Abstract
The tetrachloroethene (PCE) reductive dehalogenase (encoded by the pceA gene and designated PceA dehalogenase) of Desulfitobacterium sp. strain Y51 was purified and characterized. The expression of the enzyme was highly induced in the presence of PCE and trichloroethene (TCE). The purified enzyme catalyzed the reductive dehalogenation of PCE via TCE to cis-1,2-dichloroethene at a specific activity of 113.6 nmol · min−1 · mg of protein−1. The apparent Km values for PCE and TCE were 105.7 and 535.3 μM, respectively. Chlorinated ethenes other than PCE and TCE were not dehalogenated. However, the enzyme exhibited dehalogenation activity for various chlorinated ethanes such as hexachloroethane, pentachloroethane, 1,1,1,2-tetrachloroethane, and 1,1,2,2-tetrachloroethane. The pceA gene of Desulfitobacterium sp. strain Y51 was identified in a 2.8-kb DNA fragment and used to express the protein in Escherichia coli for the preparation of antibodies. Immunoblot analyses located PceA in the periplasm of the cell.
Recently, a number of studies have described reductive dehalogenation by bacteria which occurs during the initial attack of halogenated hydrocarbons. Such bacteria can grow by anaerobic respiration, a process that has been referred to as halorespiration or dehalorespiration (6, 9, 11). Several dehalorespiring bacteria have been reported to catalyze the reductive dehalogenation of tetrachloroethene (also referred to perchloroethene [PCE]). Most of these bacteria reduce PCE or trichloroethene (TCE) to cis-1,2-dichloroethene (cis-DCE) (5, 10, 18, 19). Dehalococcoides ethenogenes strain 195 is able to catalyze the reductive dehalogenation of PCE to ethene (16, 17).
PCE dehalogenases have been purified, and their genes were cloned from some anaerobic bacteria such as Dehalospirillum multivorans (21-23), Desulfitobacterium sp. strain PCE-S (20), Dehalobacter restrictus (26, 27), and D. ethenogenes 195 (13). The molecular characterization of the chloroethene reductive dehalogenases from phylogenetically distinct bacteria has revealed significant similarities in molecular masses (51 to 65 kDa) and functional domains, but it is also true that similarities of the entire amino acid sequences are surprisingly low among these enzymes. The PCE dehalogenase of Clostridium bifermentans DPH-1 was recently purified and sequenced (24). This enzyme is a homodimer with a molecular mass of ca. 70 kDa and exhibits dehalogenation of DCE isomers along with PCE and TCE. All the PCE dehalogenase genes except pceC from C. bifermentans DPH-1 (24) characterized to date are preceded by twin arginine signal sequences (2, 3, 25). This class of proteins contains corrinoid and two Fe/S clusters as prosthetic groups (11, 31). The PCE dehalogenase genes were found to be linked with open reading frames (ORFs) coding for small hydrophobic proteins containing two or three transmembrane helices (14, 23, 28, 30). It has been proposed that the PceB protein in D. multivorans might act as a membrane anchor that functionally links the dehalogenase to the respiratory chain (7, 11, 23).
A strict anaerobic bacterium, Desulfitobacterium sp. strain Y51, isolated in our laboratory exhibits a strong dehalogenating activity for PCE at concentrations as high as 960 μM and as low as 0.6 μM, converting it to cis-DCE via TCE (29). To characterize the PceA dehalogenase of strain Y51, we have attempted to purify, characterize, and clone the genes. Also described are the induction and localization of this enzyme using a polyclonal antibody raised against the purified PceA of strain Y51.
MATERIALS AND METHODS
Strains and cultivation.
The strains and plasmids used in this study are listed in Table 1. Desulfitobacterium sp. strain Y51 was used throughout this study. Strain Y51 was isolated from soil contaminated with chloroethenes, and its microbial properties have already been reported (29). Strain Y51 was grown anaerobically on MMYP medium (pH 7.2; K2HPO4, 7.8 g; KH2PO4, 1.2 g; sodium citrate, 0.5 g; MgSO4 · 7H2O, 0.1 g; yeast extract, 2.0 g; sodium pyruvate, 5.5 g; and resazurin sodium salt, 1.0 mg [all per liter]) with or without chloroethenes and sodium fumarate as previously described (29).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Sources or references |
|---|---|---|
| Strains | ||
| Desulfitobacterium sp. strain Y51 | Wild type; PCE-dehalogenating, anaerobic bacterium | 29 |
| E. coli BL21(DE3) pLysS | F−ompT hsdSB(rB− mB−) gal dcm(DE3)/pLysS Cmr; lysogenic for λDE3; carries the T7 RNA polymerase under lacUV5 control | Novagen |
| Plasmids | ||
| pUC19 | Apr; 2.7 kb | Takara Shuzo |
| pET32b(+) | Apr; T7lac promoter; Trx · Tag, His · Tag, S · Tag; 5.9 kb | Novagen |
| pPCE10 | pUC19 with the PCR-amplified 1.0-kb DNA; pceA from strain Y51 partially included; 3.7 kb | This study |
| pPCE28 | pUC19 with the 2.8-kb SmaI/SphI fragment, containing pceA, of strain Y51; 5.5 kb | This study |
| pET32bPceA | pET32b(+) with the entire 1.7-kb pceA gene; 7.6 kb | This study |
Cmr, chloramphenicol resistance; Apr, ampicillin resistance.
