Abstract
Previously, we have characterized the HIV-ISF2 gp120 glycopeptides using matrix-assisted laser desorption/ionization mass spectrometry (MALDI/MS) and nanospray electrospray ionization (ESI). Although we characterized 25 of 26 consensus glycosylation sites, we could not obtain any information about the extent of sialylation of the complex glycans. Sialylation is known to alter the biological activity of some glycoproteins, e.g. infectivity of some human and nonhuman primate lentiviruses is reduced when the envelope glycoproteins are extensively sialylated, and, thus, characterization of the extent of sialylation of complex glycoproteins is of biological interest. Since neither MALDI/MS nor nanospray ESI provided much information about sialylation, probably because of suppression effects inherent in these techniques, we utilized online nanocapillary high performance liquid chromatography (nHPLC) with ESI/MS to characterize the sites and extent of sialylation on gp120. Eight of the known 26 consensus glycosylation sites of HIV-ISF2 gp120 were determined to be sialylated. Two of these sites were previously uncharacterized complex glycans. Thirteen high mannose sites were also determined. The heterogeneity of four of these sites had not been previously characterized. In addition, a peptide containing two consensus glycosylation sites, which had previously been determined to contain complex glycans, was also determined to be high mannose as well.
Introduction
Human immunodeficiency virus (HIV) is the human retrovirus that causes acquired immune deficiency syndrome (AIDS). Retroviruses are enveloped, positive-stranded RNA viruses in which the viral envelope is embedded with glycoproteins, known to be involved in the early steps of infection. In HIV, the initially formed envelope glycoprotein, gp160, is cleaved into gp41 and gp120. gp41 is the transmembrane portion of the glycoprotein spike and promotes cell-cell fusion during infection. The gp120 of HIV binds to the CD4 surface molecule expressed on T-helper cells and on cells of the macrophage lineage. Chemokine receptors are needed as coreceptors for attachment of the HIV virus. The gp120 structure contains five loops with varying amino acid composition. One of these, the V3 loop, elicits neutralizing anti-HIV antibodies and has been a target for HIV vaccine development [1]. It has been shown that carbohydrates within and around the V3 loop shield binding sites for the chemokine receptors, and loss of these carbohydrates increases infectivity [2,3,4]. The N-linked glycans on this loop have been reported to strongly influence coreceptor usage, with CCR5 being used during the early stages of disease and CXCR4 being used as the disease progresses [3,4]. The V1 and V2 variable loops have been shown to undergo conformational changes upon CD4 binding [5]. The V1/V2 loops may need to alter their native conformation in order for the CCR5 coreceptor to bind with a high-affinity interaction. Also, reports show that carbohydrates on Asp197, in the V1/V2 stem, restrict this movement, thereby allowing the carbohydrates to mask the binding region [6].
HIV gp120 is highly glycosylated with a large number of consensus glycosylation sites, 26, in HIVSF2 gp120. The matrix-assisted laser desorption/ionization (MALDI) spectrum of gp120 shows that the protein is very heterogeneous with a molecular mass ranging from 90,000–110,000 Da [7]. Based on the sequence, this indicates that close to half of its mass is due to carbohydrate residues. The differences include the number of glycosylated sites and the types of glycans present. Individual glycosylation sites may be occupied by structurally different glycans, which give them the ability to cover substantial parts of the peptide surface of the viral glycoprotein [9]. The structure of gp120 reveals that many of the N-linked glycans could cover regions of gp120 that would otherwise be exposed to potentially neutralizing antibodies [10]. Also, the system used to express gp120 will have an effect on the glycosylation pattern, e.g., the glycosylation pattern when gp120 is expressed in Chinese hamster ovarian (CHO) cells will differ significantly from that expressed in baculovirus [8].
