Abstract
Caenorhabditis elegans germline cells are maintained in an undifferentiated and mitotically dividing state by Notch signaling and the FBF (for fem-3 binding factor) RNA-binding protein. Here, we report that the LIP-1 phosphatase, a proposed homolog of mitogen-activated protein (MAP) kinase phosphatases, is required for the normal extent of germline proliferation, and that lip-1 controls germline proliferation by regulating MAP kinase activity. In wild-type germ lines, LIP-1 protein is present in the proximal third of the mitotic region, consistent with its effect on germline proliferation. We provide evidence that lip-1 expression in the germline mitotic region is controlled by a combination of GLP-1/Notch signaling and FBF repression. Unexpectedly, FBF controls the accumulation of lip-1 mRNA, and therefore is likely to control its stability or 3′-end formation. In a sensitized mutant background, LIP-1 can function as a pivotal regulator of the decision between proliferation and differentiation. The control of germline proliferation by LIP-1 has intriguing parallels with the control of stem cells and progenitor cells in vertebrates.
Keywords: LIP-1, Notch, phosphatase, PUF, stem cells
Introduction
During animal development, the precise regulation of proliferation and differentiation is critical for generation of spatially patterned and correctly sized tissues and organs. The control of stem cells is central to this process. Although it is well established that stem cells are controlled by signaling from a niche (Li and Xie, 2005), the regulators that act downstream of that signaling to control self-renewal or differentiation are poorly defined.
The Caenorhabditis elegans germ line provides a simple and well-defined system for analysis of stem cell controls (Crittenden et al, 2003). Figure 1A illustrates the organization of the adult germ line. Briefly, germ cells in the mitotic cell cycle are restricted to a ‘mitotic region' located at the distal end of the germline tissue. That mitotic region includes germline stem cells. As germ cells move proximally, they leave the mitotic cell cycle and enter the meiotic cell cycle. Early stages of meiotic prophase predominate in the transition zone, and later stages of meiosis and gametogenesis proceed in a spatially ordered fashion as germ cells move proximally.
Figure 1.
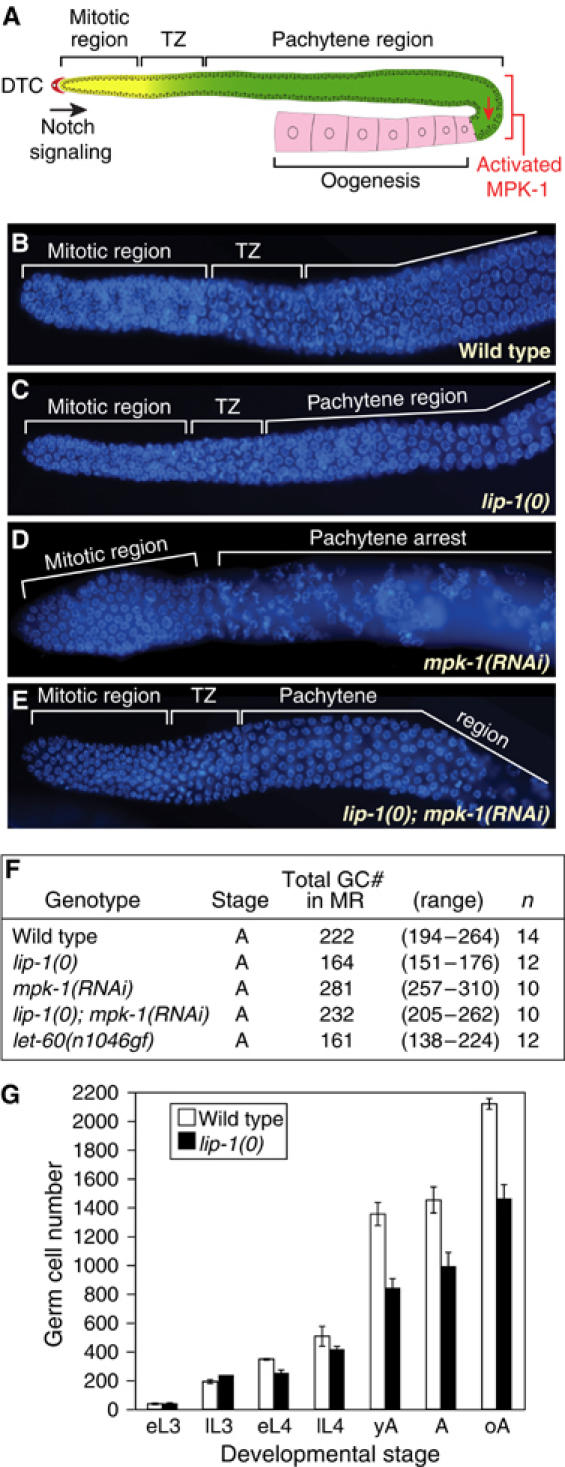
LIP-1 phosphatase promotes germline proliferation. (A) Schematic of adult hermaphrodite germ line. The somatic DTC and Notch signaling promote cell divisions in the distal mitotic region. As germ cells move proximally, they enter the ‘transition zone' (TZ), where most germ cells have entered early meiotic prophase (leptotene or zygotene); more proximally, they arrest in pachytene until stimulated by MAPK to exit pachytene and progress through meiosis. It should be noted that virtually all germ ‘cells' are part of a syncytium, including those in the mitotic region; however, mitotic cell cycles are not synchronized, suggesting ‘cell' autonomy. (B–E) Dissected adult germ lines of same stage (24 h past mid-L4) stained with DAPI to delineate stages of meiosis. Genotypes or treatment, as labeled. (F) Total number of germ cells in the mitotic region (GC# in MR). n, number of mitotic regions counted. (G) Total number of germ cells at specific stages of development in wild-type and lip-1(0) animals. L3, third larval stage; L4, fourth larval stage; e, early; l, late; yA, adult 12 h past L4; A, adult 24 h past L4; oA, adult 48 h or more past L4. Error bars were calculated from data of three independent experiments.
A single-celled somatic niche, called the distal tip cell (DTC), promotes germline proliferation during larval development and maintains germline stem cells in the adult (Kimble and White, 1981) (Figure 1A). This DTC employs the Notch signaling pathway to promote mitotic divisions in the distal germ line (Kimble and Simpson, 1997). Specifically, the GLP-1/Notch receptor receives the DTC signal and activates transcription by the LAG-1/CSL DNA-binding protein and the LAG-3 transcriptional coactivator (Crittenden et al, 2003). GLP-1/Notch signaling is restricted to the distal mitotic region in adults and maintains germline stem cells there (Crittenden et al, 1994). Recent evidence suggests that Notch signaling also acts in vertebrates to control stem and progenitor cells (Calvi et al, 2003; Fre et al, 2005). Therefore, the analysis of Notch target genes that control proliferation in the C. elegans germ line may provide insight into stem cell controls more broadly.