Purification of PceA dehalogenase.
The Y51 cells were grown on MMYP medium with 5 mM sodium fumarate and 0.6 mM PCE. The cells were harvested in the late exponential growth phase, and the cell pellets were resuspended in 25 mM imidazole-HCl buffer (pH 7.5) containing 2.5 mM dithiothreitol, 10% (vol/vol) glycerol, 0.05 mM 4-(2-aminoethyl)-benzene-sulfofluoride (p-ABSF), and 0.01 mM RNase A. The cells were disrupted by a French press (Ohtake Works). Cell extracts were centrifuged at 12,000 × g at 4°C for 20 min. After the addition of 1.9% (wt/vol) streptomycin, the cell extracts were centrifuged as above. All steps were performed under anaerobic conditions as much as possible with nitrogen gas to avoid inactivation of enzyme by air. The resulting supernatant was applied to a hydroxyapatite column (3.0 by 20 cm; Bio-Rad Laboratories) preequilibrated with 20 mM potassium phosphate buffer (KP) (pH 7.5) containing 2.5 mM dithiothreitol, 10% (vol/vol) glycerol, and 0.05 mM p-ABSF. The column was eluted with a linear gradient from 20 to 150 mM KP, and a fraction showing dehalogenation activity was eluted with approximately 80 mM KP. The fraction containing the dehalogenase activity was applied to a butyl-Toyopearl 650 M column (1.5 by 20 cm; Tosoh) preequilibrated with 1.0 M ammonium sulfate in 25 mM imidazole-HCl buffer. The column was eluted in stepwise fashion with the same buffer containing 0.5, 0.1, and 0 M ammonium sulfate. The fraction having the highest dehalogenase activity was applied to a chromatofocusing column (1.5 by 20 cm; Amersham Biosciences) preequilibrated with 25 mM imidazole-HCl buffer (pH 7.5). The dehalogenase was eluted at approximately pH 5.8 during the increasing linear gradient from pH 7.4 to 4.5 with Polybuffer74 (Amersham Biosciences).
Analytical methods.
The dehalogenase activities for PCE, TCE, cis-DCE, and various chloroalkanes were assayed as previously described (29). The reaction mixture (1.0 ml in a 6-ml serum vial) was as follows: the enzyme fraction, 25 mM imidazole-HCl buffer (pH 7.5), 3.0 mM methyl viologen, 2.5 mM dithiothreitol, and 20 mM sodium pyruvate. Methyl viologen was reduced by 2 mM titanium (III) citrate solution. The vials were sealed with butyl rubber stoppers and then crimped. The substrates were added at a concentration of 0.6 mM.
The pH dependence of the PceA dehalogenase activity was measured. Sodium acetate buffer was used in the range from pH 4.4 to 5.4, morpholinepropanesulfonic acid buffer was used from pH 5.2 to pH 6.5, and Tris-HCl buffer was used from pH 6.5 to pH 9.0. The temperature dependence of the enzyme was determined by incubation of the PceA dehalogenase (5.0 μg · ml−1 in imidazole-HCl buffer) at a temperature ranging from 16 to 50°C. The oxygen sensitivity was determined by stirring the PceA dehalogenase (5.0 μg · ml−1 in imidazole-HCl buffer) in air at 4°C.
The effect of propyl iodide on the PceA dehalogenase activity was determined under anoxic conditions in the cell extracts (8.0 μg · ml−1 in imidazole-HCl buffer). Propyl iodide (25 μM) was added to the assay mixture. After a 1-h incubation in the dark, the mixture was exposed to the light (250-W lamp).
The molecular masses of the protein were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and gel filtration using TSKgel G3000SW (Tosoh). The protein concentration was determined by the method of Bradford (3a) with the Bio-Rad protein assay (Bio-Rad Laboratories) using bovine serum albumin as the standard.
Determination of amino acid sequences.
The N-terminal amino acid sequence of the PceA dehalogenase was determined with a protein sequencer (PPSQ-21; Shimadzu). The internal amino acid sequences were determined as follows. The purified PceA dehalogenase was treated with lysylendopeptidase (Wako Pure Chemical Industries). The obtained peptides were separated by high-pressure liquid chromatography with an ODP50 column (Asahi Chemical Industry), and the N-terminal amino acid sequences of the resultant six peptides were determined.
Cloning and DNA sequencing.