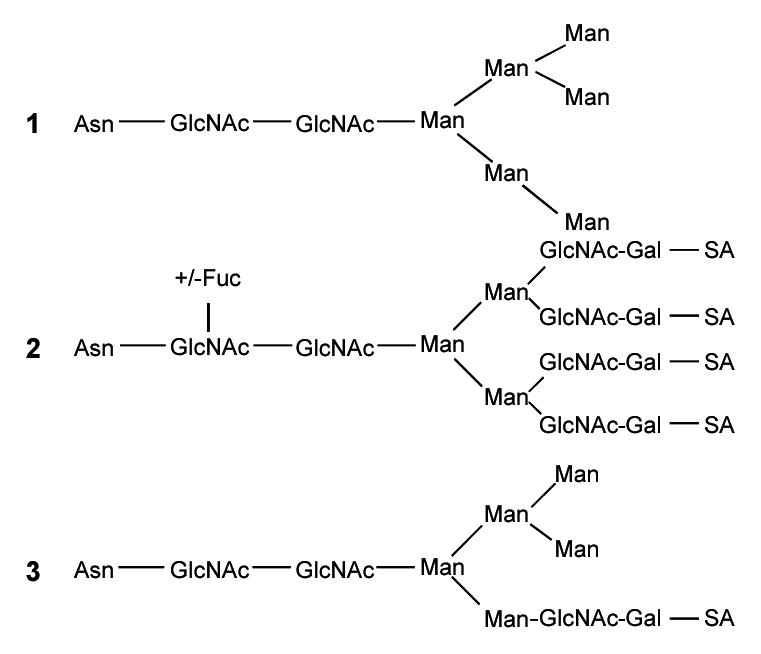
There have been several reports on the determination of glycosylation sites and of the types of glycans found on gp120. One research group found that the gp120 glycans are primarily high mannose, hybrid, and complex varieties, and more than 100 different glycans could be identified [11,12]. The major types of glycans that can be found in a glycopeptide are shown in the structures below. High mannose glycans consist primarily of mannose (Man) with a maximum number of 9 mannoses possible unless not fully processed (Structure 1), whereas the complex glycans are primarily composed of N-acetylglucosamine (GlcNAc) and galactose (Gal) with sialic acid (SA) (Structure 2). In the complex glycans, both the number of N-acetylglucosamine galactose (GlcNAc-Gal) units as well as the number of sialic acids (SA) can vary. A fucose (Fuc) may be added to the first GlcNAc in the core of the complex glycans. The hybrid glycans are composed of mannose, GlcNAc-Gal, and SA (Structure 3).
We have previously identified the major types of glycans found on 25 of 26 consensus glycosylation sites in HIV-ISF2 rgp120 expressed in CHO cells using nanoelectrospray and MALDI [13]. Using a molecular model of intact glycosylated gp120 based on the crystal structure of a truncated highly deglycosylated construct [10], high mannose glycans appear to cluster together on the surface, as do complex glycans [13]. Consistent with this model, it has been noted that mannose-binding lectin (MBL) interacts with gp120 by binding to HIV-1 via high mannose or hybrid carbohydrates on gp120 [14,15]. MBL is a serum protein that binds to carbohydrates on microorganisms and plays a role in the clearance and destruction of these microorganisms [14,16]. It has also been reported that high mannose glycans of gp120 contribute largely to the MBL-HIV interaction rather than host cell-derived glycoproteins [17].
There have been suggestions that modifications to these glycans, such as the sialylation of the complex glycans, could affect their potential biological activity by possibly reducing the infectivity of human and nonhuman primate lentivirus [18]. Sialic acids are a family of naturally occurring derivatives of neuraminic acid that play a role in biological and immunological events. Sialylation usually occurs on glycoproteins embedded in the plasma membrane, where the sialic acids stabilize the plasma membranes by their negative charge [19]. Removal of the fucose associated with sialylated glycans has been shown to shift the protein conformation, possibly disclosing epitopes that were located in the interior of gp120 [20]. Others have shown that the removal of some sialic acids from rgp120 enhanced MBL binding [15]. It has also been shown in a simian immunodeficiency virus (SIV) monkey model that SIV-mutants lacking N-linked glycosylation are more immunogenic compared to the wild type. This report suggests that carbohydrates may be responsible for masking immunodominant epitopes on external glycoproteins and, thus, protect the virus against neutralizing antibodies [21].
