Abstract
SlyA is a transcriptional regulator of Escherichia coli, Salmonella enterica, and other bacteria belonging to the Enterobacteriaceae. The SlyA protein has been shown to be involved in the virulence of S. enterica serovar Typhimurium, but its role in E. coli is unclear. In this study, we employed the proteome technology to analyze the SlyA regulons of enteroinvasive E. coli (EIEC) and Salmonella serovar Typhimurium. In both cases, comparative analysis of the two-dimensional protein maps of a wild-type strain, a SlyA-overproducing derivative, and a corresponding slyA mutant revealed numerous proteins whose expression appeared to be either positively or negatively controlled by SlyA. Twenty of the putative SlyA-induced proteins and 13 of the putative SlyA-repressed proteins of the tested EIEC strain were identified by mass spectrometry. The former proteins included several molecular chaperones (GroEL, GroES, DnaK, GrpE, and CbpA), proteins involved in acid resistance (HdeA, HdeB, and GadA), the “starvation lipoprotein” (Slp), cytolysin ClyA (HlyE or SheA), and several enzymes involved in metabolic pathways, whereas most of the latter proteins proved to be biosynthetic enzymes. Consistently, the resistance of the EIEC slyA mutant to heat and acid stress was impaired compared to that of the wild-type strain. Furthermore, the implication of SlyA in the regulation of several of the identified E. coli proteins was confirmed at the level of transcription with lacZ fusions. Twenty-three of the Salmonella serovar Typhimurium proteins found to be affected by SlyA were also identified by mass spectrometry. With the exception of GroEL these differed from those identified in the EIEC strain and included proteins involved in various processes. The data suggest that gene regulation by SlyA might be crucial for intracellular survival and/or replication of both EIEC and Salmonella serovar Typhimurium in phagocytic host cells.
The coordinated regulation of genes enables bacteria to adapt to new environmental conditions and is, in particular, a prerequisite for bacterial pathogens to respond to the diverse conditions encountered during infection of the host. In the past decade considerable progress in the elucidation of the regulatory networks that control the expression of virulence-associated genes in various pathogenic bacteria has been made.
Recently, a novel regulatory protein, SlyA, has been identified within the family of Enterobacteriaceae. The slyA gene was first isolated from the chromosome of Salmonella enterica serovar Typhimurium and recognized as a gene that confers a hemolytic phenotype on Escherichia coli K-12 when introduced in trans on a plasmid. In addition, slyA was found to be required for the virulence of Salmonella serovar Typhimurium in mice and for the survival of salmonellae in murine peritoneal macrophages (26). Initially, slyA was presumed to encode a hemolytic/cytolytic protein (Salmonella cytolysin). Further studies demonstrated, however, that the 16.7-kDa SlyA protein itself does not exhibit hemolytic activity but rather acts as a regulator that induces, when overproduced in E. coli K-12, the expression of a novel E. coli hemolysin not related to α-hemolysin (HlyA) (27). In addition, slyA homologues were identified in other Enterobacteriaceae including E. coli and Shigella flexneri, and it has been shown that SlyA from E. coli is also capable of activating the expression of the novel hemolysin in E. coli K-12 (27, 35). The SlyA-controlled chromosomal hemolysin gene of E. coli K-12, clyA (also referred to as sheA and hlyE), has recently been identified and has been shown to encode a pore-forming protein of 34 kDa (15, 20, 28, 36, 45).
Sequence alignments revealed that SlyA is a member of a large family of bacterial transcriptional regulators which control a diverse range of physiological processes in human, animal, and plant pathogens. Within this protein family, SlyA is particularly closely related to RovA of Yersinia enterocolitica and Yersinia pseudotuberculosis, Rap of Serratia marcescens, and Hor of Erwinia carotovora (32, 38, 42), while it is more distantly related to EmrR (MprA) and MarR of E. coli, PecS of Erwinia chrysanthemi, Hpr of Bacillus subtilis, and several other regulatory proteins (14). RovA has been shown to be required for the expression of invasin, the primary invasion factor of Y. enterocolitica and Y. pseudotuberculosis, which is encoded by the inv gene (32, 38). RovA was also found to be essential for the virulence of Y. enterocolitica although an inv mutant was not significantly attenuated in a mouse model, which suggested that RovA probably regulates other virulence genes in addition to inv (38).
The significance of SlyA for the pathogenicity of Salmonella has been established in several studies. In addition to the initial finding that SlyA is required for the virulence of Salmonella serovar Typhimurium and for the capability of these bacteria to survive in professional phagocytes (26), further investigations indicated that SlyA is required for the destruction of murine M cells following uptake of Salmonella serovar Typhimurium and for intracellular survival and/or intracellular replication of the salmonellae in murine Peyer's patches (11). In addition, a role for SlyA in the resistance of Salmonella serovar Typhimurium to oxidative stress has been proposed (6). Recently, Watson et al. (46) suggested that SlyA particularly regulates virulence genes of Salmonella involved in systemic, but not enteric, salmonellosis.
Polyacrylamide gel electrophoresis (PAGE) of proteins from a Salmonella serovar Typhimurium wild-type strain and from a slyA mutant derivative indicated that SlyA regulates the synthesis of several Salmonella proteins during stationary phase and during infection of macrophages (6). These SlyA-regulated proteins have, however, not yet been identified. At present, the only gene that is known to be controlled by SlyA is clyA of E. coli (28, 47). clyA homologues have recently been identified in S. enterica serovar Typhi and serovar Paratyphi-A (45) but not in several tested Salmonella serovar Typhimurium strains (29; Ludwig et al., unpublished data).
The two-dimensional PAGE (2D-PAGE) analyses presented in this study suggest that in enteroinvasive E. coli (EIEC) and Salmonella serovar Typhimurium SlyA positively regulates the synthesis of several proteins and negatively regulates that of others. We have identified a number of these putative SlyA-controlled proteins by mass spectrometry and have confirmed for some of these proteins that SlyA is indeed involved in the regulation of their structural genes. Several of the proteins found to be SlyA induced proved to be stress response proteins. The tight regulation of these proteins by SlyA might be significant for the capability of EIEC and Salmonella serovar Typhimurium to survive and/or replicate within professional phagocytes.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
EIEC wild-type strain 12860 (40) was obtained from the Institute of Hygiene and Microbiology, University of Würzburg, Würzburg, Germany. Salmonella serovar Typhimurium ATCC 14028slyA is a Salmonella serovar Typhimurium ATCC 14028s slyA mutant carrying a derivative of suicide plasmid pGP704 (Ampr) (31) inserted in the 5′-terminal region of the chromosomal slyA gene. This slyA mutant was previously referred to as SLYAII (11). E. coli SY327λpir and E. coli SM10λpir (31) were used as host strains for pGP704 and pGP704 derivatives. E. coli DH5α [F− φ80 dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 phoA hsdR17 (rK− mK+) supE44 λ− thi-1 gyrA96 relA1] was used for the propagation of all other plasmids. Plasmid pAL102 is a derivative of pBluescript II SK(+) (Ampr; Stratagene) carrying slyA from Salmonella serovar Typhimurium ATCC 14028s (slyAST) on a 0.84-kb ClaI-SacII insert (27). pAL105 carries slyA from E. coli K-12 (slyAEC) on a 0.86-kb EcoRI-PstI fragment inserted in pBluescript II SK(+) (27). The sequences upstream of slyA included in the inserts of pAL102 and pAL105 comprise 170 and 167 bp, respectively. In both plasmids, slyA is exclusively under the control of its native 5′-flanking regulatory region. Plasmid pAL108 is a derivative of pACYC184 (Cmr; New England Biolabs) carrying slyAEC as an insert (28). pβgal-Basic (Ampr) was obtained from Clontech. All bacterial strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (39) or on LB medium solidified with 1.5% (wt/vol) agar, if not indicated otherwise. Antibiotics were used at the following final concentrations: ampicillin, 100 μg/ml; streptomycin, 30 μg/ml; chloramphenicol, 30 μg/ml.
DNA manipulations.
All DNA cloning procedures were performed as described by Sambrook et al. (39). PCR amplification of DNA fragments was conducted with Deep Vent DNA polymerase (New England Biolabs), in accordance with the recommendations of the manufacturer. Nucleotide sequences were determined by automated cycle sequencing.