Isolation of the genomic DNA of strain Y51, plasmid DNA isolation, and other standard DNA manipulations were performed according to established procedures (1). The N-terminal amino acid sequence (ADIVAPITETSEFPYKVDAK) from the PceA dehalogenase of strain Y51 was used to design the degenerate oligonucleotide NTERM (5′-CIGA(C/T)ATIGTIGCICCIAT-3′). The internal peptide sequence (peptide14; VYTDLELAPDK) was used to design the degenerate oligonucleotide INT14 (5′-(C/T)TT(A/G)TCIGGIGCIA (A/G)(C/T)TC-3′). PCR was performed with a total volume of 50 μl, which contained the reaction buffer (Promega), 500 pmol of each primer, a 0.2 mM concentration of each deoxynucleotide triphosphate, 2.0 mM MgCl2, 2 U of Taq DNA polymerase (Promega), and 1.0 μg of genomic DNA of strain Y51 as a DNA template. Amplification of the PceA dehalogenase gene by PCR was carried out for 30 cycles under the following conditions: a preheating step of 94°C for 5 min; 30 cycles of 94°C for 1 min (denaturation), 46°C for 1.5 min (primer annealing), and 72°C for 2 min (primer extension); and a final extension step of 72°C for 10 min. The PCR-amplified 1.0-kb DNA fragment was cloned into SmaI-digested pUC19 (Toyobo). The resulting plasmid, named pPCE10, was sequenced with a Thermo Sequence kit (Amersham Biosciences) and DNA sequencer 4000L (Li-Cor).
The genomic DNA was digested with several restriction endonucleases. The 2.5- to 3.0-kb SmaI/SphI fragments were isolated from the agarose gels, ligated into the SmaI/SphI-digested pUC19 vector, and transformed into Escherichia coli JM109 to construct the genomic library of strain Y51. Positive clones were screened for the genomic library, using the 1.0-kb PCR product as the probe. Computer-assisted DNA and protein sequence analyses, alignments, and hydropathic plots were performed with the software package Genetyx-Mac (Software Development). Searches for sequence homology were done with BLASTP.
Expression of the PceA dehalogenase.
First we failed to express pceA and pceB using vector pBluescript II KS+ (Stratagene) in E. coli JM109. Therefore, we used the pET system (Novagen) as follows. The pceA gene of strain Y51 was amplified from pPCE28 by PCR using the following primers: as a forward primer, PCEAF, 5′-GGCGGGGATCCAATGGGAGAAATCAAC-3′, where the BamHI site is underlined and the start codon is in bold type; as a reverse primer, PCEAR, 5′-GGCGGGTCGACTTGTTTTATAGACTCAG-3′, where the SalI site is underlined. The amplification of pceA was carried out for 25 cycles under the conditions described above. The PCR product (a 1.7-kb fragment) was digested with BamHI and SalI and ligated into BamHI/SalI-digested pET32b(+) (Novagen), which contains an isopropyl-d-thiogalactopyranoside-inducible T7 promoter. The pceA gene product was designed to be fused toTrx (thioredoxin protein) · Tag, S · Tag, and His · Tag at the 5′ end and His · Tag at the 3′ end. The resulting plasmid, pET32b-pceA, was introduced into E. coli BL21(DE3)/pLysS (Novagen).
Immunoblot analysis.
The expression of the pceA gene in E. coli BL21(DE3)/pLysS and purification of the PceA fusion protein were carried out according to the method recommended by the supplier (Novagen). The PceA dehalogenase fused with Trx · Tag, S · Tag, and His · Tag purified by SDS-PAGE was subjected to an electroeluter (Bio-Rad Laboratories). The purified PceA dehalogenase fusion protein was used to raise anti-PceA antibody. Western blot analyses using the anti-PceA antibody were carried out to measure the production of PceA. The same antibody was also used to detect the localization of PceA in strain Y51 according to the method described by Imajoh-Ohmi et al. (12).
Fractionation of cell components.
The preparations of the periplasmic fraction, cytoplasm, inner membrane, and outer membrane were done according to the method of Grahn et al. (8).
Nucleotide sequence accession number.
The DNA sequence determined in this study was deposited in GenBank under accession no. AB070709.
RESULTS
Induction of PceA dehalogenase activity.
The growth of strain Y51 reached an optical density of 0.06 at 660 nm in MMYP medium (control). When PCE, TCE, or sodium fumarate was added to MMYP medium at a concentration of 0.6 mM, the growth reached to 0.1, 0.21, and 0.34, respectively. On the other hand, the growth was 0.02 when 0.6 mM cis-DCE was added. These results indicated that PCE, TCE, and sodium fumarate serve as energy-generating electron acceptors for cell growth but cis-DCE does not.