Within the germ line, the FBF (for fem-3 binding factor) RNA-binding protein is required for maintenance of germline stem cells (Crittenden et al, 2002). FBF is a collective term for two nearly identical RNA-binding proteins, FBF-1 and FBF-2, which belong to the PUF (for Pumilio and FBF) family of RNA regulators. PUF proteins bind specifically to regulatory elements in the 3′ untranslated region (3′UTR) of target mRNAs and repress their expression (Wickens et al, 2002). Furthermore, PUF proteins have been implicated broadly in stem cell controls during animal development (Wickens et al, 2002). FBF promotes mitosis by repressing a battery of mRNAs that encode regulators that drive germ cells into the meiotic cell cycle (Zhang et al, 1997; Crittenden et al, 2002; Eckmann et al, 2004; Thompson et al, 2005). The fbf-2 gene is a direct target of GLP-1/Notch signaling (Lamont et al, 2004). Therefore, Notch signaling promotes mitosis and the undifferentiated state, at least in part, by increasing fbf-2 expression.
The lip-1 (for lateral signaling-induced phosphatase) gene was initially identified as a direct target of LIN-12/Notch signaling in somatic tissues (Berset et al, 2001). The LIP-1 protein is homologous to phosphatases of the mitogen-activated protein kinase phosphatase (MKP) family, which are dual-specificity phosphatases that directly inhibit the activity of Ras/mitogen-activated protein kinase (MAPK) (Camps et al, 1998; Berset et al, 2001). Although LIP-1 has not yet been tested in vitro for direct MAPK inhibition, it acts upstream of MAPK as a negative regulator (Berset et al, 2001; Hajnal and Berset, 2002). In C. elegans, the primary MAPK is called MPK-1 (Lackner and Kim, 1998). During vulval development, LIN-12/Notch signaling activates lip-1 and thereby inactivates MPK-1 to induce secondary vulval fates (Berset et al, 2001). Given this vulval circuitry and the importance of Notch signaling for germline development, one might suspect that lip-1 would also regulate germline proliferation. However, lip-1 null mutants have no dramatic defect in germline proliferation, but instead display defects in progression through meiosis (Hajnal and Berset, 2002). The role of LIP-1 in meiotic progression is consistent with its role as an inhibitor of MAPK activity, because MPK-1 is required for progression from pachytene to diplotene and also controls oocyte maturation (Church et al, 1995; Lackner and Kim, 1998; Miller et al, 2001; Page et al, 2001).
In this paper, we demonstrate that LIP-1 controls the extent of germline proliferation rather than proliferation per se: lip-1 null mutants have fewer germ cells than wild type, but do have proliferating germ cells. Furthermore, LIP-1 protein is present in the mitotic region. Several lines of evidence support the idea that lip-1 is activated by GLP-1/Notch signaling, but repressed in the distal-most germ line by FBF. We suggest that LIP-1 promotes mitosis in the proximal part of the germline mitotic region and thereby extends mitotic divisions and delays the transition from the mitotic cell cycle into the meiotic cell cycle.
Results
lip-1 is required for the normal extent of germline proliferation
To ask if lip-1 null mutants affect germline proliferation, we first compared the number of germ cells present in the adult mitotic region of wild-type and lip-1(0) germ lines. The mitotic region extends from the distal tip of the germ line tissue to the distal border of the transition zone (Figure 1A); in 4′, 6-diamidino-2-phenylindole (DAPI)-stained germ lines, transition zone nuclei are easily distinguished by their crescent-shaped chromatin (Figure 1B). The wild-type mitotic region possesses ∼225 cells (Figures 1B and F) (Eckmann et al, 2004; Hansen et al, 2004), but in lip-1(0) mutants, the mitotic region contained only ∼165 cells (Figures 1C and F). Therefore, lip-1 is required to maintain the normal number of germ cells within the mitotic region.
We also compared the total number of germ cells in staged wild-type animals and lip-1 null mutants during development. In larvae, germ cell numbers were similar in the two strains, but during adulthood, lip-1(0) mutants had fewer total germ cells than wild type (Figure 1G). Therefore, LIP-1 does not control germline proliferation per se, but instead it is required for the robust proliferation typical of the adult germ line.
LIP-1 and MPK-1 have opposite effects on germline proliferation
To ask if the lip-1 defect in mitotic region size might depend on MAPK activity, we used RNA interference (RNAi) to achieve a partial loss of mpk-1 function. These mpk-1(RNAi) germ lines contained both mitotic and pachytene regions in the correct spatial order, but they had no transition zone (Figure 1D). Most relevant to this work, the mpk-1 mitotic region consistently possessed more germ cells than normal (Figure 1F). The simplest explanation is that MAPK functions in wild-type germ lines to reduce the number of cells in the mitotic region and establish a transition zone. Consistent with this idea, the germline mitotic region of let-60 ras gain-of-function mutants is smaller than normal (Figure 1F).
To ask if lip-1 might affect germline proliferation by inhibiting MAPK activity, we used RNAi to deplete mpk-1 in lip-1 null mutants and examined their germ lines. The mitotic region in lip-1(0); mpk-1(RNAi) germ lines was restored to the wild-type cell number and a typical transition zone was reinstated (Figures 1E and F). This reciprocal suppression is best explained by mpk-1 RNAi reducing rather than eliminating mpk-1 expression, a phenomenon typical of RNAi experiments. By this reasoning, the double mutant co-suppression is expected: elimination of LIP-1 activity (the effect of the lip-1 null mutant) permits residual mpk-1 mRNA to make some active enzyme, while reduction in mpk-1 expression (the effect of mpk-1(RNAi)) ameliorates damage caused by removing the LIP-1 inhibitor. Consistent with this idea, activated MAPK is not present in the wild-type distal germ line, but appears to extend abnormally into the distal germ line of lip-1 null mutants (Hajnal and Berset, 2002; Figure 6B). We conclude that LIP-1 normally promotes germline proliferation by inhibiting MPK-1.
Figure 6.