Disruption of the chromosomal slyA gene of EIEC 12860.
The slyA gene of EIEC 12860 was amplified by PCR using forward primer 5′-CTTAGCAAGCTAATTATAAGGAG-3′ and reverse primer 5′-CAGGTGACCGTTTCTCC-3′, which correspond to sequences flanking slyA from E. coli K-12. The 534-bp PCR product was sequenced and was shown to be identical to the slyA-carrying genomic DNA fragment of E. coli K-12. To disrupt slyA in EIEC 12860, a SalI-KpnI fragment carrying codons 7 to 37 of slyAEC was isolated by PCR using primers 5′-GAAATTGGAGTCGACACTAGGTTC-3′ and 5′-GATATTGTGTAACGGTACCCAATGG-3′ and a KpnI-SphI fragment carrying the 3′-terminal 22 codons of slyAEC (i.e., codons 125 to 146), the slyAEC stop codon, and the following 12 nucleotides was amplified by PCR using primers 5′-CTCCGCAGAGGTACCGGAGCAACTG-3′ and 5′-GTTACTGACCGCATGCCCCCTTC-3′. The restriction sites at the ends of these DNA fragments were introduced by the PCR primers (underlined sequences), and the sticky ends were generated by cutting the PCR products with the corresponding restriction enzymes. Both fragments were inserted in series between the SalI and SphI restriction sites of pGP704. The resulting suicide plasmid, pGP704CT1, thus carried a truncated slyAEC gene missing its 5′-terminal sequence (nucleotides 1 to 16) as well as codons 38 to 124 (internal in-frame deletion).
pGP704CT1 was transferred by conjugation from E. coli SM10λpir into a spontaneous streptomycin-resistant derivative of EIEC 12860. Transconjugant clones were selected on agar plates containing streptomycin and ampicillin, and insertion of the plasmid into the chromosomal slyA gene via homologous recombination was tested by PCR analysis. Plasmid insertion by recombination between the SalI-KpnI insert fragment of pGP704CT1 and the 5′-terminal region of slyA of EIEC 12860 resulted in the disruption of this gene, as confirmed by sequencing the corresponding PCR products, and generated slyA knockout mutant EIEC 12860slyA, used in this study. In EIEC 12860slyA, the truncated slyA sequence preceding the inserted pGP704 DNA has an in-frame deletion of codons 38 to 124, while the slyA sequence following the suicide vector DNA lacks the 5′-terminal 16 nucleotides of the slyA open reading frame (ORF) and the entire 5′-flanking region. Theoretically, a second homologous recombination between the KpnI-SphI fragment of the inserted pGP704CT1 and the corresponding genomic DNA fragment would result in the excision of the suicide plasmid and consequently would generate a slyA mutant containing only the truncated slyA sequence with the in-frame deletion of codons 38 to 124. However, this event has not been observed upon examination of about 15,000 clones grown in the absence of ampicillin.
Isolation and 2D-PAGE of proteins.
Bacteria from 20 ml of an overnight culture, grown for 16 to 17 h in LB medium supplemented with appropriate antibiotics, were harvested by centrifugation, washed with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH was adjusted to 7.4 with HCl]) and resuspended in 400 μl of lysis buffer containing 8 M urea, 80 mM dithiothreitol, and 5% ampholyte (pH 3 to 10; Amersham Biosciences). After addition of phenylmethylsulfonyl fluoride to give a final concentration of 5 mM, the cells were lysed by ultrasonication (five times for 10 s each) and the lysate was mixed with 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). To degrade the bacterial DNA, the lysate was mixed with 50 U of Benzonase (Merck) and incubated for 5 min at room temperature (RT). Subsequently, the cell debris was removed by centrifugation, and the protein concentration in the aqueous supernatant was determined by using the DC protein assay from Bio-Rad. Equal amounts of protein from strains to be compared to each other (up to 100 μg) were separated by 2D-PAGE according to the method of O'Farrell (34), with modifications described by Hochstrasser et al. (23, 24). Briefly, the proteins were separated in the first dimension according to their isoelectric point (pI) by isoelectric focusing in a pH gradient ranging from 3 to 10. In the second dimension, the proteins were separated according to their molecular weights by sodium dodecyl sulfate (SDS)-PAGE and visualized by Coomassie blue or silver staining.
The cellular proteins of each tested strain were isolated in at least three separate experiments, and 2D-PAGE of each protein preparation was performed at least in triplicate. Only proteins which were reproducibly and clearly found to be relatively increased or decreased in abundance based on the intensity of the protein spots were further analyzed.
Identification of proteins by MS.
Protein spots of interest were precisely excised from Coomassie blue-stained 2D gels on which about 100 μg of protein was separated. The gel pieces were cut into small cubes, rinsed several times with 100 μl of water for 15 to 30 min each, and washed three times with 100 μl of acetonitrile-water at a ratio of 1:1 for 10 to 20 min. To shrink the gel and to extract residual water, pure acetonitrile was added for 10 min. After removal of the acetonitrile, 30 to 50 μl of digestion buffer (50 mM N-methylmorpholine [pH 8.1]) and 0.5 μg of trypsin were added. Trypsin digestion of the protein was performed at 37°C for 6 to 12 h. The supernatant containing the generated peptides was recovered, and the gel pieces were extracted twice with 0.1% trifluoroacetic acid for 20 to 30 min. The volume of the combined extracts was reduced to 5 μl in a Speedvac concentrator. Liquid chromatography-mass spectrometry (LC-MS) and collision-induced fragmentation (CID) spectra were recorded on a Finnigan LCQ ion trap mass spectrometer equipped with an electrospray ionization source. The grouping of fragment ion (CID) spectra that originated from the same precursor ion and cross-correlation analysis of the data were performed by using the Sequest program (16). The Sequest algorithm compares the measured fragment ion spectra of all selected peptides to the predicted spectra of tryptic peptides that are contained in protein databases (NCBI, OWL, and NRDB) and that exhibit the same molecular weight. Identification of multiple peptides derived from the same protein and evaluation of their cross-correlation scores result in unambiguous identification of the protein.
Construction of transcriptional fusions with reporter gene lacZ.
The DNA sequence carrying the putative promoter of the E. coli hdeAB operon (2) was isolated by PCR on a 0.21-kb KpnI-BglII fragment using genomic DNA from E. coli K-12 (strain CC118) (28) as the template and oligonucleotides 5′-GAAAATCCCCTGGTACCAATCTATGCC-3′ and 5′-TTTCATCGTAAGATCTTCAACTATAAAG-3′ as forward and reverse primers, respectively. A 0.34-kb KpnI-HindIII fragment carrying the promoter region of the E. coli K-12 slp gene was amplified by PCR using upstream primer 5′-GCCTTATTTAAAGGTACCACTGCCTAATG-3′ and downstream primer 5′-GTTCATGTTACTAAGCTTATCAACAAA-3′. This DNA fragment includes the putative slp promoter predicted by Blattner et al. (5) (GenBank accession no. AE000427), which is referred here to as slpp1, as well as the promoter proposed by Alexander and St John (1), which is termed slpp2 in this study. slpp2 is located downstream from slpp1 and overlaps with the 5′ end of the slp ORF; it may control the expression of a 5′-terminally shortened form of slp starting at an alternative ATG codon. A 0.29-kb KpnI-BglII fragment containing slpp1 but not slpp2 was amplified by PCR from E. coli K-12 using primers 5′-GCCTTATTTAAAGGTACCACTGCCTAATG-3′ and 5′-CTTTATAGTTTAGATCTGATTCTGAGG-3′. The proposed promoter sequences of the E. coli mopBA operon (5, 21, 30) (GenBank accession no. AE000487, U00096 X07850, and X07899) were amplified from E. coli K-12 on a 0.21-kb KpnI-BglII fragment using upstream primer 5′-CCAAATTTTGGGTACCTGCGTAGATTTTC-3′ and downstream primer 5′-GAGAAAGTCCAGATCTGTTATGGGTG-3′. The restriction sites at the ends of these DNA fragments were introduced by the PCR primers (underlined sequences) and were exposed by cutting the PCR products with the corresponding restriction enzymes. The isolated DNA fragments were individually cloned into β-galactosidase (β-Gal) reporter vector pβgal-Basic, which carries a promoterless lacZ gene downstream from its multiple cloning site, resulting in transcriptional fusion of the promoter (p) regions with lacZ. The inserts of the generated plasmids (pβgal-hdeABp, pβgal-slpp1, pβgal-slpp1p2, and pβgal-mopBAp) were identical in sequence to the corresponding genomic DNA fragments of E. coli K-12 MG1655 (5), as confirmed by DNA sequencing.