In our previous work, we were able to detect complex and high mannose glycans at 25 of the 26 consensus sites of HIV-ISF2 rgp120 [13], however, we could not detect the presence of sialic acid residues on the complex glycopeptides using nanospray/MS or MALDI/MS, presumably due to suppression effects associated with these two ionization techniques. Harvey and co-workers used MALDI/MS and normal-phase HPLC to investigate oligosaccharides released from gel-separated proteins by in-gel deglycosylation [22]. This method was used because previous methods required the sialic acids of the glycopeptides or oligosaccharides be removed in order for the glycans to be identified [9,23]. Although various MALDI/MS methods have been used to determine the structure of sialylated glycans [24], successful applications of these methods to the characterization of complex mixtures of sialylated glycopeptides have not been reported. Gibson and co-workers characterized sialylated complex glycans using negative ion MALDI-TOF MS, but, in our initial study, we were unsuccessful in the characterization of sialylated complex glycopeptides with this approach. If the sensitivity of the sialylated glycopeptides was reduced due to suppression by other components in the complex proteolytic digests, we hypothesized that the nHPLC/ESI/MS approach should reduce the complexity of the mixture presented to the MS at any given time during the analysis and, thus, reduce these suppression effects. We, therefore, have analyzed the proteolytic digest of gp120 using nHPLC/ESI/MS as a means of characterizing the sialylation of the complex glycopeptides from gp120.
Methods
gp120 Sample Preparation
The HIV-ISF2 env gp120 (100 μg) (Austral Biologicals, San Ramon, CA), expressed in Chinese hamster ovary (CHO) cells, was diluted in 152 μL of 0.36 M Tris-HCl buffer (pH 8.6), containing 8 M urea, 3 mM ethylenediaminetetraacetic acid (EDTA), and 10 mM dithiothreitol (DTT) and incubated under nitrogen for 4 hours to reduce disulfide bonds. Carboxymethylation was performed by adding iodoacetic acid (Sigma, St. Louis, MO) to a final concentration of 25 mM and incubating in the dark for 45 minutes. The reaction was quenched by adding DTT to a final concentration of 40 mM. The carboxymethylated gp120 was purified by HPLC on an Aglient model 1100 HPLC system (Palo Alto, CA) using a 25 cm × 4.6 mm i.d. Vydac 214TP54 C4 column (Hesperia, CA). Elution from the column was achieved using solvents A (water/0.1% trifluoroacetic acid (TFA)) and B (acetonitrile/0.1% TFA) with a linear gradient from 20% B to 60% B over 50 minutes, followed by a ramp from 60% B to 90% B over 10 minutes at a flow rate of 1 mL/min with fractions collected at 1.5 minute intervals. Collected fractions were lyopholized, reconstituted in 20 μL of 1:1 acetonitrile:0.1% formic acid, and analyzed for the presence of gp120 by MALDI/MS (as described below). Those fractions containing gp120 were pooled into one fraction and lyophilized to dryness. The lyophilized gp120 was then reconstituted in 100 μL of 50 mM ammonium bicarbonate buffer (pH 7.95) and reanalyzed by MALDI/MS to confirm the presence of the gp120.
Tryptic Digestion of gp120
The 100 μL sample of gp120 was digested with 5 μL of 1 μg/μL sequencing grade modified trypsin reconstituted in the supplied buffer of 50 mM acetic acid (Promega, Madison, WI) at a ratio of 20:1 gp120:trypsin (w/w). The tryptic digest was then analyzed by MALDI/MS to ensure complete digestion.
MALDI-TOF/MS
The MALDI/MS used was an Applied Biosystems Voyager- SUPER DE STR (Framingham, MA). MALDI/MS analyses of the lyopholized gp120 fractions were performed in the positive linear mode using an accelerating voltage of 25 kV, a grid voltage of 93%, and a delay time of 400 nsec. MALDI/MS analysis of the tryptic digest was performed using an accelerating voltage of 20 kV, a grid voltage of 95%, and a delay time of 400 nsec in the positive linear mode. Analysis of the tryptic digest in the negative ion linear mode was performed using an accelerating voltage of −25 kV, a grid voltage of 95%, and a delay time of 150 nsec. All analyses in the positive ion mode were performed using a saturated solution of α-cyano-4-hydroxycinnamic acid in 45:45:10 water:ethanol:formic acid (v:v:v) as the matrix. The analyses in the negative ion mode were performed using a 100 mM solution of 2,5-dihydroxybenzoic acid in 50% acetonitrile as the matrix. External calibration was performed using the Applied Biosystems (Framingham, MA) calibration mixes.