We examined the PceA dehalogenase activities for the cell extracts using 0.6 mM PCE as the substrate. The enzyme activity was 4.7 ± 0.4 nmol · min−1 · mg of protein−1 in the MMYP medium-grown cells (control). Addition of PCE, TCE, and sodium fumarate resulted in enhanced PceA dehalogenase activities as follows: 44.3 ± 3.9 nmol · min−1 · mg of protein−1 for PCE, 130.9 ± 10.1 nmol · min−1 · mg of protein−1 for TCE, and 13.8 ± 4.3 nmol · min−1 · mg of protein−1 for sodium fumarate. On the other hand, addition of cis-DCE reduced the enzyme activity to 1.0 ± 0.3 nmol · min−1 · mg of protein−1. The enhanced or decreased enzyme production was also demonstrated using anti-PceA antibody (Fig. 1).
FIG. 1.
Effects of various chloroethenes on production of PceA dehalogenase. Each chloroethene was added to the culture medium at a concentration of 0.6 mM. Western blot analyses using anti-PceA antibody were done for the crude extracts. Lane 1, no addition (control); lane2, cis-DCE; lane 3, TCE; lane 4, PCE; lane 5, sodium fumarate.
Purification and some properties of PceA dehalogenase.
In three chromatography steps described in Materials and Methods, the PceA dehalogenase was purified to 2.55-fold with a yield of 0.2%, as summarized in Table 2. The specific activity of the purified PceA dehalogenase toward PCE was 113.6 nmol · min−1 · mg of protein−1 at 37°C and pH 7.5. The enzyme was oxygen sensitive and lost approximately 50% of its activity during incubation and stirring for 330 min at 4°C in the presence of air. The dehalogenation activity was observed at temperatures between 25 and 37°C. The highest dehalogenation activity was observed at 37°C, but the activity was very low at 42°C (8.1 nmol · min−1 · mg of protein−1). The pH optimum was between 7.0 and 7.5. At pH 6.0 or 9.5, the dehalogenase activities were extremely low (7.5 and 2.3 nmol · min−1 · mg of protein−1, respectively).
TABLE 2.
Purification of PceA dehalogenase from Desulfitobacterium sp. strain Y51
| Purification step | Protein (mg) | Total activity (Ua) | Sp act (Ua/mg of protein) | Yield (%) | Purifica- tion factor (fold) |
|---|---|---|---|---|---|
| Crude extracts | 291.06 | 12992.92 | 44.64 | 100.0 | 1.00 |
| Hydroxyapatite | 85.44 | 4838.46 | 56.63 | 37.2 | 1.27 |
| Butyl-Toyopearl | 5.44 | 377.70 | 69.43 | 2.9 | 1.56 |
| Chromatofocusing | 0.26 | 29.54 | 113.62 | 0.2 | 2.55 |
One nanomole of PCE dehalogenated per min (0.6 mM PCE initially, pH 7.5, 37°C).
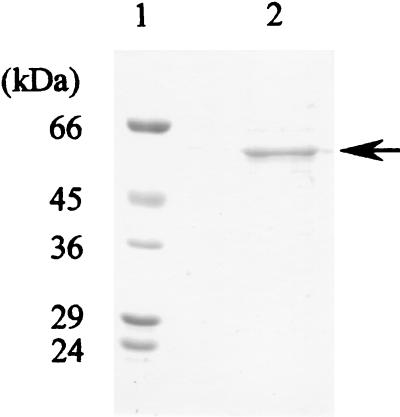
SDS-PAGE of the purified PceA dehalogenase revealed a single protein band (Fig. 2). The molecular mass of the enzyme was estimated to be approximately 58 kDa. Gel filtration on TSK gel G3000SW indicated that the molecular mass of the native PceA dehalogenase is about 67 kDa.
FIG. 2.
SDS-PAGE with the PceA dehalogenase from strain Y51. Proteins were stained with silver. Lane 1, molecular mass markers. The standard proteins were bovine serum albumin (66 kDa), ovalbumin (45 kDa), glyceraldehyde-3-phosphate dehydrogenase (36 kDa), carbonic anhydrase (29 kDa), and trypsinogen (24 kDa); lane 2, PceA dehalogenase (2 μg). The arrow indicates PceA dehalogenase, with a molecular mass of ca. 58 kDa.
The kinetic parameters were measured for various substrates, using the purified enzyme. The assay mixture contained 4.6 μg of enzyme and various concentrations (from 100 to 1,000 μM) of substrates. The apparent Km values for PCE and TCE were determined to be 105.7 ± 24.8 and 535.3 ± 47.8 μM, respectively. The corresponding Vmax values were 164.4 ± 27.2 nmol · min−1 · mg of protein−1 for PCE and 811.3 ± 39.0 nmol · min−1 · mg of protein−1 for TCE. On the other hand, 1,1-DCE, cis-DCE, and trans-1,2-DCE were not the substrates for this enzyme. Besides PCE and TCE, the same enzyme was also capable of dehalogenating various chloroethanes. Hexachloroethane was dehalogenated to cis-DCE via PCE and TCE. Pentachloroethane was dehalogenated to cis-DCE via TCE. 1,1,2,2-Tetrachloroethane was dehalogenated to cis-DCE. 1,1,1,2-Tetrachloroethane was dehalogenated to 1,1-DCE. These results indicate that at least two halogen atoms are reductively removed from the polychloroethanes to form chloroethenes. On the other hand, trichloroethane isomers and chlorobenzoate isomers were not attacked. The Km and Vmax values for these compounds are presented in Table 3. The Km and Vmax values for hexachloroethane were comparable to those for PCE. Thus, the PceA dehalogenase of strain Y51 exhibited a broad range of dehalogenation capabilities for not only PCE and TCE but also for various highly halogenated (from four to six chlorides) ethanes.