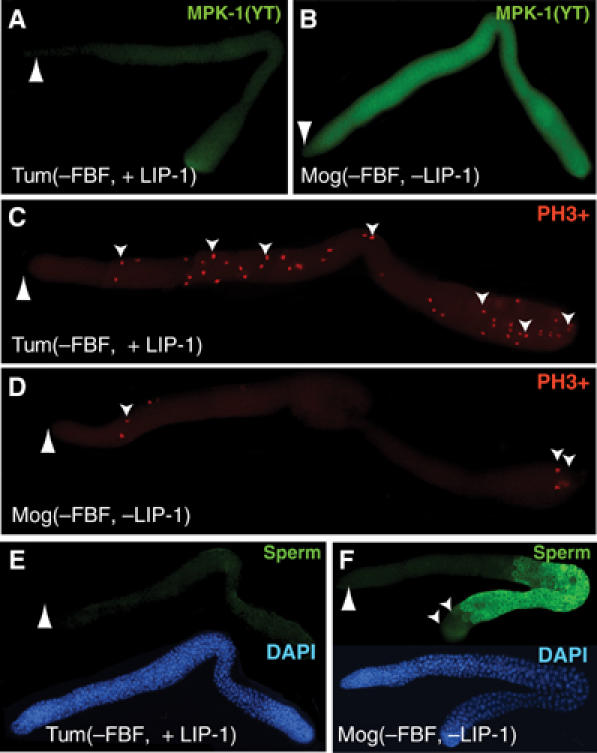
LIP-1 is a pivotal regulator of proliferation when tested in a sensitized background. (A–F) Adult germ lines dissected from either quadruple mutants that possess a wild-type copy of lip-1 (Tum(−FBF), fbf-1 fbf-2 gld-3 nos-3) or quintuple mutants that carry the lip-1 null mutant (Mog(−FBF, -LIP-1), fbf-1 fbf-2 gld-3 nos-3; lip-1(0)). Large arrowhead, distal end. (A, B) Staining with antibody specific for activated form of MAPK(YT). Images treated identically so that MAPK(YT) levels can be compared. (C, D) Staining with antibody to phosphohistone H3 to visualize mitotic germ cells. Small arrowheads indicate some of the mitotic germ cells. (E, F) Upper image, staining with sperm-specific antibody (green); lower image, same germ line stained with DAPI.
Chromatin immunoprecipitation of lip-1 promoter with LAG-3 antibodies is germline dependent
A previous study showed that the lip-1 gene is a direct target of LIN-12/Notch signaling during vulval development (Berset et al, 2001). To investigate whether lip-1 is also activated by GLP-1/Notch signaling in the germ line, we carried out chromatin immunoprecipitation (ChIP) assays (Figure 2). Notch signaling is mediated by a ternary complex containing the LAG-3 transcriptional coactivator (Petcherski and Kimble, 2000). To detect activated Notch signaling-specific transcriptional complexes, we raised a rat polyclonal antibody to a LAG-3-specific peptide. The specificity of the α-LAG-3 antibody was confirmed using lag-3(RNAi) (see Materials and methods). We immunoprecipitated LAG-3 from wild-type animals or from glp-1 mutants that had no germ line, and then performed PCR to assay the presence of specific DNA sequences in the precipitate. We tested the lip-1 promoter region and as controls tested the lip-1 coding region or the promoter of a different gene (Figure 2A). The lip-1 promoter region co-immunoprecipitated with LAG-3 antibodies from wild-type adults, but did not immunoprecipitate from glp-1 adults that had no germ line (Figure 2B). The lack of a signal in glp-1 adults was expected, because all known Notch signaling between somatic cells occurs during embryogenesis and larval development—not during adulthood. The lip-1 promoter failed to immunoprecipitate with preimmune serum, and control regions failed to immunoprecipitate with LAG-3 antibodies (Figure 2B). We conclude that LAG-3 selectively associates with the lip-1 promoter, and that its association is dependent on the presence of the germ line. The simplest explanation is that lip-1 is a direct target of GLP-1/Notch signaling in the adult C. elegans germ line.
Figure 2.
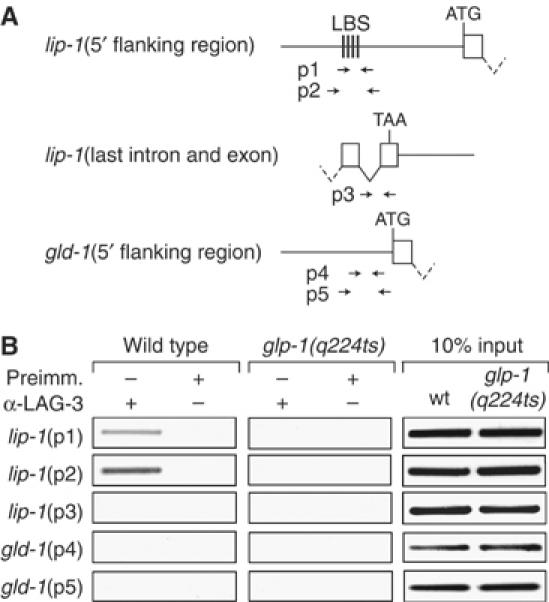
Germline-dependent association of lip-1 promoter with LAG-3, a transcriptional coactivator specific to Notch signaling. (A) Genome regions assayed for ChIP. Straight line, intergenic DNA; box, exon; V-shaped line, intron; ATG, initiation codon; TAA, termination codon; LBS, LAG-1-binding site; arrow sets below, positions of primers used to assay specific region. (B) Representative example of PCR analysis of DNA from ChIPs performed with affinity-purified LAG-3 antibodies. All animals were grown at 25°C, the restrictive temperature for glp-1(q224ts). The lip-1 5′ flanking region is specifically immunoprecipitated in LAG-3 ChIP from wild-type animals, which possess about 2000 germ cells (left column). The lip-1 5′ flanking region is not immunoprecipitated from glp-1 adults, which possess essentially no germ cells (middle column). Input was the same from wild-type and glp-1 extracts (right column). Similar results were obtained in three additional independent experiments.
LIP-1 protein in the mitotic region
We next asked if lip-1 is expressed in mitotic germ cells. To this end, we generated a rat polyclonal antibody that specifically recognizes C. elegans LIP-1, and stained dissected adult germ lines with both the LIP-1 antibody and DAPI. As previously described (Hajnal and Berset, 2002), LIP-1 staining was concentrated in membrane-associated bright puncta, which were easily detectable throughout the pachytene region (Figure 3A). We found similar LIP-1 puncta extending into the transition zone and the proximal-most germ cells of the mitotic region (Figure 3A). A closer examination revealed LIP-1 puncta extending distally into the proximal one-third of the mitotic region (Figure 3B), but these bright foci were absent from lip-1 null mutants (Figure 3C). We conclude that LIP-1 protein is present at a low level in the proximal mitotic region, but that it does not extend to the distal end. The mitotic region is defined by the presence of mitotically dividing germ cells (Crittenden et al, 1994; Hansen et al, 2004; Lamont et al, 2004). Therefore, the presence of LIP-1 in the proximal mitotic region is consistent with its role in controlling the extent of germline proliferation.