β-Gal assay.
Bacteria were grown at 37°C in 2× yeast extract-tryptone (YT) medium (28) supplemented with appropriate antibiotics for selection of plasmids. Cells from either 100 μl of an overnight culture (grown for 16 to 17 h) or from 1 ml of a culture grown to the log phase (optical density at 600 nm [OD600] ≈ 0.7) were harvested by centrifugation, washed with PBS, and resuspended in 500 μl of lysis buffer (0.25 M Tris [pH 7.4], 0.25% [vol/vol] NP-40, 2.5 mM EDTA). After addition of lysozyme (final concentration, 1 mg/ml), the cell suspension was incubated for 10 min at RT. Lysis of the bacteria was subsequently achieved by repeated freezing (−75°C) and thawing. To degrade the bacterial DNA, the lysate was mixed with 10 mM (final concentration) MgCl2 and 30 U of DNase I and incubated for 15 min at RT. Finally, the cell debris was removed by centrifugation. The total protein concentration in the cell lysates was measured by using the DC protein assay from Bio-Rad.
The specific β-Gal activity in bacterial cell lysates was determined by using the High Sensitivity β-Gal assay kit (Stratagene) in accordance with the recommendations of the manufacturer. This assay system employs chlorophenol red-β-d-galactopyranoside (CPRG) as the substrate for β-Gal. The enzyme converts the yellow-orange CPRG into galactose and chromophore chlorophenol red, which has an absorbance maximum at 570 nm. To quantitate the β-Gal activity, the amount of substrate converted is therefore determined by measuring the absorbance of the samples at a wavelength of 570 nm.
RESULTS
Detection and identification of SlyA-regulated proteins of EIEC strain 12860.
Whole-cell proteins isolated from overnight cultures of EIEC 12860, SlyAEC-overexpressing derivative EIEC 12860/pAL105, and slyA mutant EIEC 12860slyA were separated by 2D-PAGE. Comparison of the patterns of protein spots on the 2D gels reproducibly revealed more than 40 proteins that appeared to be differentially expressed in these strains (Fig. 1). About one-half of these proteins were found in significantly higher amounts in EIEC 12860 and EIEC 12860/pAL105 than in EIEC 12860slyA (some of them were hard to detect on the 2D gels obtained from the slyA mutant), while the others were present in higher levels in the slyA mutant than in the wild-type and the SlyAEC-overproducing strains. SlyA therefore appeared to control synthesis of the former proteins positively and that of the latter negatively. Most of the putative SlyA-induced proteins were found in similar amounts in EIEC 12860 and EIEC 12860/pAL105. Only a few proteins were found in higher levels in EIEC 12860/pAL105 than in the wild-type strain.
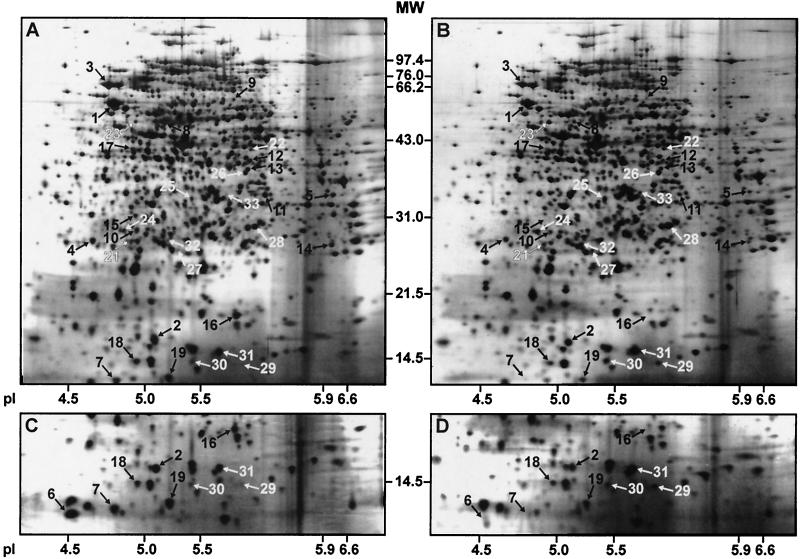
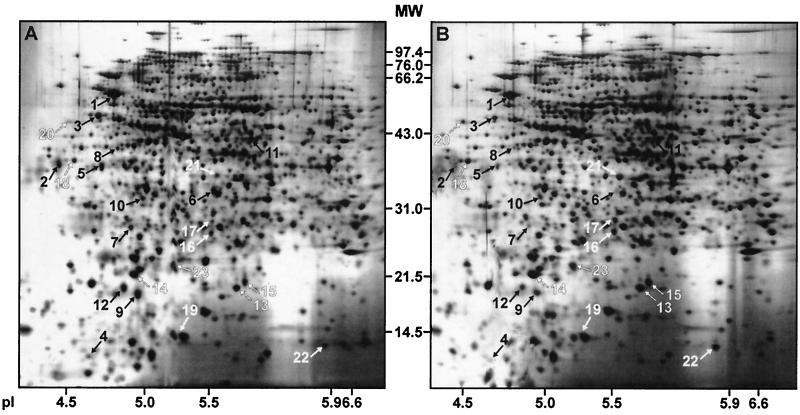
FIG. 1.
2D maps of whole-cell lysate proteins from EIEC 12860 (A and C) and EIEC 12860slyA (B and D) grown to the stationary phase. In each case, 50 μg of protein was separated by 2D-PAGE and visualized by silver staining. Separation in the second dimension was performed using 12% (A and B) and 15% (C and D) polyacrylamide gels. In panels C and D, only the lower parts of the 2D gels are shown. Proteins identified by LC-MS are indicated by numbers that correspond to the spot numbers given in Table 1. Black and white arrows, putative SlyA-induced and SlyA-repressed proteins, respectively. The positions of molecular weight (MW) marker proteins and their molecular masses (in kilodaltons) are indicated. pI markers were obtained by running a 2D-PAGE standard under conditions identical to those applied for sample separation.
Proteins clearly showing differential expression patterns in the three tested strains were isolated from the 2D gels and analyzed by LC-MS. The fragment ion spectra of 20 of the putative SlyA-induced proteins and of 13 of the putative SlyA-repressed proteins matched with those of E. coli proteins accessible in the NCBI, OWL, and NRDB protein databases. Some additional proteins presumed to be controlled by SlyA could not be identified because the available amount of protein was too small for LC-MS. The identified proteins are listed in Table 1, and their positions in the 2D maps of EIEC 12860 and EIEC 12860slyA are shown in Fig. 1.
TABLE 1.