TABLE 3.
Substrate specificity profile of purified PceA dehalogenase from Desulfitobacterium sp. strain Y51
| Substrate | Km (μM) | Vmax (nmol/min/mg protein) |
|---|---|---|
| Chloroethenes | ||
| PCE | 105.7 ± 24.8 | 164.4 ± 27.2 |
| TCE | 535.3 ± 47.8 | 811.3 ± 39.0 |
| Chloroethanes | ||
| Hexachloroethane | 125.7 ± 14.6 | 148.7 ± 10.4 |
| Pentachloroethane | 619.9 ± 37.6 | 876.2 ± 43.4 |
| 1,1,2,2-Tetrachloroethane | 336.5 ± 35.3 | 42.4 ± 7.2 |
| 1,1,1,2-Tetrachloroethane | 785.0 ± 71.4 | 772.9 ± 59.7 |
Corrinoid enzymes are known to be inactivated by alkyl halides such as propyl iodide and to be reactivated by light. Also, sulfite is known to bind and form sulfito-cobalamin. In the presence of propyl iodide (25 mM), the PceA dehalogenase activity was decreased to 13%. When the mixture was then exposed to light, the activity was restored to 59% after 6 h and 90% after 24 h. The addition of 20 mM sulfite resulted in inhibition of the dehalogenase activity to 53%. These results suggest that a corrinoid is involved in the PceA dehalogenase of strain Y51, similar to the other PCE/TCE dehalogenases.
Cloning of the PceA dehalogenase gene.
The N terminus of the purified PceA dehalogenase of strain Y51 was determined to be ADIVAPITETSEFPYKVDAK. The enzyme was digested with lysylendopeptidase, and the amino acid sequences of the six resulting peptides were determined as follows: peptide10, TFDPEANK; peptide11, YAGFK; peptide12, FHYDDVSK; peptide14, VYTDLELAPDK; peptide17, DLPTLNAERLGIK; and peptide19, LPWDLPK. For PCR amplification with the Y51 genomic DNA as the template, a pair of degenerate oligonucleotides, NTERM and INT14, were used to obtain a 1.0-kb PCR product. Three of 3,700 clones from the genomic DNA library of strain Y51 hybridized with this 1.0 kb-DNA as a probe. One such positive clone, pPCE28, contained a SmaI/SphI insert of about 2.8-kb DNA.
Sequence analysis of the PceA dehalogenase gene.
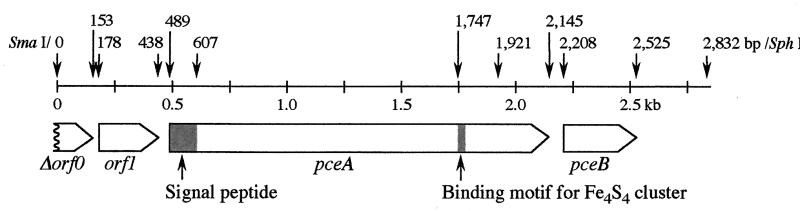
The nucleotide sequence of the 2,832-bp SmaI/SphI fragment from pPCE28 revealed three ORFs (GenBank accession no. AB070709). An overview of the DNA region comprising the genes for the PceA dehalogenase is given in Fig. 3. The pceA gene codes for a protein containing the N terminus and all six internal peptides of the purified PceA dehalogenase. The deduced amino acid sequence of this ORF started 39 amino acids upstream of the N terminus of the PceA dehalogenase isolated from strain Y51. In this region the Tat (twin arginine translocation) consensus sequence RRXFXK (2, 3, 25) was detected. The molecular masses of the deduced 551-amino-acid protein (nonprocessed) and of the processed 512-amino-acid protein were calculated to be 61,283 and 57,444 Da, respectively.
FIG. 3.
Physical map of the pceA gene region of Desulfitobacterium sp. strain Y51.