Figure 3.
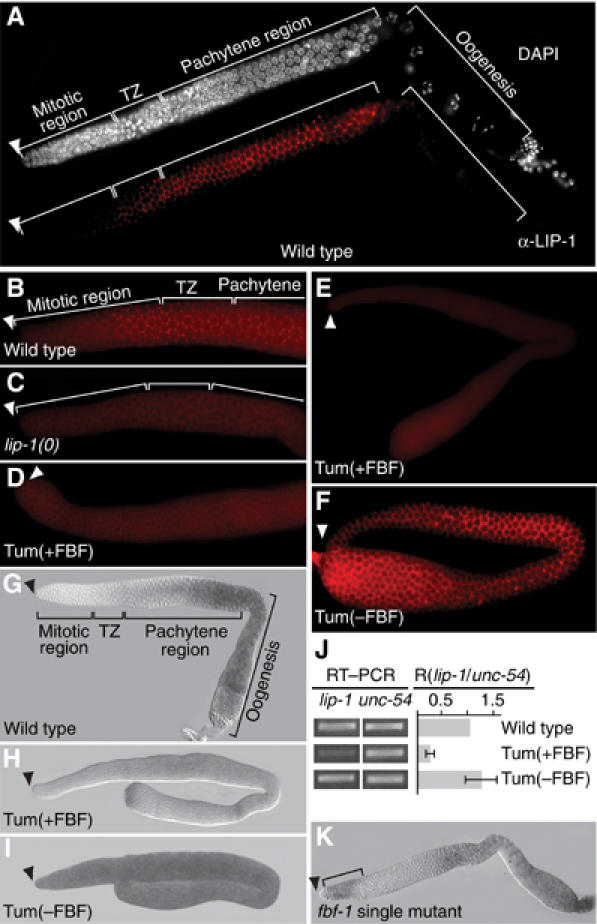
LIP-1 protein in the proximal mitotic region and FBF repression of lip-1 expression. (A–I, K) Adult germ lines dissected from animals of genotype, as labeled. Arrowhead, distal end; TZ, transition zone; A–F, staining with LIP-1-specific antibody; G–I, K, staining with antisense probe specific for lip-1 mRNA. Images in panels B–D were treated identically; same for panels E and F and for panels H and I. Tum(+FBF), gld-3 nos-3; Tum(−FBF), fbf-1 fbf-2 gld-3 nos-3. (A) LIP-1 protein in wild-type germ line. Upper image, DAPI staining used to assess nuclear morphology in the mitotic region, transition zone (TZ), and pachytene region; lower image, same germ line stained with LIP-1 antibody. LIP-1 is membrane associated and punctate, as shown previously (Hajnal and Berset, 2002). (B) LIP-1 protein in the mitotic region and transition zone of wild-type germ line. LIP-1 staining is membrane associated and punctate in proximal-most cells of the mitotic region and in transition zone (TZ). Blurry staining in cytoplasm is background (see C). (C) Background staining of LIP-1 antibody in germ line dissected from lip-1(zh15) deletion mutant. (D) Distal end of Tum(+FBF) germ line. Only background staining was detected (compare to C). (E) Complete Tum(+FBF) germ line. Staining with LIP-1 antibody is low relative to Tum(−FBF) and likely to be background. (F) Complete Tum(−FBF) germ line. LIP-1 staining is bright compared to Tum(+FBF). Also, LIP-1 is membrane associated and punctate throughout the germ line. (G) lip-1 mRNA in wild-type germ line. Distribution is similar to that of LIP-1 protein, but mRNA is not detected in the mitotic region, presumably owing to a lack of sensitivity. (H, I) In Tum(+FBF) germ lines, lip-1 mRNA level is lower than in Tum(−FBF). (J) Semiquantitative RT–PCR to compare abundance of lip-1 and unc-54 mRNAs in animals of genotype, as labeled. PCR cycles set within the linear range and amounts normalized to unc-54 product in wild type. (K) fbf-1 single mutant germ line. lip-1 mRNA is present in distal-most germ cells (bracket), which are being signaled by GLP-1/Notch signaling from the DTC.
FBF represses lip-1 expression
The lack of LIP-1 protein from the distal mitotic region seemed paradoxical. The somatic DTC promotes mitotic divisions by GLP-1/Notch signaling (Figure 1A), and one GLP-1/Notch target gene, fbf-2, is expressed in the distal-most germ line as expected (Lamont et al, 2004). As lip-1 also appears to be a direct target of GLP-1/Notch signaling (see above), we expected that lip-1 would also be expressed distally. One possible explanation is that lip-1 is repressed post-transcriptionally in the distal germ line. FBF-1 and FBF-2 are both localized to the mitotic region (Crittenden et al, 2002; Lamont et al, 2004), and therefore are in the right location to repress lip-1 expression.
To ask if FBF might repress lip-1 expression in mitotic germ cells, we decided to compare the abundance of LIP-1 protein in germ lines with and without FBF. An fbf-1 fbf-2 double mutant germ line does not have mitotic germ cells, but instead makes only sperm (Crittenden et al, 2002), and therefore is not useful for this experiment. To circumvent this problem and to analyze germ lines of similar cellular fate, we used tumorous germ lines. Specifically, we employed gld-3 nos-3 and fbf-1 fbf-2 gld-3 nos-3, two strains with germ lines that are composed largely of mitotic germ cells (Eckmann et al, 2004). For simplicity, we refer to gld-3 nos-3 as Tumorous plus FBF (Tum(+FBF)) and fbf-1 fbf-2 gld-3 nos-3 as Tumorous minus FBF (Tum(−FBF)). In Tum(+FBF) germ lines, LIP-1 was not detected above background (compare Figures 3C and D). However, much more LIP-1 was present in Tum(−FBF) than Tum(+FBF) germ lines (compare Figures 3E and F). A similar difference was seen in other germline tumorous strains (gld-1 versus fbf-1 fbf-2; gld-1 as well as gld-1 gld-2 versus fbf-1 fbf-2; gld-1 gld-2 tumorous germ lines (data not shown)). Therefore, FBF is critical for maintaining the low level of LIP-1 protein typical of the distal germ line.
