monomeric and dimeric forms of the fusion protein were detected, but none of these bands reacted against the anti-c-myc antibody (Fig. 3B, lane 3). A similar pattern was observed when Western blotting was performed with strain QC4(pM46Lbla), which lacks PBP1a (Fig. 3B, lane 1). Thus, it appears that these additional bands do not correspond to a putative PBP1a/PBP1b-TEM heterodimer but probably result from protein degradation. The use of the anti-c-myc antibody confirmed the presence of the monomeric and dimeric forms of PBP1a in QC4(pY1a/pM46Lbla) cells and did not reveal any additional band corresponding to a PBP1a/PBP1b-TEM heterodimer (Fig. 3B, lane 3). Since PBP1b-TEM has a higher molecular mass than PBP1a, such an heterodimer would be expected to migrate at a mass higher than that of the PBP1a dimer. Together, these experiments strongly suggest that PBP1a and PBP1b are not able to form heterodimers, confirming the two-dimensional electrophoresis experiments which failed to detect PBP1a-associated proteins.
The functional complementation of PBP1a and PBP1b in cell growth and division imply that both of them interact with the other components involved in the cell wall synthesis process. Therefore, it can be hypothesized that they present a similar three-dimensional structure and thus act as a PBP1a/PBP1b heterodimer in peptidoglycan synthesis. Our data argue against this hypothesis, since no such heterodimers could be detected. This negative result is in line with previous observations that the large and small forms of PBP1b (α and γ) form α-α or γ-γ homodimers but not heterodimers (3, 28). Another hypothesis is that PBP1a and PBP1b act as independent dimers in different multienzymatic complexes, the two proteins having different functions in cell wall synthesis, as suggested by various observations (4, 6, 7). Consistent with this is the fact that overexpression of either PBP1a or PBP1b in an E. coli strain yields cells with different shapes. Indeed, bacteria overexpressing PBP1a grow as very short cells, whereas those overexpressing PBP1b grow as longer cells (unpublished observations). Sequence analysis also shows that these two PBPs fall into different family clusters, suggesting that they are true paralogs (8). Taken together, these data suggest different interactions between the various PBP1 and the other proteins involved in the cell wall synthesis machinery.
These results, which demonstrate that PBP1a and PBP1b can be found in a separate but similar oligomerization state, support the idea that the peptidoglycan synthesis process at least requires the formation of dimeric bifunctional PBPs. However, the connection of PBP1a and PBP1b with other enzymes involved in the control of bacterial morphology, elongation, and cell division remains unclear, and no additional interactions have been detected in our experiments. Further investigations will be necessary to elucidate the nature of these interactions and to experimentally demonstrate the biological role of these dimers in the peptidoglycan synthesis. Recently, dimerization of PBP1 in Neisseria gonorrhoeae, the PBP1a E. coli homolog, has been reported (20), and it would be interesting to extend the study to other bacterial species to check if dimerization of bifunctional PBPs occurs as a general and essential process in eubacteria.
REFERENCES
- 1.Broome-Smith, J. K., A. Edelman, S. Yousif, and B. G. Spratt. 1985. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding protein 1A and 1B of Escherichia coli K12. Eur. J. Biochem. 147:437-446. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, A. M., P. D. Kessler, and D. M. Fambrough. 1992. The alternative carboxyl termini of avian cardiac and brain sarcoplasmic reticulum/endoplasmic reticulum Ca2+-ATPases are on opposite sides of the membrane. J. Biol. Chem. 267:9321-9325. [PubMed] [Google Scholar]
- 3.Chalut, C., M. H. Rémy, and J. M. Masson. 1999. Disulfide bridges are not involved in penicillin-binding protein 1b dimerization in Escherichia coli. J. Bacteriol. 181:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalut, C., X. Charpentier, M. H. Rémy, and J. M. Masson. 2001. Differential responses of Escherichia coli cells expressing cytoplasmic domain mutants of penicillin-binding protein 1b after impairment of penicillin-binding proteins 1a and 3. J. Bacteriol. 183:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García del Portillo, F., and M. A. de Pedro. 1990. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J. Bacteriol. 172:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto-Gotoh, T., F. C. Franklin, A. Nordheim, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene 16:227-235. [DOI] [PubMed] [Google Scholar]
- 10.Henderson, T. A., P. M. Dombrosky, and K. D. Young. 1994. Artifactual processing of penicillin-binding proteins 7 and 1b by the OmpT protease of Escherichia coli. J. Bacteriol. 176:256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Höltje, J. V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 12.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishino, F., K. Mitsui, S. Tamaki, and M. Matsuhashi. 1980. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem. Biophys. Res. Commun. 97:287-293. [DOI] [PubMed] [Google Scholar]
- 14.Kato, J., H. Suzuki, and Y. Hirota. 1984. Overlapping of the coding regions for alpha and gamma components of penicillin-binding protein 1 b in Escherichia coli. Mol. Gen. Genet. 196:449-457. [DOI] [PubMed] [Google Scholar]
- 15.Kato, J., H. Suzuki, and Y. Hirota. 1985. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol. Gen. Genet. 200:272-277. [DOI] [PubMed] [Google Scholar]
- 16.Lefèvre, F., M. H. Rémy, and J. M. Masson. 1997. Topographical and functional investigation of Escherichia coli penicillin-binding protein 1b by alanine stretch scanning mutagenesis. J. Bacteriol. 179:4761-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massova, I., and S. Mobashery. 1998. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 42:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Generalized transduction; use of P1 in strain construction, p. 201-205. In Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ropp, P. A., and R. A. Nicholas. 1997. Cloning and characterization of the ponA gene encoding penicillin-binding protein 1 from Neisseria gonorrhoeae and Neisseria meningitidis. J. Bacteriol. 179:2783-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiffer, G., and J. V. Höltje. 1999. Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031-32033. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki, H., Y. van Heijenoort, T. Tamura, J. Mizoguchi, Y. Hirota, and J. van Heijenoort. 1980. In vitro peptidoglycan polymerization catalysed by penicillin binding protein 1b of Escherichia coli K-12. FEBS Lett. 110:245-249. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, H., J. Kato, Y. Sakagami, M. Mori, A. Suzuki, and Y. Hirota. 1987. Conversion of the alpha component of penicillin-binding protein 1b to the beta component in Escherichia coli. J. Bacteriol. 169:891-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 25.Vollmer, W., M. von Rechenberg, and J. V. Höltje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 26.Wilson, I. A., H. L. Niman, R. A. Houghten, A. R. Cherenson, M. L. Connolly, and R. A. Lerner. 1984. The structure of an antigenic determinant in a protein. Cell 37:767-778. [DOI] [PubMed] [Google Scholar]
- 27.Yousif, S. Y., J. K. Broome-Smith, and B. G. Spratt. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131:2839-2845. [DOI] [PubMed] [Google Scholar]
- 28.Zijderveld, C. A., M. E. Aarsman, T. den Blaauwen, and N. Nanninga. 1991. Penicillin-binding protein 1B of Escherichia coli exists in dimeric forms. J. Bacteriol. 173:5740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zijderveld, C. A., M. E. Aarsman, and N. Nanninga. 1995. Differences between inner membrane and peptidoglycan-associated PBP1B dimers of Escherichia coli. J. Bacteriol. 177:1860-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]