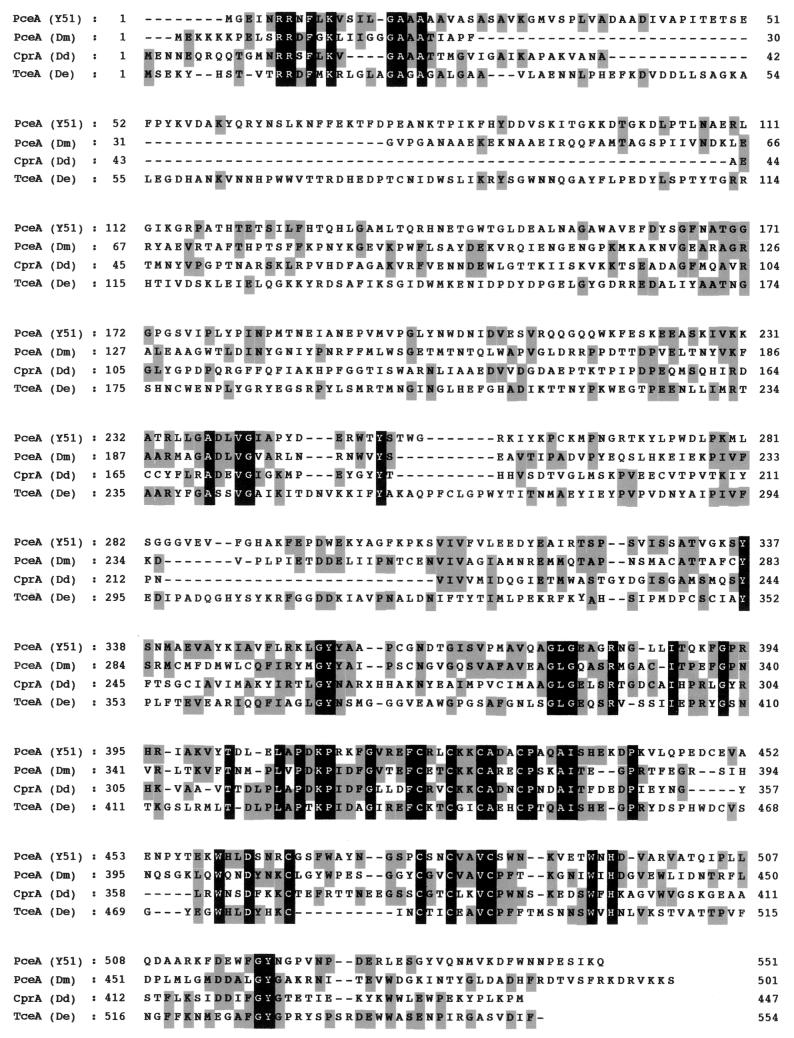
The consensus sequence similar to that for the Fe4S4 ferredoxin-like cluster (CXXCXXCXXXCP) (4) was identified from amino acids 420 to 431 (Fig. 4). In addition, a stretch (GXXCXXCXXXCS) was also identified from amino acids 475 to 486 (Fig. 4). The difference in the two consensus sequences is a glycine instead of a cysteine at amino acid position 475 and a serine instead of a proline at amino acid position 486. A consensus sequence for the binding of a corrinoid (DXHXXG) (15) could not be detected. The primary sequence alignment of the PceA of strain Y51, PceA from D. multivorans, ortho-chlorophenol reductase (CprA) from D. dehalogenans, and trichloroethene dehalogenase (TceA) from D. ethenogenes 195 is shown in Fig. 4. The identities of the amino acid sequence of the PceA of strain Y51 with the PceA of D. multivorans, the CprA of D. dehalogenans, and the TceA of D. ethenogenes 195 were 28.0, 26.3, and 23.9%, respectively.
FIG. 4.
Primary sequence alignment for the PceA dehalogenase from strain Y51 and other reductive dehalogenases. The alignment was performed using BLASTP and the program Genetyx-Mac 10.1. The gray boxes mark identical amino acid residues. Residues highlighted in black indicate the conserved sequences. PceA (Y51), PceA dehalogenase from Desulfitobacteium sp. strain Y51 (GenBank accession no. AB070709); PceA (Dm), PCE dehalogenase from D. multivorans (GenBank accession no. AF022812); CprA (Dd), ortho-chlorophenol reductase from D. dehalogenans (GenBank accession no. AF115542); TceA (De), TCE dehalogenase form D. ethenogenes 195 (GenBank accession no. AF228507).
Downstream of pceA, one ORF (pceB, 315 bp) was detected (Fig. 3). The 105-amino-acid protein encoded by this gene has a calculated molecular mass of 11,843 Da. Three hydrophobic regions in PceB were detected by the hydropathic plot, indicating the presence of three membrane-spanning helices. Upstream of the pceA gene, one ORF (ORF1) with no significant sequence similarities to genes in the databases was detected.
By BLASTP analysis upstream of Orf1 (positions 1 to 153; a partial orf0 gene product), an amino acid sequence similar to the C-terminal region of the transposases from IS1601 of Mycobacterium avium, IS406 of Burkholderia cepacia, and IS1081 of Mycobacterium tuberculosis was found. The identities of the amino acid sequence between the partial C-terminal sequence of Orf0 of strain Y51 and the corresponding regions of the transposases from IS1601, IS406, and IS1081 were 26.0, 31.6, and 26.0%, respectively.