FBF controls lip-1 mRNA accumulation
To ask whether FBF affects lip-1 mRNA accumulation or translation, we assayed lip-1 mRNA by in situ hybridization. Little or no lip-1 mRNA was present in the mitotic region of wild-type germ lines (Figure 3G). Instead, lip-1 mRNA began to increase in the transition zone and became abundant in the pachytene and oogenic regions, except in the last oocyte (Figure 3G). The graded distribution of lip-1 mRNA in the distal arm of the gonad is similar to that of LIP-1 protein, but the in situ probes appeared less sensitive than LIP-1 antibodies. The uniform and strong distribution of lip-1 mRNA in the oogenic region (all except the last oocyte) is dramatically different from the apparent absence of LIP-1 protein in that same region, suggesting the existence of either translational repression or protein instability.
We next tested whether FBF influences the abundance of lip-1 mRNA. We again used germline tumors with or without FBF for reasons outlined above. lip-1 mRNA was present at a low level in Tum(+FBF) germ lines (Figure 3H), but it was abundant in Tum(−FBF) germ lines (Figure 3I). To confirm this effect, we used semiquantitative RT–PCR to compare lip-1 mRNA levels in total RNA extracted from Tum(+FBF) or Tum(−FBF) animals. Specifically, we compared the relative amounts of PCR products generated from lip-1 mRNA and unc-54 mRNA, which encodes the major body wall myosin (MacLeod et al, 1977). Consistent with the in situ results, Tum(+FBF) possessed less lip-1 mRNA than Tum(−FBF) (Figure 3J). Therefore, both by in situ hybridization and by semiquantitative PCR, the presence of FBF correlates with a lowered accumulation of lip-1 mRNA.
Finally, we examined lip-1 mRNA in fbf-1 and fbf-2 single mutants. For both single mutants, lip-1 mRNA was detected in the pachytene region, as expected. More importantly, lip-1 mRNA was also detected at the distal end in about 10% of dissected germ lines (Figure 3K; data not shown). This distally localized lip-1 mRNA is strong support for the idea that GLP-1/Notch signaling activates lip-1 transcription in the distal germ line. Furthermore, the fact that this distal lip-1 mRNA was only detected in germ lines compromised for FBF (i.e. fbf-1 or fbf-2 single mutants) supports the idea that FBF represses lip-1 expression. We suggest that lip-1 mRNA is normally generated in the distal germ line in response to GLP-1/Notch signaling, but that FBF prevents its accumulation in that same region.
FBF binding elements in variant lip-1 3′UTRs
FBF binds 3′UTR regulatory elements to repress RNA expression (Wickens et al, 2002). To delineate the lip-1 3′UTR, we used ‘circular PCR' (Figure 4A). Briefly, we decapped and ligated total mRNA, and then performed PCR using primers designed to span the 3′UTR and poly(A) tail. To our surprise, we identified multiple products among RNAs extracted from wild-type, gld-3 nos-3 (Tum(+FBF)), and fbf-1 fbf-2 gld-3 nos-3 (Tum(−FBF)) animals (Figure 4B). We sequenced 20 representatives from each major PCR product, and found a range of lip-1 3′UTR lengths (Figure 4C and data not shown). The major lip-1 3′UTR in wild-type germ lines, dubbed lip-1M, was intermediate in length between the primary lip-1 3′UTRs in tumorous germ lines, which we dub lip-1L and lip-1S for the longer and shorter 3′UTRs, respectively (Figure 4C). We next designed primers to specifically detect the lip-1S and lip-1L 3′UTRs (Figure 4C). Wild-type and Tum(+FBF) animals possessed the lip-1S 3′UTR, but did not have lip-1L (Figure 4D). By contrast, Tum(−FBF) animals possessed both lip-1S and lip-1L mRNAs (Figure 4D). We then scanned the lip-1 3′UTR sequences for potential FBF binding elements (FBEs) that conform to the consensus UGURHHAUW (Bernstein et al, 2005 and references therein). Two potential FBEs were identified: lip-1–FBEa and lip-1–FBEb (Figure 4C). FBEa occurred in all lip-1 3′UTRs, but FBEb was present only in the lip-1L variant that was specific to Tum(−FBF) RNAs. Therefore, lip-1L RNA does not accumulate in the presence of wild-type FBF. This lack of accumulation may be due to effects on mRNA stability or 3′-end formation.
Figure 4.
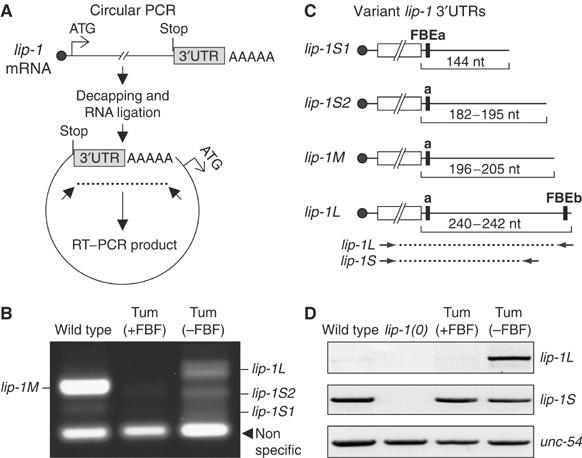
Identification of lip-1 3′UTRs harboring potential FBEs. (A) Schematic of circular RT–PCR method. See text for explanation. (B) Identification of multiple lip-1 3′UTRs. Tum(+FBF), gld-3 nos-3 double mutant; Tum(−FBF), fbf-1 fbf-2 gld-3 nos-3 quadruple mutant. Products of circular RT–PCR were generated from total RNA from animals of genotype noted above lane, and separated on agarose gel. (C) Schematic of variant lip-1 3′UTRs. Solid circle, 5′ cap; open box, coding region, not to scale; black line, 3′UTR; black bar, potential FBE. Numbers below 3′UTR indicate their lengths in nucleotides (nt). ‘M' is the major form in wild-type germ lines, which are largely meiotic and contain few mitotic cells; by contrast, ‘S1' and ‘S2' are the major species in Tum(+FBF) and ‘L' was found only in Tum(−FBF) mutants. (D) Products of semiquantitative RT–PCR from total RNA prepared from animals of genotype noted above lane. Primers were specific to lip-1S, lip-1L, or unc-54 mRNAs (see Materials and methods and arrows in panel C). Note that the PCR conditions used in panels B and D were different, and therefore that products are not comparable.