Putative SlyA-regulated proteins of EIEC 12860 identified by MSa
| Putative SlyA regulation | Spot | Protein | Gene | Mol. mass (kDa)b | pIb |
|---|---|---|---|---|---|
| Inductione | 1 | Chaperonin GroEL (chaperone Hsp60) | mopA | 57.14 | 4.85 |
| 2 | GroES (cochaperonin of GroEL) | mopB | 10.39 | 5.15 | |
| 3 | DnaK (chaperone Hsp70) | dnaK | 68.98 | 4.83 | |
| 4 | GrpE (cochaperone of DnaK) | grpE | 21.80 | 4.68 | |
| 5 | Curved DNA-binding protein (CbpA) | cbpA | 34.46 | 6.33 | |
| 6 | HdeA (periplasmic protein involved in acid resistance) | hdeA | 11.86/9.74d | 5.06/4.68 | |
| 7 | HdeB (periplasmic protein involved in acid resistance) | hdeB | 12.04/9.07 | 5.73/4.94 | |
| 8 | Glutamate decarboxylase alpha (GadA) | gadA | 52.69 | 5.22 | |
| 9 | Hydrogenase 1, large subunit (HyaB) | hyaB | 66.25 | 5.61 | |
| 10 | SAICAR synthetase (PurC) | purC | 26.99 | 5.07 | |
| 11 | Aspartate transcarbamoylase, catalytic subunit (PyrB) | pyrB | 34.30 | 6.13 | |
| 12 | GAPDH-C (GapC) | gapC | 35.75 | 5.45 | |
| 13 | Transaldolase A (TalA) | talA | 35.66 | 5.89 | |
| 14 | Periplasmic molybdate-binding protein (ModA) | modA | 27.36/24.92 | 7.81/6.38 | |
| 15 | YbaS (putative glutaminase) | ybaS | 32.90 | 4.81 | |
| 16 | Starvation lipoprotein (Slp; outer membrane protein) | slp | 20.96/19.09 | 6.82/6.32 | |
| 17 | YhiU (putative membrane protein) | yhiU | 41.19/38.89 | 5.73/5.12 | |
| 18 | YfiD (function unknown) | yfiD | 14.28 | 5.09 | |
| 19 | YjgF (function unknown) | yjgF | 13.48 | 5.36 | |
| 20 | Cytolysin A (ClyA, HlyE, SheA)c | clyA | 33.76 | 5.08 | |
| Repressionf | 21 | HisA (phosphoribosylformimino-5-amino-1-phosphoribosyl-4-imidazole carboxamide isomerase) | hisA | 26.03 | 4.94 |
| 22 | HisB (imidazoleglycerolphosphate dehydratase; histidinol phosphatase) | hisB | 40.28 | 5.76 | |
| 23 | HisD (histidinol dehydrogenase) | hisD | 45.98 | 5.19 | |
| 24 | HisF (cyclase) | hisF | 28.45 | 5.03 | |
| 25 | HisG (ATP phosphoribosyltransferase) | hisG | 33.37 | 5.47 | |
| 26 | Phenylalanyl-tRNA synthetase, α-subunit (PheS) | pheS | 36.83 | 5.79 | |
| 27 | UPRTase | upp | 22.53 | 5.32 | |
| 28 | Periplasmic ribose-binding protein (RbsB) | rbsB | 30.95/28.47 | 6.85/5.99 | |
| 29 | RbsD (membrane-associated protein of the high-affinity ribose transporter) | rbsD | 16.70 | 6.36 | |
| 30 | YkfE (function unknown) | ykfE | 16.87/14.10 | 6.27/5.51 | |
| 31 | YnaF (function unknown) | ynaF | 16.02 | 5.60 | |
| 32 | YcaC (function unknown) | ycaC | 23.10 | 5.20 | |
| 33 | YfeU (function unknown) | yfeU | 31.22 | 5.70 |
Whole-cell lysate proteins from bacteria grown to the stationary phase were analyzed.
Theoretical molecular (mol.) masses and pI values of the E. coli proteins were calculated from the corresponding amino acid sequences deposited in the Swiss-Prot database by using ExPASy proteomics tool Compute pI/Mw (http://www.expasy.ch).
ClyA was detected only on the 2D protein map of EIEC 12860/pAL105, not on the maps of EIEC 12860 and EIEC 12860slyA.
Value for the precursor protein/value for the processed protein (after removal of the signal peptide).
Proteins show higher expression levels in EIEC 12860 and EIEC 12860/pAL105 than in EIEC 12860slyA.
Proteins show higher expression levels in EIEC 12860slyA than in EIEC 12860 and EIEC 12860/pAL105.
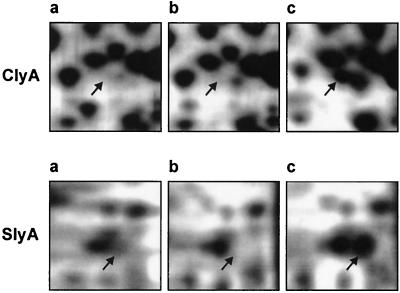
The proteins found in significantly higher levels in EIEC 12860 and EIEC 12860/pAL105 than in EIEC 12860slyA included several molecular chaperones, i.e., chaperonin GroEL (Hsp60) and its cochaperonin GroES, DnaK (Hsp70) and its cochaperone GrpE, and the “curved DNA-binding protein” (CbpA), which is a functional analog of DnaK cochaperone DnaJ. Another set of proteins presumed to be positively controlled by SlyA comprised proteins involved in the acid resistance of E. coli, particularly periplasmic proteins HdeA and HdeB and cytoplasmic glutamate decarboxylase alpha (GadA). Furthermore, the expression of several proteins involved in anabolic and catabolic processes was found to be induced in the presence of SlyA. These included (i) HyaB, the catalytic large subunit of E. coli hydrogenase 1, which catalyzes the oxidation of hydrogen gas; (ii) phosphoribosylaminoimidazole-succinocarboxamide (SAICAR) synthetase, the enzyme catalyzing the synthesis of SAICAR from 5′-phosphoribosyl-5-aminoimidazole-4-carboxylic acid in the de novo purine nucleotide synthesis pathway; (iii) PyrB, the catalytic subunit of aspartate transcarbamoylase, which mediates the first reaction unique to de novo pyrimidine synthesis; (iv) glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase C (GAPDH-C); (v) transaldolase A (TalA), an enzyme of the pentosephosphate pathway; (vi) ModA, the molybdate-specific periplasmic binding protein of the molybdate transporter, which is encoded by the modABCD operon; and (vii) YbaS, a putative glutaminase. The E. coli outer membrane “starvation lipoprotein” (Slp), putative membrane protein YhiU, and two E. coli proteins of unknown function, YfiD and YjgF, were also more strongly expressed in EIEC 12860 and EIEC 12860/pAL105 than in EIEC 12860slyA. Cytolysin ClyA was detected on the 2D gels obtained from EIEC 12860/pAL105, but it was not found in significant levels in wild-type EIEC 12860 and EIEC 12860slyA. Another protein detected only in EIEC 12860/pAL105 was identified as SlyA (Fig. 2).
FIG. 2.
Production of ClyA and SlyA in EIEC 12860 (a), EIEC 12860slyA (b), and EIEC 12860/pAL105 (c).Whole-cell lysate proteins (50 μg) from cultures grown to the stationary phase were separated by 2D-PAGE (12% polyacrylamide gels in the second dimension) and visualized by silver staining. ClyA and SlyA were identified by LC-MS. Only details of the 2D gels are shown. Arrows, positions of ClyA and SlyA.
The proteins showing increased levels in the EIEC 12860 slyA mutant compared to levels in the wild-type and the SlyAEC-overproducing strains were mainly enzymes involved in anabolic pathways. Among these proteins were five of the eight enzymes of the histidine biosynthesis pathway, i.e., HisA, HisB, HisD, HisF, and HisG. Since the structural genes encoding these five proteins are part of the hisGDCBHAFI operon, it is likely that the entire his operon is induced in the absence of SlyA. Production of the α-subunit of phenylalanyl-tRNA synthetase, PheS, was also found to be relieved in the EIEC 12860 slyA mutant. The proteins found in higher levels in EIEC 12860slyA than in the wild-type strain and EIEC 12860/pAL105 further included pyrimidine salvage enzyme uracil phosphoribosyltransferase (UPRTase), which catalyzes the synthesis of UMP from uracil and phosphoribosyldiphosphate, two components of the E. coli high-affinity ribose transporter (RbsB and RbsD), and several E. coli proteins of unknown function (YkfE, YnaF, YcaC, and YfeU) that were predicted from ORFs in the E. coli K-12 genome (5).
Influence of SlyA on the resistance of EIEC 12860 to heat and acid stress.