nHPLC/ESI/MS
For the nHPLC/ESI/MS analyses, HPLC separations were performed using an Agilent 1100 Series Capillary LC (Palo Alto, CA). For LC separations acquired by positive ion ESI/MS, a 15 cm × 75 μm i.d. PepMap™ C18 column (LC Packings, San Francisco, CA) with a flow rate of 400 nL/min was used. An 8 μL aliquot of the tryptic digest was injected. The solvents used with this column were the following: A, water/0.1% formic acid; and B, acetonitrile/0.1% formic acid. A gradient of 8% B to 40% B over 50 minutes, followed by a ramp from 40% B to 90% B over 10 minutes was used to elute the tryptic peptides. For LC separations acquired by negative ion ESI/MS, a 10 cm × 300 μm i.d. ZORBAX 300Extend-C18 column (Agilent Technologies, Palo Alto, CA) with a flow rate of 1.0 μL/min was used. The solvents used with this column were the following: A, water/1.0% NH4OH; and B, acetonitrile/1.0% NH4OH. The injection amount and gradient used were the same as in the positive ion ESI/MS analyses.
The ESI spectra were obtained using either the Q-Tof1 mass spectrometer (Micromass, Altrincham, UK) or the Q-Tof Ultima Global mass spectrometer (Waters/Micromass, Milford, MA), both of which are equipped with a nanoflow z-spray source. The Q-Tof1 mass spectrometer was operated at a FWHM resolution of 7,500. The observed mass accuracy of high abundance glycosylated peaks was approximately 0.008%, while that of peaks of low abundance was lower, < 0.07%. Spectra were acquired in the positive ion mode over the mass range of m/z 300–2500 with the source temperature at 80 °C, a capillary voltage of 3.00 kV, a cone voltage of 30 V, a collision energy of 4.0 eV, and a scan time of 1.9 seconds. The collision gas was argon at a gauge pressure of 20 psi. The Q-Tof Ultima Global mass spectrometer was operated at a FWHM resolution of 10,000 in V mode and a mass accuracy of 0.02%. The spectra were acquired in the positive ion mode over the mass range of m/z 300–2500 with the source temperature at 80 °C, a capillary voltage of 3.05 kV, a cone voltage of 45 V, a collision energy of 10.0 eV, and a scan time of 1.0 seconds. The collision gas was argon at a gauge pressure of 25 psi. MS/MS analyses were performed on the Q-Tof Ultima Global with all parameters the same as in MS mode, except the collision energy was 30.0 eV and the scan time was 1.9 seconds. The negative ion ESI/MS spectra were acquired over the mass range of m/z 100–3000 with the source temperature at 80 °C, a capillary voltage of 3.0 kV, a cone voltage of 100 V, a collision energy of 10.0 eV, and a scan time of 1.9 seconds. The collision gas was argon at a pressure of 25 psi. Peaks with a signal:noise greater than five were considered. External calibration was performed using renin substrate tetradecapeptide (Sigma, St. Louis, MO).