To assess FBF binding to the potential lip-1–FBEs, we used both yeast three-hybrid assays and gel retardation assays (Figure 5). Yeast three-hybrid interactions were monitored by a growth assay (Figure 5B) and by production of β-galactosidase from a reporter (Figure 5C). Both FBEs interacted specifically with FBF-1 and FBF-2 in three-hybrid assays (Figures 5B and C) and bound specifically to purified FBF-2 in gel shifts (Figure 5D). Wild-type lip-1–FBEa and lip-1–FBEb bound FBF, but not PUF-8 (Figures 5B and C) or PUF-5 (data not shown); by contrast, mutant FBEs (FBE*) with an altered consensus (Figure 5B; UGU changed to ACA) did not interact with either FBF-1 or FBF-2 (Figures 5B–D). The apparent Kd for FBEa is lower in the three-hybrid system than in vitro, perhaps owing to aberrant folding of the synthetic RNA (not shown). We conclude that lip-1 3′UTRs possess FBEs, and suggest that the FBF repression of lip-1 expression is direct.
Figure 5.
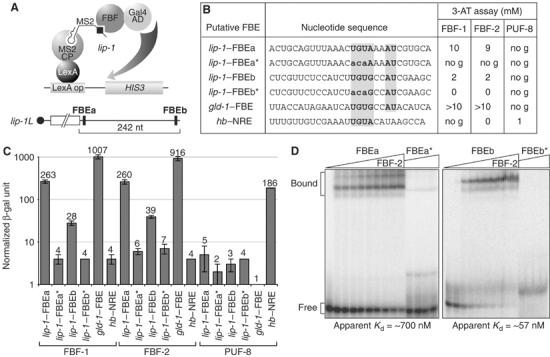
FBF binds specifically to FBEs in the lip-1 3′UTR. (A) Schematic of three-hybrid assay. (B) Three-hybrid interactions assayed by growth in 3-AT. FBF-1 and FBF-2 interact with lip-1–FBEa and lip-1–FBEb, but not with mutants (*) that have an altered consensus sequence. Critical nucleotides of consensus are bold faced and shaded in gray. PUF-8 does not interact with either FBE, but interacts with the hunchback (hb) nanos response element (NRE). no g, no growth. (C) Three-hybrid interactions assayed by β-galactosidase activity. Nomenclature and conventions are same as in panel B. Number at the top of the bar indicates average units of normalized β-galactosidase activity for RNA tested; error bars are calculated from three independent experiments. (D) Purified FBF-2 binds lip-1–FBEa and lip-1–FBEb in gel mobility assays, but it does not bind mutants (*) with an altered consensus as detailed in panel B. Apparent affinities are shown below.
LIP-1 can function as a pivotal regulator of germline proliferation
The lip-1 effect on proliferation is modest in the wild-type germ line (Figure 1). To confirm the ability of LIP-1 to promote mitotic divisions, we sought a sensitized mutant background to see a more prominent effect. For reasons outlined above, we compared Tum(−FBF) germ lines with and without LIP-1. Remarkably, removal of lip-1 transformed these germ lines from a tumorous to a Mog phenotype (Mog, for masculinization of the germ line). Thus, whereas fbf-1 fbf-2 gld-3 nos-3 quadruple mutants had tumorous germ lines, fbf-1 fbf-2 gld-3 nos-3; lip-1(0) quintuple mutants had a Mog germline. Therefore, the uncontrolled mitosis in Tum(−FBF) germ lines requires wild-type LIP-1.
We next assayed the Tum and Mog germ lines for molecular markers of activated MAPK (Figures 6A and B), mitotic divisions (Figures 6C and D), and spermatogenesis (Figures 6E and F). To detect activated MAPK, we used a monoclonal antibody that specifically recognizes the dual phosphorylation (YT) of the active enzyme (Yung et al, 1997). Activated MAPK was generally low in Tum(−FBF, +LIP-1) germ lines (Figure 6A), but high in Mog(−FBF, −LIP-1) germ lines (Figure 6B). Therefore, LIP-1 is required to maintain a low level of activated MAPK, consistent with previous studies in more normal germ lines (Hajnal and Berset, 2002). To visualize mitotically dividing germ cells, we used a monoclonal antibody that recognizes phosphohistone H3, an epitope present in the condensed chromatin of dividing cells. In Tum(−FBF) germ lines, the number of mitotic divisions was high (Figure 6C), as reported previously (Eckmann et al, 2004), but in Mog(−FBF, −LIP-1) germ lines, the number of mitotic divisions was sharply reduced (Figure 6D). Therefore, in this sensitized background, LIP-1 has a profound effect on germline proliferation. Finally, we visualized sperm with the sperm-specific polyclonal antibody, SP56 (Ward et al, 1986). In Tum(−FBF) germ lines, virtually no sperm were made (Figure 6E), as reported previously (Eckmann et al, 2004), but in Mog(−FBF, −LIP-1) germ lines, the proximal half of the germ line began to actively differentiate and produce sperm (Figure 6F). Together, these experiments indicate that LIP-1 promotes mitosis and prevents spermatogenesis. However, not all germ cells were able to differentiate in Tum(−FBF, −LIP-1) mutants, despite the uniform distribution of activated MAPK, suggesting the existence of additional controls that suppress differentiation distally. We conclude that LIP-1 can function as a pivotal regulator of the decision between proliferation and differentiation.
Discussion
This paper focuses on the regulation and function of the LIP-1 phosphatase in the distal C. elegans germ line. Our results support three major conclusions. First, LIP-1 is essential for the robust proliferation typical of wild-type adult germ lines. Second, lip-1 expression is controlled in the germ line by both Notch signaling and FBF repression. Third, LIP-1 controls germline proliferation, at least in part, by inhibiting MAPK activity. This regulatory circuitry controlling C. elegans germline proliferation has important parallels with the control of stem cells and progenitor cells in vertebrates.
Figure 7 proposes a simplified model for how lip-1 expression is regulated in the distal C. elegans germ line. Previous work showed that the somatic DTC niche promotes germline proliferation by GLP-1/Notch signaling (Kimble and Simpson, 1997), and that GLP-1/Notch signaling activates fbf-2 expression (Lamont et al, 2004). The finding that lip-1 is a direct target of LIN-12/Notch signaling in somatic tissues (Berset et al, 2001) was the initial impetus that led us to explore its role in the distal germ line. Here, we show that lip-1 is also a target of GLP-1/Notch signaling. Technical difficulties precluded analysis in the germ line of transgenes lacking LAG-1-binding sites. However, the lip-1 promoter associates with Notch-activated transcription complexes in a germline-dependent manner (Figure 2), and lip-1 RNA is present in the distal-most germ cells that are signaled by the DTC (Figure 3K). Therefore, the lip-1 gene appears to be a direct target of GLP-1/Notch signaling.
Figure 7.