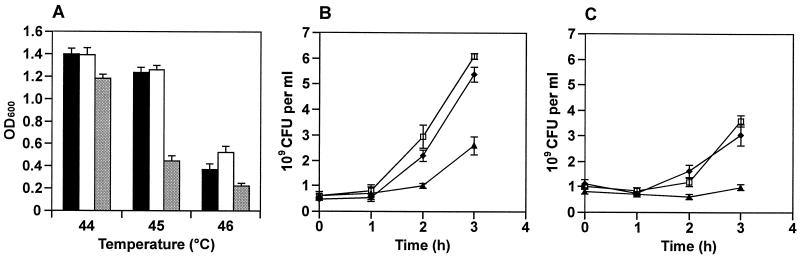
The finding that the expression of several stress response proteins, including molecular chaperones and proteins involved in acid resistance, is positively controlled in EIEC 12860 by SlyA prompted us to study the growth characteristics of EIEC 12860, EIEC 12860/pAL105, and EIEC 12860slyA at elevated temperatures and under conditions of acid stress. As shown in Fig. 3A, the wild-type strain and the SlyAEC-overexpressing strain exhibited a higher resistance to heat than the slyA mutant. In particular, growth of EIEC 12860 and EIEC 12860/pAL105 at 45°C was only slightly inhibited compared to the growth at 37°C, and only at temperatures of 46°C and higher was the propagation of both strains markedly impaired. Growth of EIEC 12860slyA, on the other hand, was already strongly inhibited at 45°C and was almost completely suppressed at 46°C.
FIG. 3.
Growth characteristics of EIEC 12860 and derivative strains at elevated temperatures and under conditions of acid stress. (A) Aliquots (5 ml) of LB medium were inoculated 1:100 with overnight cultures (grown at 37°C) of EIEC 12860 (white bars), EIEC 12860/pAL105 (black bars), and EIEC 12860slyA (grey bars). The diluted cultures were incubated either at 44, 45, or 46°C. Growth of the bacteria was determined 14 h after inoculation by measuring the OD600 of the cultures. (B and C) EIEC 12860 (□), EIEC 12860/pAL105 (⧫), and EIEC 12860slyA (▴) were grown at 37°C in LB medium to an OD600 of approximately 1.0. Bacteria from 20 ml of the culture were then harvested by centrifugation, resuspended in 1 ml of PBS, and inoculated into 19 ml of LB medium-10 mM Tris-HCl adjusted to a pH of either 4.4 (B) or 4.2 (C). The numbers of viable bacteria were determined after incubation for 1, 2, and 3 h at 37°C. Error bars, standard deviations of the means from three separate experiments.
EIEC 12860 and EIEC 12860/pAL105 also displayed stronger resistance to acid stress than the slyA mutant. When the three strains were grown at 37°C in LB medium to the late log phase (OD600 ≈ 1.0) and then shifted into fresh medium supplemented with 10 mM Tris and adjusted to pH 4.4 with HCl, efficient replication was observed for the wild-type strain and the SlyAEC-overproducing strain, whereas growth of the slyA mutant was significantly retarded (Fig. 3B). When the cells were shifted into medium that was adjusted to pH 4.2, replication of EIEC 12860slyA was almost completely inhibited within the first 3 h of incubation, while significant albeit reduced growth was still observed for EIEC 12860 and EIEC 12860/pAL105 (Fig. 3C). The wild-type and the SlyAEC-overproducing strains thus adapted more readily to acidic growth conditions than the slyA mutant.
Transcriptional analysis of putative SlyA-regulated E. coli genes.
To confirm the proteomic data presented above, we studied the influence of SlyA on the regulation of E. coli genes encoding some of the identified putative SlyA-controlled proteins. We chose the hdeAB operon and the slp gene for these analyses, because synthesis of HdeA, HdeB, and Slp was strongly induced in the presence of SlyA (Fig. 1, spots 6, 7, and 16). In addition, the mopBA (groES-groEL) operon was included because GroES and GroEL, like a number of other proteins, seemed to be less strongly, albeit clearly, induced by SlyA (Fig. 1, spots 1 and 2). DNA fragments carrying the putative promoter sequences of these genes and operons were isolated from E. coli K-12 and cloned into β-Gal reporter vector pβgal-Basic, resulting in transcriptional fusion with the plasmid-encoded lacZ gene (see Materials and Methods for details). For the slp promoter region, two recombinant pβgal-Basic derivatives were constructed, one carrying both predicted slp promoters, slpp1 and slpp2, and the other carrying only slpp1.
To analyze whether SlyA affects transcription from the cloned DNA sequences, we determined the specific β-Gal activity in lysates of E. coli DH5α harboring either pβgal-Basic or one of the constructed pβgal-Basic derivatives (pβgal-hdeABp, pβgal-slpp1, pβgal-slpp1p2, and pβgal-mopBAp), as well as in DH5α double transformants each harboring slyAEC-carrying plasmid pAL108 in addition to one of these pβgal-Basic derivatives.
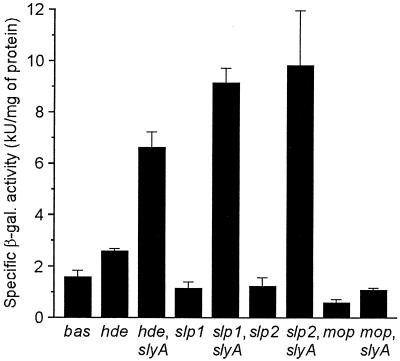
As shown in Fig. 4, the specific β-Gal activity measured in DH5α/pβgal-hdeABp grown to the stationary phase was increased by a factor of about 1.6 compared to the basal activity found under the same conditions in DH5α/pβgal-Basic. On the other hand, the activity detected in the DH5α clones carrying pβgal-slpp1 and pβgal-slpp1p2 was reduced on average by about 28 and 22%, respectively, and that measured in DH5α/pβgal-mopBAp was reduced on average by approximately 62% compared to DH5α/pβgal-Basic activity. However, introduction of pAL108 into the strains harboring the recombinant pβgal-Basic constructs caused in each case a significant increase of the specific β-Gal activity, confirming that SlyA activates the expression of hdeAB, slp, and mopBA in E. coli. In DH5α/pβgal-hdeABp/pAL108 grown to the stationary phase, the specific β-Gal activity was about 2.5 times as high as in DH5α carrying only pβgal-hdeABp. The activity detected in the double transformants harboring pAL108 in combination with either pβgal-slpp1 or pβgal-slpp1p2 was, under the same conditions, about eight times as high as that measured in the DH5α clones harboring only pβgal-slpp1 and pβgal-slpp1p2, respectively. SlyA thus activated transcription from slpp1 to a similar extent as from the DNA fragment carrying both slpp1 and slpp2, suggesting that only slpp1 is controlled by SlyA. Transformation of pAL108 into DH5α/pβgal-mopBAp increased the specific β-Gal activity of this strain by a factor of about 1.8, which indicated that SlyA does not activate transcription of the mopBA operon as strongly as that of slp and that of the hdeAB operon, consistent with the protein data obtained by 2D-PAGE (see above). Introduction of pAL108 into DH5α harboring only pβgal-Basic did not significantly affect the basal β-Gal activity of this strain.
FIG. 4.
SlyA-dependent transcription of hdeAB, slp, and mopBA of E. coli. Transcription was quantitated by using fusions of the 5′-flanking regulatory regions of these genes and operons with reporter gene lacZ on plasmid pβgal-Basic. The specific β-Gal activities of the following strains were determined as described in Materials and Methods: bas, E. coli DH5α/pβgal-Basic; hde, DH5α/pβgal-hdeABp; hde slyA, DH5α/pβgal-hdeABp/pAL108; slp1, DH5α/pβgal-slpp1; slp1 slyA, DH5α/pβgal-slpp1/pAL108; slp2, DH5α/pβgal-slpp1p2; slp2 slyA, DH5α/pβgal-slpp1p2/pAL108; mop, DH5α/pβgal-mopBAp; mop slyA, DH5α/pβgal-mopBAp/pAL108. The results shown were obtained with bacteria grown for 16 to 17 h at 37°C in 2× yeast extract-tryptone medium supplemented with appropriate antibiotics. Error bars, standard deviations of the means from three independent experiments.
Detection and identification of SlyA-regulated proteins of Salmonella serovar Typhimurium.