Expression of the PceA dehalogenase genes in E. coli.
After induction of E. coli carrying pET32b-pceA with 1.0 mM isopropyl-β-d-thiogalactopyranoside, a PceA dehalogenase tagged with Trx, S, and (His)6 with a molecular mass of approximately 77 kDa was detected in the insoluble fraction of the cell extracts (not shown). The PceA dehalogenase fusion protein was verified by N-terminal and internal amino acid sequencing. However, dehalogenase activity for PCE could not be detected in the cell extracts of the recombinant E. coli irrespective of the conditions of aerobic and anaerobic growth. Denaturation followed by refolding of the PceA dehalogenase fusion protein failed to recover the dehalogenase activity. We used the solubilized fusion protein to raise antibody.
Localization of PceA in Desulfitobacterium sp. strain Y51.
Immunoblotting with the anti-PceA antibody demonstrated that a protein with a molecular mass of 58 kDa corresponding to mature PceA was present in the periplasmic fraction (Fig. 5). Proteins with molecular masses of 61 and 58 kDa were detected in the cytoplasmic fraction. The 61- and 58-kDa proteins were considered to be the unprocessed and processed PceAs, respectively. Since the periplasmic fraction could not be completely removed from the cytoplasmic fraction, the 58-kDa protein in the cytoplasmic fraction could be the processed protein from the periplasmic fraction. In the inner and outer membranes, protein cross-reacted with the anti-PceA antibody could not be detected. These results demonstrated that the mature PceA is located in the periplasm of the strain Y51 cell.
FIG. 5.
Localization of the PceA dehalogenase in the cell of strain Y51 by Western blot analysis with anti-PceA antibody. Lane 1, cytoplasmic fraction; lane 2, inner membrane fraction; lane 3, periplasmic fraction; lane 4, outer membrane fraction.
DISCUSSION
In this study we report molecular characterization of the PceA dehalogenase of Desulfitobacterium sp. strain Y51. The PceA dehalogenase activity of strain Y51 was highly enhanced when PCE or TCE was added to the culture medium; in particular the addition of TCE at a concentration of 0.6 mM led to 25-fold-higher enzyme activity compared to the control (no addition). These data were confirmed at the protein level by Western blot analysis (Fig. 1). The addition of PCE or TCE to the culture medium also greatly enhanced the growth of strain Y51, indicating that these two compounds serve as terminal electron acceptors for dehalorespiration. The addition of sodium fumarate led to good growth of strain Y51, and the induction of PceA dehalogenase. However, the extent of induction was much lower compared with TCE and PCE. On the other hand, the addition of cis-DCE reduced not only the growth of strain Y51 but also the PceA dehalogenase activity. Thus, cis-DCE seems to be toxic to the Y51 cells, although the reason remains to be elucidated.
The enzyme exhibits some features that are functionally similar to those of other enzymes. The similarities of the amino acid sequence of the Y51 enzyme and others are between 23 and 28% (Fig. 4). These values are significantly low when considering the similar function of the enzymes. With the exception of the signal sequence and the binding sequence of the Fe4S4 cluster, no significant similarities were found with the reductive dehalogenases between strain Y51, D. multivorans (23), D. ethenogenes (14), and D. dehalogenans (30)(Fig. 4). However, the N-terminal amino acid sequence of the PceA dehalogenase of strain Y51 showed a high degree of similarity to that of strain PCE-S (20), where 19 of 20 amino acids were identical, although the entire sequence of the PCE dehalogenase of strain PCE-S has not been published yet.
The half-life of the strain Y51 PceA dehalogenase was 330 min at 4°C in the presence of air. The half-life values of the other dehalogenases were reported to be 50 min (Desulfitobacterium sp. strain PCE-S [20]), 90 min (D. dehalogenans [30]), 120 min (D. multivorans [22]), and 280 min (D. restrictus [26]). Thus, the PceA dehalogenase of strain Y51 is more resistant to oxygen than reported similar enzymes.
Another feature of the PceA dehalogenase of strain Y51 is wide substrate specificity for various chlorinated ethanes besides PCE and TCE. The mode of dehalogenation by the PceA dehalogenase of strain Y51 for polychloroethanes was the reductive elimination of two halide substituents to form an alkene, resulting in the formation of cis-DCE or 1,1-DCE. It is interesting that 1,1,1,2-tetrachoroethane can be converted to 1,1-DCE, but the 1,1,1-tri- and 1,1,2-trichloroethanes cannot be attacked at all (Table 3). The difference between these three compounds is only one chloride substitution. Thus, the substrate specificity of the PceA dehalogenase of strain Y51 is relaxed toward highly chlorinated alkanes such as the tetra-, penta-, and hexachloroethanes but rather strict for non-highly chlorinated ones, as seen for the cis-DCE and chloroalkanes with three chlorines. The PCE dehalogenase from D. multivorans showed limited substrate ranges for PCE, TCE, and tetraiodoethene but not for chloroethanes and chloromethanes (22). The PCE dehalogenase from D. ethenogenes showed dehalogenation only toward PCE (13). On the other hand, the TCE dehalogenase from the same strain catalyzed a wide range of dehalogenation for TCE, the DCE isomers, and haloalkanes with two and three chlorines except for 1,1,1-trichloroethane (14). The PCE dehalogenase of C. bifermentans also exhibited dehalogenation for DCE isomers as well as for PCE and TCE (5, 24).