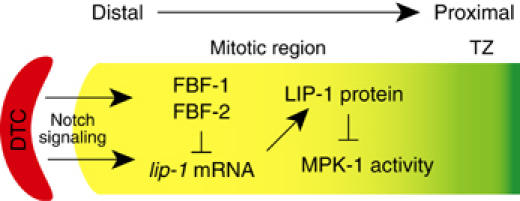
LIP-1 controls the normal extent of germline proliferation. Model for regulation of lip-1 expression by both Notch signaling and FBF. The lip-1 gene is activated by GLP-1/Notch signaling, but accumulation of the lip-1 mRNA is repressed by FBF in the distal-most germ cells. As germ cells progress away from the DTC niche, lip-1 mRNA begins to generate LIP-1 protein, which ensures the downregulation of MAPK in the proximal mitotic region. We suggest that LIP-1 activity in the proximal mitotic region enhances the mitotic capacity of germ cells and delays entry into meiosis. See text for further explanation.
The absence of lip-1 mRNA from the distal germ line of wild-type animals was initially puzzling. That paradox was resolved by finding that the FBF RNA-binding protein represses lip-1 expression post-transcriptionally. Thus, lip-1 3′UTRs possess specific FBEs, and lip-1 expression increases dramatically upon removal of FBF. It is striking that FBF regulates the accumulation of lip-1 mRNA. This finding was unexpected, because metazoan PUF proteins had been thought to act by regulating translation (Wickens et al, 2002). However, yeast PUF proteins control mRNA stability by promoting deadenylation and mRNA decay (Olivas and Parker, 2000). Perhaps metazoan PUFs can repress mRNAs by a similar mechanism. In the case of lip-1, a lip-1L variant mRNA was not detected in wild-type animals, but was seen in animals lacking FBF. Importantly, the lip-1L 3′UTR carried an extension harboring an FBE with high affinity for FBF. In principle, FBF could control the accumulation of lip-1L mRNA at the level of RNA degradation or regulated 3′-end formation. Important challenges for the future are to elucidate the molecular mechanism by which FBF controls this variant lip-1 mRNA and to learn whether that mechanism is FBF specific or mRNA specific.
LIP-1 appears to promote mitotic divisions in the proximal mitotic region of the wild-type germ line by decreasing MAPK activity. This idea is based on several lines of evidence. First, LIP-1 protein is present in the proximal mitotic region. Second, the mitotic region contains fewer germ cells than normal in lip-1 null mutants. Third, activated MAPK appears to extend abnormally into the proximal mitotic region of lip-1 null mutants (Hajnal and Berset, 2002). And finally, depletion of mpk-1 by RNAi suppresses the loss of germ cells from the mitotic region in lip-1 null mutants. Other models in which LIP-1 and MAPK activities act more proximally, but feed back onto the mitotic region, are plausible, but unnecessarily complex. We favor the straightforward explanation that LIP-1 functions in the proximal mitotic region to extend mitotic proliferation and delay the transition into the meiotic cell cycle.
A possible role for MAPK activity in control of early meiotic prophase is thought provoking. By RNAi, we found that a reduction in MAPK activity leads to an increased number of germ cells in the mitotic region and abolishes the transition zone. One speculative idea is that the spatially patterned progression from mitosis through the transition zone and into later stages of meiosis is controlled, at least in part, by a gradient of MAPK activity. However, activated MAPK has not been detected in wild-type germ lines in this distal region (Miller et al, 2001; Page et al, 2001; Hajnal and Berset, 2002), so if it does work there, it must function at a low level. Additional experiments will be required to assess the importance of MAPK activity to distal events of germline development.
The LIP-1 control of robust germline proliferation in C. elegans may provide insight into how the extent of proliferation is regulated in vertebrates. The control of proliferation extent is a significant problem. As animals age, regeneration potential decreases, but the underlying mechanisms remain unknown. A simple model is that stem or progenitor cells become less capable of robust proliferation, which appears to be the case in at least some tissues (Morrison et al, 1996; Iakova et al, 2003). The LIP-1 phosphatase is proposed to be the C. elegans homolog of MKPs (Berset et al, 2001). Indeed, mammalian MKPs have been implicated in stem cell self-renewal (Burdon et al, 2002) and are often upregulated in tumor cell lines (Vogt et al, 2005). By analogy with our finding that LIP-1 functions during normal development to increase the pool of mitotically dividing cells, we speculate that MKPs may function in vertebrate development to promote the robust proliferation of progenitor cells.
Materials and methods
ChIP assay
ChIP assay was performed by a slightly modified version of the procedures described previously (Shang et al, 2000; Chu et al, 2002). Briefly, wild-type and glp-1(q224ts) adults were fixed in M9 buffer with 2% formaldehyde at room temperature for 30 min. Excess formaldehyde was quenched and washed with a 125 mM glycine solution. Worms were washed sequentially with ice-cold PBS and ChIP buffer (5 mM Pipes, pH 8.0, 85 mM KCl, 0.5% NP-40). Worms were resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.1, 1 × protease inhibitor cocktail (Roche Molecular Biochemicals)), frozen in liquid nitrogen, and then homogenized with a cooled mortar and pestle. The frozen extracts were thawed on ice and sonicated three times for 10 s at the maximum setting (Fisher Scientific, Sonic Dismembrator Model 100), followed by centrifugation for 10 min. For each reaction, 3 mg of total protein was diluted in buffer (16.7 mM Tris–HCl, pH 8.1, 150 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA) followed by immunoclearing with 2 μg sheared salmon sperm DNA and 50 μl protein G-agarose (Sigma) for 2 h at 4°C. Polyclonal antibodies were generated against the N-terminal fragment of LAG-3 (amino acids 6–80) fused to HIS. The specificity of the LAG-3 antibody was confirmed by Western blots with recombinant LAG-3 protein and by immunostaining of lag-3(RNAi) animals. ChIP was performed overnight at 4°C with 5 μg of affinity-purified LAG-3 antibodies or preimmune rat sera. After this, 50 μl protein G-agarose and 2 μg of salmon sperm DNA were added and the incubation continued for another 1 h. Precipitates were washed sequentially for 10 min each in washing buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 150 mM NaCl), buffer 2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 500 mM NaCl), and buffer 3 (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1), and then formaldehyde crosslinks were reversed by incubation at 65°C overnight in 1% SDS and 0.1 M NaHCO3. Proteins were removed by proteinase K digestion and DNA fragments were purified using the Qiagen PCR purification kits. We carried out PCR for 35 cycles using primers specific to lip-1 and gld-1, which were designed to produce similar size of PCR products, and resolved amplified products on 1.0% agarose gels.