To analyze the SlyA regulon of Salmonella serovar Typhimurium, whole-cell proteins isolated from overnight cultures of (i) Salmonella serovar Typhimurium ATCC 14028s, (ii) Salmonella serovar Typhimurium ATCC 14028s harboring slyAST-carrying plasmid pAL102, and (iii) slyA mutant Salmonella serovar Typhimurium ATCC 14028slyA were subjected to 2D-PAGE. When the obtained 2D protein maps of the wild-type strain and of the slyA mutant were compared to each other, differences in the protein patterns were not as obvious as when maps for EIEC 12860 and EIEC 12860slyA were compared. However, clear differences were seen upon comparison of the 2D gels obtained from Salmonella serovar Typhimurium ATCC 14028s/pAL102 and Salmonella serovar Typhimurium ATCC 14028slyA (Fig. 5). More than 20 proteins exhibited significantly higher levels in the SlyAST-overproducing strain than in the slyA mutant, suggesting that their synthesis is positively regulated at the transcriptional level by SlyA. In addition, at least 20 proteins were reproducibly found in larger amounts in Salmonella serovar Typhimurium ATCC 14028slyA than in Salmonella serovar Typhimurium ATCC 14028s/pAL102, which indicated that transcription of the corresponding structural genes is induced in the absence of SlyA. Most of these putative SlyA-regulated proteins were isolated from the 2D gels and analyzed by LC-MS.
FIG. 5.
Silver-stained 2D maps of whole-cell lysate proteins from Salmonella serovar Typhimurium ATCC 14028s/pAL102 (A) and Salmonella serovar Typhimurium ATCC 14028slyA (B) grown to the stationary phase. In both cases, 50 μg of protein was separated by 2D-PAGE. Twelve percent polyacrylamide gels were used for separation in the second dimension. Proteins identified by LC-MS are indicated by numbers corresponding to those listed in Table 2. Black and white arrows, putative SlyA-induced and SlyA-repressed proteins, respectively. The positions of molecular weight (MW) and pI markers are indicated. Molecular masses of size markers are given in kilodaltons.
Cross-correlation analysis of the LC-MS data using the NCBI protein database led to the identification of 12 of the putative SlyA-induced proteins (Table 2; Fig. 5). These included chaperonin GroEL, outer membrane porin OmpD, flagellar filament structural protein (flagellin) FliC, 50S ribosomal subunit protein L7/L12, PotD, which functions as the periplasmic substrate-binding protein of the spermidine-preferential polyamine uptake system, a putative cytoplasmic protein of unknown function (YhcH), and several proteins representing homologues of known bacterial enzymes: NanA, the protein subunit of homotetrameric N-acetylneuraminate lyase (sialic acid aldolase), which is involved in the catabolism of sialic acid; a putative N-acetylmannosamine-6-phosphate (ManNAc-6P) epimerase (NanE); the NAD+-dependent glycerol dehydrogenase (GldA), which catalyzes the oxidation of glycerol to dihydroxyacetone; a thiol peroxidase (Tpx); a putative oxidoreductase (UcpA); and a putative glutamate dehydrogenase (product of STM1795).
TABLE 2.
Putative SlyA-regulated proteins of Salmonella serovar Typhimurium ATCC 14028s identified by MSa
| Putative SlyA regulation | Spot | Protein | Gene | Mol. mass (kDa)b | pIb |
|---|---|---|---|---|---|
| Inductionc | 1 | GroEL (chaperone Hsp60) | mopA | 57.29 | 4.85 |
| 2 | OmpD (outer membrane porin) | nmpC | 39.68/37.63e | 4.66/4.57 | |
| 3 | FliC (flagellin, flagellar filament structural protein) | fliC | 51.61 | 4.79 | |
| 4 | 50S ribosomal subunit protein L7/L12 | rplL | 12.30 | 4.60 | |
| 5 | PotD (spermidine/putrescine-binding periplasmic protein) | potD | 39.02/36.65 | 5.17/4.84 | |
| 6 | N-acetylneuraminate lyase (aldolase) subunit (NanA) | nanA | 32.46 | 5.56 | |
| 7 | Putative ManNAc-6P epimerase | nanE | 24.03 | 5.07 | |
| 8 | Glycerol dehydrogenase, NAD+ dependent (GldA) | gldA | 38.74 | 4.88 | |
| 9 | Thiol peroxidase | tpx | 18.03 | 4.93 | |
| 10 | Putative oxidoreductase | ucpA | 27.87 | 5.05 | |
| 11 | Putative glutamate dehydrogenase | STM1795 | 48.04 | 6.23 | |
| 12 | Putative cytoplasmic protein | yhcH | 17.06 | 4.93 | |
| Repressiond | 13 | Periplasmic Cu,Zn-superoxide dismutase (SodCII) | sodCII | 17.74/15.81 | 6.03/5.69 |
| 14 | OsmY (hyperosmotically and carbon starvation-inducible periplasmic protein) | osmY | 21.45/18.55 | 5.78/5.14 | |
| 15 | LuxS (quorum-sensing protein; autoinducer synthase) | luxS | 19.31 | 5.71 | |
| 16 | Purine nucleoside phosphorylase (DeoD) | deoD | 25.98 | 5.56 | |
| 17 | Putative oxidoreductase | yohF | 26.83 | 5.50 | |
| 18 | Putative catalase | ORF G57 (STM1731) | 31.85 | 4.62 | |
| 19 | YgaU (putative LysM domain protein) | ygaU | 16.12 | 5.44 | |
| 20 | Putative outer membrane or exported protein | STM4242 | 46.98/44.45 | 4.69/4.56 | |
| 21 | Periplasmic l-asparaginase II (AnsB) | ansB | 36.93/34.61 | 5.84/5.61 | |
| 22 | YbdQ (function unknown) | ybdQ | 15.90 | 6.18 | |
| 23 | YciF (putative cytoplasmic protein) | yciF | 18.65 | 5.24 |
Cellular proteins from bacteria grown to the stationary phase were analyzed.
Theoretical molecular (mol.) masses and pI values of the Salmonella serovar Typhimurium proteins were calculated from the corresponding amino acid sequences deposited in the NCBI protein database (http://www.ncbi.nlm.nih.gov/Entrez) by using ExPASy proteomics tool Compute pI/Mw (http://www.expasy.ch). Putative signal peptide cleavage sites in the precursors of the identified periplasmic and outer membrane (or exported) Salmonella serovar Typhimurium proteins were predicted as described by Nielsen et al. (33) with ExPASy proteomics tool SignalP.
Proteins show higher expression levels in Salmonella serovar Typhimurium ATCC 14028s/pAL102 than in slyA mutant Salmonella serovar Typhimurium ATCC 14028slyA.
Proteins show higher expression levels in Salmonella serovar Typhimurium ATCC 14028slyA than in Salmonella serovar Typhimurium ATCC 14028s/pAL102.
Value for precursor form/value for predicted mature form.
In addition, 11 of the proteins found in significantly higher levels in the slyA mutant than in the SlyAST-overproducing derivative of Salmonella serovar Typhimurium ATCC 14028s were identified by LC-MS: periplasmic Cu,Zn-cofactored superoxide dismutase SodCII, which catalyzes the conversion of superoxide radicals to hydrogen peroxide; OsmY, a periplasmic protein of unknown function that is expressed during transition into the stationary phase and in response to increased medium osmolarity; autoinducer synthase LuxS, which is involved in quorum sensing; purine nucleoside phosphorylase (DeoD), an enzyme involved in purine metabolism catalyzing the reversible conversion of guanosine to guanine; a putative oxidoreductase (YohF); a putative catalase (product of ORFG57/STM1731); a putative LysM domain protein (YgaU); a putative outer membrane or exported protein (product of STM4242); AnsB, the subunit of periplasmic, homotetrameric enzyme l-asparaginase II, which catalyzes the conversion of l-asparagine to l-aspartate and ammonium; and two proteins of unknown function that are encoded by ybdQ and yciF.
DISCUSSION
The data presented in this paper demonstrate that SlyA differentially affects the expression of various proteins in E. coli and Salmonella serovar Typhimurium. 2D-PAGE analyses particularly suggested that in EIEC strain 12860 and in Salmonella serovar Typhimurium ATCC 14028s transcriptional regulator SlyA positively controls the expression of several proteins and negatively controls that of others. We have identified most of these putative SlyA-controlled proteins by MS and have confirmed for some of them that SlyA is indeed implicated in the regulation of the corresponding structural genes.