Using the polyclonal antibody raised against PceA, it was clearly demonstrated that the mature PceA dehalogenase is localized in the periplasm and that the unprocessed enzyme is localized in the cytoplasm (Fig. 5). These findings are in agreement with the feature of the amino acid sequence deduced from the pceA gene, where a hydrophobic stretch of 39 N-terminal amino acids (containing the consensus sequence RRXFXK) may act as a signal peptide. The rest of the 512 amino acids can be exported into the periplasm by the Tat pathway (2, 3, 25). The PCE dehalogenase of the gram-negative bacterium D. multivorans was purified from the soluble fraction, which suggests that the enzyme is localized in the cytoplasm (22). In contrast, the PCE dehalogenases of strain PCE-S (20), D. restrictus (27), and D. ethenogenes (13) were purified from the membrane fraction and were present as membrane-associated forms.
PceB coding for the putative integral membrane protein contained three membrane-spanning helices, as is the case for the proteins of the other strains. The PceB of D. multivorans (23) possesses two, and the CprB of D. dehalogenans (30) and TceB of D. ethenogenes (14) possess three membrane-spanning helices. It is suggested that these proteins act as a membrane anchor for the reductive dehalogenases. However, no significant similarities were revealed in the amino acid sequences between the PceB of strain Y51 and the above-mentioned PceB, CprB, and TceB.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates, Inc., and John Wiley & Sons, Inc., New York, N.Y.
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bruschi, M., and F. Guerlesquin. 1988. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol. Rev. 54:155-176. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. C., B. C. Okeke, M. Hatsu, and K. Takamizawa. 2001. In vitro dehalogenation of tetrachloroethylene (PCE) by cell-free extracts of Clostridium bifermentans DPH-1. Bioresour. Technol. 78:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Damborsky, J. 1999. Tetrachloroethene-dehalogenating bacteria. Folia Microbiol. 44:247-262. [DOI] [PubMed] [Google Scholar]
- 7.Fetzner, S., and F. Lingens. 1994. Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications. Microbiol. Rev. 58:641-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holliger, C., and W. Schumacher. 1994. Reductive dehalogenation as a respiratory process. Antonie van Leeuwenhoek 66:239-246. [DOI] [PubMed] [Google Scholar]
- 10.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Vazquez, N. Weiss, and A. J. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 11.Holliger, C., G. Wohlfarth, and G. Diekert. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 12.Imajoh-Ohmi, S., K. Tsujimura, and M. Inagaki. 1994. Koupeputidokoutaijikken purotokohru, p. 47-118. Shujunsha, Tokyo, Japan. (In Japanese.)
- 13.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh, E. N., and D. E. Holloway. 1992. Cloning and sequencing of glutamate mutase component S from Clostridium tetanomorphum. FEBS Lett. 310:167-170. [DOI] [PubMed] [Google Scholar]
- 16.Maymo-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 17.Maymo-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1, 2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, E., G. Wohlfarth, and G. Diekert. 1996. Studies on tetrachloroethene respiration in Dehalospirillum multivorans. Arch. Microbiol. 166:379-387. [DOI] [PubMed] [Google Scholar]
- 19.Miller, E., G. Wohlfarth, and G. Diekert. 1997. Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 168:513-519. [DOI] [PubMed] [Google Scholar]
- 20.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 21.Neumann, A., H. Scholz-Muramatsu, and G. Diekert. 1994. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch. Microbiol. 162:295-301. [DOI] [PubMed] [Google Scholar]
- 22.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 23.Neumann, A., G. Wohlfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 25.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher, W., and C. Holliger. 1996. The proton/electron ratio of the menaquinone-dependent electron transport from dihydrogen to tetrachloroethene in “Dehalobacter restrictus”. J. Bacteriol. 178:2328-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher, W., C. Holliger, A. J. Zehnder, and W. R. Hagen. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409:421-425. [DOI] [PubMed] [Google Scholar]
- 28.Smidt, H., M. van Leest, J. van Der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suyama, A., R. Iwakiri, K. Kai, T. Tokunaga, N. Sera, and K. Furukawa. 2001. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65:1474-1481. [DOI] [PubMed] [Google Scholar]
- 30.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 31.Wohlfarth, G., and G. Diekert. 1997. Anaerobic dehalogenases. Curr. Opin. Biotechnol. 8:290-295. [DOI] [PubMed] [Google Scholar]