In situ mRNA hybridization and immunohistochemistry
In situ hybridization was performed as described (Jones et al, 1996), using an antisense digoxigenin-labeled cDNA (nucleotides 42–559 in the lip-1 cDNA sequence). Rat polyclonal antibodies specific to LIP-1 were made by standard methods, using LIP-1 amino acids 5–175. For immunohistochemistry, gonads were extruded, fixed with 3% formaldehyde and 0.1 M K2HPO4 (pH 7.2) for 1 h, and postfixed with cold (−20°C) 100% methanol for 5 min. Antibody incubations and washes were performed as described (Jones et al, 1996). Di-phosphorylated MAPK, dividing nuclei, and sperm were detected with the monoclonal antibody MAPK-YT (Sigma), phosphohistone H3 (PH3+, Upstate Biotechnology), and SP56 (a gift from S Ward), respectively. DAPI staining followed standard methods.
Three-hybrid and gel retardation assays
Three-hybrid assays were performed as described (Bernstein et al, 2002). Levels of 3-aminotriazole (3-AT) resistance were determined by assaying multiple transformants at 12 different concentrations of 3-AT, up to 11 mM. For β-galactosidase assays, cells were grown in selective media to an OD600 of 1.0 and mixed with an equal volume of β-Glo (Promega) reagent. Luminescence was measured after 1 h. Gel shift assays were performed as described (Hook et al, 2005).
RT–PCR
Circular RT–PCR was performed according to the procedure of Couttet et al (1997) with modifications (N Suh, unpublished). For semiquantitative RT–PCR, 20 adult worms were transferred into 50 μl RNase-free ddH2O and 200 μl of GibcoBRL Trizol was added. Samples were placed at −80°C for 15 min, at 65°C for 90 s, and at room temperature for 15 min. Total RNA was extracted by CHCl3, precipitated with isopropanol and 70% ethanol solution, and then resuspended in RNase-free ddH2O. After reverse transcription, we carried out PCR for 35 cycles using primers specific to lip-1 and unc-54 and analyzed PCR products at six time points during the reaction. After examination of PCR products, cycle 30 was found to be within the linear range. We resolved amplified products on 1.0% agarose gels and scored band intensity using 1D image analysis software (Kodak).
Acknowledgments
We thank members of the Kimble and Wickens laboratories for comments on the manuscript, Sungtae Kim and J Wesley Pike for ChIP protocols, Nayoung Suh for advice on circular PCR, and Anne Helsley-Marchbanks and Laura Vanderploeg for assistance in preparation of the manuscript and figures. LBL was supported by National Institutes of Health Predoctoral Training Grant T32 GM07215 in Molecular Biosciences; MW and JK were supported by the National Institutes of Health; JK is an investigator of the Howard Hughes Medical Institute.
References
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M (2005) Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DS, Buter N, Stumpf C, Wickens M (2002) Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods 26: 123–141 [DOI] [PubMed] [Google Scholar]
- Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A (2001) Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P (2002) Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 12: 432–438 [DOI] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT (2003) Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425: 841–846 [DOI] [PubMed] [Google Scholar]
- Camps M, Nichols A, Gillieron C, Antonsson B, Muda M, Chabert C, Boschert U, Arkinstall S (1998) Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science 280: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Chu DS, Dawes HE, Lieb JD, Chan RC, Kuo AF, Meyer BJ (2002) A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev 16: 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ (1995) Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121: 2525–2535 [DOI] [PubMed] [Google Scholar]
- Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA 94: 5628–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J (2002) A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663 [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Eckmann CR, Wang L, Bernstein DS, Wickens M, Kimble J (2003) Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos Trans R Soc London B 358: 1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J (1994) GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120: 2901–2911 [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Crittenden SL, Suh N, Kimble J (2004) GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S (2005) Notch signals control the fate of immature progenitor cells in the intestine. Nature 435: 964–968 [DOI] [PubMed] [Google Scholar]
- Hajnal A, Berset T (2002) The C. elegans MAPK phosphatase LIP-1 is required for the G2/M meiotic arrest of developing oocytes. EMBO J 21: 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D, Hubbard EJ, Schedl T (2004) Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol 268: 342–357 [DOI] [PubMed] [Google Scholar]
- Hook B, Bernstein D, Zhang B, Wickens M (2005) RNA–protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11: 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova P, Awad SS, Timchenko NA (2003) Aging reduces proliferative capacities of liver by switching pathways of C/EBPα growth arrest. Cell 113: 495–506 [DOI] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T (1996) GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol 180: 165–183 [DOI] [PubMed] [Google Scholar]
- Kimble J, Simpson P (1997) The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol 13: 333–361 [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG (1981) On the control of germ cell development in Caenorhabditis elegans. Dev Biol 81: 208–219 [DOI] [PubMed] [Google Scholar]
- Lackner MR, Kim SK (1998) Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics 150: 103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J (2004) FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell 7: 697–707 [DOI] [PubMed] [Google Scholar]
- Li L, Xie T (2005) Stem cell niche: structure and function. Annu Rev Cell Dev Biol 21: 605–631 [DOI] [PubMed] [Google Scholar]
- MacLeod AR, Waterston RH, Brenner S (1977) An internal deletion mutant of a myosin heavy chain in Caenorhabditis elegans. Proc Natl Acad Sci USA 74: 5336–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D (2001) A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147 [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL (1996) The aging of hematopoietic stem cells. Nat Med 2: 1011–1016 [DOI] [PubMed] [Google Scholar]
- Olivas W, Parker R (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J 19: 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Guedes S, Waring D, Priess JR (2001) The C. elegans E2F- and DP-related proteins are required for embryonic asymmetry and negatively regulate Ras/MAPK signaling. Mol Cell 7: 451–460 [DOI] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J (2000) LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405: 364–368 [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843–852 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Bernstein DS, Bachorik JL, Petcherski AG, Wickens M, Kimble J (2005) Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development 132: 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Tamewitz A, Skoko J, Sikorski RP, Giuliano KA, Lazo JS (2005) The benzo[c]phenanthridine alkaloid, sanguinarine, is a selective, cell-active inhibitor of mitogen-activated protein kinase phosphatase-1. J Biol Chem 280: 19078–19086 [DOI] [PubMed] [Google Scholar]
- Ward S, Roberts TM, Strome S, Pavalko FM, Hogan E (1986) Monoclonal antibodies that recognize a polypeptide antigenic determinant shared by multiple Caenorhabditis elegans sperm-specific proteins. J Cell Biol 102: 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R (2002) A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 18: 150–157 [DOI] [PubMed] [Google Scholar]
- Yung Y, Dolginov Y, Yao Z, Rubinfeld H, Michael D, Hanoch T, Roubini E, Lando Z, Zharhary D, Seger R (1997) Detection of ERK activation by a novel monoclonal antibody. FEBS Lett 408: 292–296 [DOI] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484 [DOI] [PubMed] [Google Scholar]