We chose an EIEC strain instead of E. coli K-12 for the analysis of the SlyA regulon of E. coli, since we were interested to study whether SlyA controls not only housekeeping genes but also E. coli genes involved in virulence. EIEC appeared to be particularly promising in this context since, for Salmonella serovar Typhimurium, SlyA has already been shown to be required for virulence and intracellular survival in macrophages (26).
Clearly, none of the putative SlyA-regulated proteins of EIEC 12860 identified in this study represents a known virulence factor; all of them are also found in E. coli K-12. Nevertheless, several of these proteins might play a role in pathogenicity. For example, cytolysin ClyA, which is positively controlled by SlyA, may be considered a potential virulence factor, and although the clyA gene of E. coli K-12 is silent under standard laboratory conditions, it is possible that expression of clyA is induced in EIEC (and other groups of pathogenic E. coli) during infection of the host. In addition, several of the putative SlyA-induced proteins of EIEC 12860 proved to be stress response proteins, which might play an important role in the course of an infection by helping the bacteria to survive under different stress conditions encountered in the host. The identified stress response proteins particularly included several molecular chaperones (8), which assist in the correct folding of other proteins (GroEL, GroES, DnaK, GrpE, and CbpA), proteins supporting the acid resistance of E. coli (HdeA, HdeB, and GadA), the outer membrane starvation lipoprotein (Slp), and the catalytic subunit of hydrogenase 1 (HyaB). GroEL, GroES, DnaK, and GrpE are heat shock proteins that are controlled at the transcriptional level by alternative sigma factor σ32 (RpoH) (4, 43), whereas the synthesis of CbpA, HdeA, HdeB, GadA, and HyaB is controlled by σS (RpoS, KatF, σ38), the master regulator of general stress response in E. coli (2, 3, 9, 10, 12, 22, 48). The slp gene is induced during carbon starvation and when cells enter the stationary phase, but it is apparently not controlled by σS (1).
The finding that SlyA positively affects the cellular levels of several heat shock proteins and of proteins involved in the acid resistance of E. coli was consistent with the observation that the resistance of the EIEC 12860 slyA mutant to heat and acid stresses is impaired compared to that of the wild-type strain and to that of the SlyAEC-overproducing strain. In addition, β-Gal assays performed with transcriptional fusions between lacZ and the promoter regions of E. coli slp, hdeAB, and mopBA (groES-groEL), respectively, confirmed that SlyA activates transcription of the genes and operons encoding Slp, HdeA, HdeB, GroES, and GroEL.
GroEL was also found among the putative SlyA-induced proteins of Salmonella serovar Typhimurium ATCC 14028s. All other putative SlyA-regulated proteins of this strain that were identified by MS differed, however, from those of EIEC 12860 that were detected. The identified Salmonella serovar Typhimurium proteins are involved in different cellular processes, and, although none of these proteins represents a typical virulence factor, some of them might play a role in the virulence of Salmonella serovar Typhimurium, as already shown for Cu,Zn-superoxide dismutase SodCII (17). Interestingly, ompD, one of the genes found to be positively regulated by SlyA in Salmonella serovar Typhimurium, is absent from the genome of E. coli (5, 25, 29), while several of the putative SlyA-induced genes of E. coli (hdeA, hdeB, clyA, and gadA) do not have homologues in Salmonella serovar Typhimurium (19, 29, 41), which is consistent with the presumption that the SlyA regulons of E. coli and Salmonella serovar Typhimurium are not identical. Nevertheless, these regulons overlap partially, as exemplified by GroEL.
Induction of the synthesis of stress response proteins such as GroEL may be crucial for the capability of EIEC and Salmonella serovar Typhimurium to survive and replicate within macrophages, which are major host cells for both pathogens. Buchmeier and colleagues (6) have shown that slyA is expressed in Salmonella serovar Typhimurium when the bacteria are in the intracellular environment of J774 macrophages. The specific in vivo conditions causing production of SlyA are presently unknown, but we have recently observed that the synthesis of SlyA is induced in E. coli and Salmonella serovar Typhimurium when these bacteria are grown in acidified media (Ludwig et al., unpublished data). Based on these data it seems likely that the expression of slyA is activated in EIEC and Salmonella serovar Typhimurium following uptake into macrophages by acidification of the vacuole. The ensuing SlyA-induced synthesis of stress response proteins may then help these pathogens overcome the hostile environment of this compartment. Consistently, GroEL and DnaK have been shown to be among the major proteins that are selectively induced in Salmonella serovar Typhimurium during infection of macrophages, and it has been suggested that these proteins may stabilize bacterial macromolecular complexes after exposure to the macrophage's toxic and degradative products (7).
In contrast to Salmonella serovar Typhimurium, EIEC evades the phagosome and replicates in the host cell cytosol. The mechanism mediating the escape of EIEC into the cytosol has not yet been elucidated, but, for facultative intracellular pathogen Listeria monocytogenes, cytolysin listeriolysin O has been shown to be required for the disruption of the phagosomal membrane (37). Whether ClyA plays a similar role in EIEC is presently unknown, but in this context it is interesting that tested Salmonella serovar Typhimurium strains do not possess a clyA-homologous gene. In the host cell cytosol EIEC may be better protected from bactericidal mechanisms of macrophages but probably needs specific metabolic prerequisites for efficient proliferation. The synthesis of purine and pyrimidine bases seems to be one of these requirements. Hence the finding that the synthesis of key enzymes for the de novo biosynthesis of these components is induced by SlyA is in accord with this requirement, as is the apparent SlyA-mediated down-regulation of genes involved in histidine biosynthesis, since biosynthesis of both purine and histidine competes with phosphoribosyl pyrophosphate for a common precursor. Histidine seems to belong to those amino acids which can be provided by the host cell.
It is remarkable that for EIEC 12860 most of the identified SlyA-regulated proteins were detected by comparing the 2D maps of the wild-type strain and of the slyA mutant, while most of the proteins presumed to be positively or negatively controlled by SlyA in Salmonella serovar Typhimurium ATCC 14028s were clearly detected only when the 2D maps of the SlyAST-overproducing derivative and of the slyA mutant were compared to each other. The reason for this difference is presently unknown, but a possible explanation could be that in E. coli and Salmonella serovar Typhimurium different concentrations of SlyA are required for regulation. Clearly, variations in protein abundance observed only with SlyA overexpression may not necessarily be physiologically relevant, but it is conceivable that the synthesis of SlyA in Salmonella serovar Typhimurium is indeed strongly induced under certain in vivo conditions. Furthermore, the 2D-PAGE data suggested that SlyA differentially induces the production of different proteins in EIEC 12860. In particular, proteins such as Slp, HdeA, and HdeB appeared to be more strongly induced by SlyA then others, an observation that was also confirmed at the level of transcription with the lacZ fusions. Induction of the synthesis of ClyA in EIEC 12860 required higher concentrations of SlyA. This is consistent with the findings regarding the regulation of clyA in E. coli K-12 (27, 28, 35) and suggests that SlyA interacts less efficiently with the clyA promoter region than with the regulatory regions of other SlyA-induced genes and operons.
DNA band shift assays performed by Oscarsson et al. (35) indicated that SlyAEC activates clyA expression by binding to the clyA promoter region. Nevertheless, we do not know as yet whether the other SlyA-regulated genes are also directly or only indirectly controlled by this transcription factor. SlyA might, for example, induce the expression of genes encoding other transcriptional regulators, which themselves induce or repress the production of some of the proteins identified in this study. An indirect role for SlyA in gene regulation might be particularly relevant for genes presumed to be repressed by SlyA, because so far it has been shown merely that SlyA acts only as a transcriptional activator. It is not known whether SlyA is also capable of directly repressing the transcription of genes. In addition, some of the proteins found in larger amounts in the EIEC and Salmonella serovar Typhimurium slyA mutants than in the corresponding wild-type and slyA-overexpressing strains might not be negatively controlled by SlyA but rather might increase in steady-state abundance as a stress response to the slyA mutation. This could, for example, be true for SodCII, which has been proposed to play a role in the defense of Salmonella serovar Typhimurium against phagocyte-derived reactive oxygen species (17). Since SlyA appears to be involved in the resistance of Salmonella serovar Typhimurium to oxidative stress (6), perhaps enhanced sodCII expression in the Salmonella serovar Typhimurium ATCC 14028s slyA mutant is simply a compensatory phenomenon. On the other hand, negative control of the expression of sodCII by SlyA would not necessarily be inconsistent with the observed importance of SlyA for the resistance of Salmonella to oxidative damage, because Salmonella serovar Typhimurium is capable of producing several different superoxide dismutases, including an additional Cu,Zn-superoxide dismutase (SodCI), which contributes to the virulence of Salmonella serovar Typhimurium (13, 17, 18, 44).
Obviously, the putative SlyA-controlled genes identified in this study do not represent the complete SlyA regulons of EIEC and Salmonella serovar Typhimurium. For example, a number of these genes are located within operons, but in several instances other genes present in the same operons were not identified. In E. coli, this is exemplified by the hyaABCDEF operon (only HyaB was identified), the modABCD operon (ModA was identified), the pyrBI operon (PyrB was identified), the hisGDCBHAFI operon (HisA, HisB, HisD, HisF, and HisG were identified), the pheST-himA operon (PheS was identified), the upp-uraA operon (UPRTase was identified), and the rbsDACBK operon (RbsB and RbsD were identified). Therefore, it seems likely that other genes of these operons are also regulated by SlyA.
There are several conceivable reasons why we were unable to identify additional SlyA-regulated, possibly virulence-associated proteins of these pathogens. (i) Only cellular proteins that are abundant, stable, and soluble could be detected by 2D-PAGE and identified by MS. (ii) Although we used a pH gradient ranging from 3 to 10 for isoelectric focusing, proteins with pIs below about 4 and above 7 were hard to detect on the 2D gels and consequently escaped further analysis. (iii) Several SlyA-activated genes may not be expressed under the in vitro cultivation conditions used in this study; they may require other stimuli for induction, which might, for example, be encountered during infection of phagocytic cells. One-dimensional SDS-PAGE analysis indeed suggested that the pattern of SlyA-regulated proteins of Salmonella serovar Typhimurium in the stationary phase is different from that during infection of macrophages (6).
Thus, additional SlyA-regulated proteins of E. coli and Salmonella serovar Typhimurium remain to be identified. Further investigations including transcriptome analyses will be necessary to completely unravel the SlyA regulons of these pathogens and to elucidate the regulatory mechanism of SlyA.
Acknowledgments
We thank Helge Karch for providing the EIEC wild-type strain 12860 and Claudia Tengel for constructing slyA mutant EIEC 12860slyA.
This work was supported by grants from the Deutsche Forschungsgemeinschaft. A.S. received a fellowship from the University of Würzburg (Graduiertenkolleg 'Infektiologie').
REFERENCES
- 1.Alexander, D. M., and A. C. St John. 1994. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol. Microbiol. 11:1059-1071. [DOI] [PubMed] [Google Scholar]
- 2.Arnqvist, A., A. Olsen, and S. Normark. 1994. σS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., K. Knudsen, L. Heerfordt, and L. Brøndsted. 1997. Effects of σS and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J. Bacteriol. 179:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi, A. A., and F. Baneyx. 1999. Hyperosmotic shock induces the σ32 and σE stress regulons of Escherichia coli. Mol. Microbiol. 34:1029-1038. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Buchmeier, N., S. Bossie, C.-Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730-732. [DOI] [PubMed] [Google Scholar]
- 8.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 9.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colland, F., N. Fujita, D. Kotlarz, J. A. Bown, C. F. Meares, A. Ishihama, and A. Kolb. 1999. Positioning of σS, the stationary phase σ factor, in Escherichia coli RNA polymerase-promoter open complexes. EMBO J. 18:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels, J. J. D., I. B. Autenrieth, A. Ludwig, and W. Goebel. 1996. The gene slyA of Salmonella typhimurium is required for destruction of M cells and intracellular survival but not for invasion or colonization of the murine small intestine. Infect. Immun. 64:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 13.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehoux, P., and P. Cossart. 1995. Homologies between salmolysin and some bacterial regulatory proteins. Mol. Microbiol. 15:591-592. [DOI] [PubMed] [Google Scholar]
- 15.Del Castillo, F. J., S. C. Leal, F. Moreno, and I. del Castillo. 1997. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 16.Eng, J. K., A. L. McCormak, and J. R. Yates, Jr. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 17.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Bäumler, U. Ochsner, T. Testerman, S. Bearson, J.-C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrant, J. L., A. Sansone, J. R. Canvin, M. J. Pallen, P. R. Langford, T. S. Wallis, G. Dougan, and J. S. Kroll. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol. Microbiol. 25:785-796. [DOI] [PubMed] [Google Scholar]
- 19.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 20.Green, J., and M. L. Baldwin. 1997. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology 143:3785-3793. [DOI] [PubMed] [Google Scholar]
- 21.Hemmingsen, S. M., C. Woolford, S. M. van der Vies, K. Tilly, D. T. Dennis, C. P. Georgopoulos, R. W. Hendrix, and R. J. Ellis. 1988. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333:330-334. [DOI] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R. 1996. Back to log phase: σS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 23.Hochstrasser, D. F., V. Augsburger, M. Funk, R. Appel, C. Pelegrini, and A. F. Müller. 1986. Immobilized pH gradients in capillary tubes and two-dimensional gel electrophoresis. Electrophoresis 7:505-511. [Google Scholar]
- 24.Hochstrasser, D. F., M. G. Harrington, A. C. Hochstrasser, M. J. Miller, and C. R. Merril. 1988. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal. Biochem. 173:424-435. [DOI] [PubMed] [Google Scholar]
- 25.Lee, D. R., and C. A. Schnaitman. 1980. Comparison of outer membrane porin proteins produced by Escherichia coli and Salmonella typhimurium. J. Bacteriol. 142:1019-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig, A., C. Tengel, S. Bauer, A. Bubert, R. Benz, H.-J. Mollenkopf, and W. Goebel. 1995. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474-486. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig, A., S. Bauer, R. Benz, B. Bergmann, and W. Goebel. 1999. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31:557-567. [DOI] [PubMed] [Google Scholar]
- 29.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 30.Miki, T., T. Orita, M. Furuno, and T. Horiuchi. 1988. Control of cell division by sex factor F in Escherichia coli. III. Participation of the groES (mopB) gene of the host bacteria. J. Mol. Biol. 201:327-338. [DOI] [PubMed] [Google Scholar]
- 31.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 34.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 35.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 36.Oscarsson, J., Y. Mizunoe, L. Li, X.-H. Lai, A. Wieslander, and B. E. Uhlin. 1999. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol. Microbiol. 32:1226-1238. [DOI] [PubMed] [Google Scholar]
- 37.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomson, N. R., A. Cox, B. W. Bycroft, G. S. A. B. Stewart, P. Williams, and G. P. C. Salmond. 1997. The Rap and Hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531-544. [DOI] [PubMed] [Google Scholar]
- 43.Tomoyasu, T., T. Ogura, T. Tatsuta, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30:567-581. [DOI] [PubMed] [Google Scholar]
- 44.Tsolis, R. M., A. J. Bäumler, and F. Heffron. 1995. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect. Immun. 63:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace, A. J., T. J. Stillman, A. Atkins, S. J. Jamieson, P. A. Bullough, J. Green, and P. J. Artymiuk. 2000. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100:265-276. [DOI] [PubMed] [Google Scholar]
- 46.Watson, P. R., S. M. Paulin, A. P. Bland, S. J. Libby, P. W. Jones, and T. S. Wallis. 1999. Differential regulation of enteric and systemic salmonellosis by slyA. Infect. Immun. 67:4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashino, T., M. Kakeda, C. Ueguchi, and T. Mizuno. 1994. An analogue of the DnaJ molecular chaperone whose expression is controlled by σs during the stationary phase and phosphate starvation in Escherichia coli. Mol. Microbiol. 13:475-483. [DOI] [PubMed] [Google Scholar]