Results and Discussion
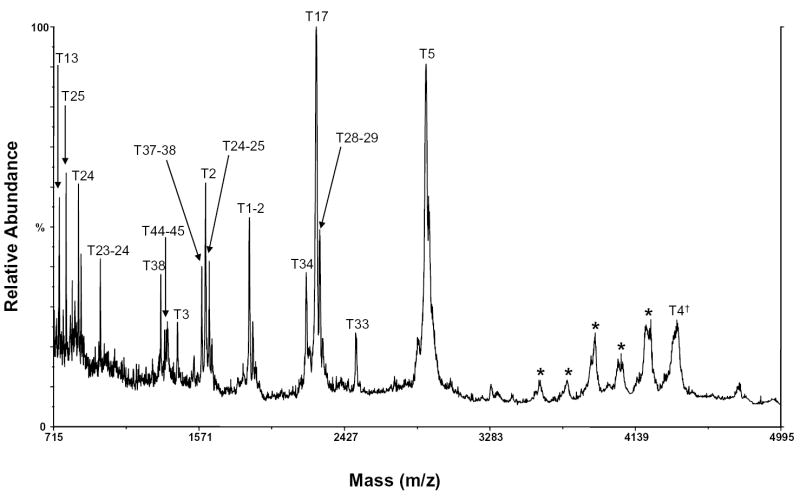
HIV-ISF2 gp120 contains glycans primarily of high mannose, hybrid, and complex varieties (See Structures 1, 2, and 3). A MALDI/MS spectrum of the carboxymethylated gp120 tryptic digest is shown in Figure 1. Only one tryptic peptide, m/z 4386, which corresponds in mass to a potentially sialylated complex tryptic glycopeptide, T4, was observed. This peptide contains the consensus glycosylation site N57. Bozue et al. have previously shown that under negative ion MALDI/MS conditions using a 100 mM solution of 2,5-dihydroxybenzoic acid as a matrix, sialylated complex oligosaccharides could be readily detected [24]. When this method was used to analyze the gp120 tryptic digest, however, sialylated glycopeptides were not detected (results not shown). This may be due to suppression of the sialylated glycopeptides in the complex gp120 digest mixture.
Figure 1.
MALDI/MS spectrum of the tryptic digest of gp120 with peptides labeled. The dagger (†) indicates a sialylated peptide. The asterisk (*) indicates a peak that could not be identified.
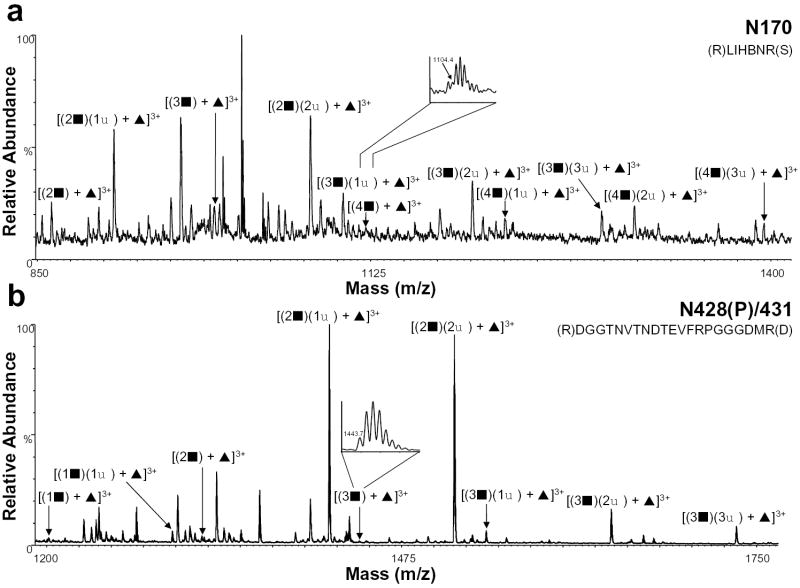
The HIV-ISF2 gp120 tryptic digest was then analyzed by nHPLC/ESI/MS in both the positive and negative ion mode in an effort to identify and characterize the sialylated complex glycopeptides. The extent of sialylation on complex glycopeptides was determined by first calculating the mass of the peptide with the glycan core with or without fucose. The masses of the GlcNAc-Gal and SA units were then added sequentially based on the previous glycosylation characterization [13] with the possible addition of one SA per GlcNAc-Gal. The nHPLC/ESI/MS data were then searched for these corresponding masses. Table 1 shows the amino acid on which a sialylated glycan was found, the tryptic peptides containing the sites, and the masses of the glycopeptides. The table also includes the number of GlcNAcGals (GlcNAc-Gal) residues observed and the extent of sialylation (SA residues) along with the relative abundance of each. The relative abundances of the summed intensities of all isotope peaks over the elution window of specific glycosylated peptides were normalized with the most abundant ion being 100%. It should be noted that the relative abundances correspond to the abundance ratios observed in the raw data, and that differences in sensitivity and ionization efficiency were not considered. Figure 2 shows the positive ion nHPLC/ESI/MS spectra of two tryptic peptides in which the extent of complex glycans had been previously identified [13] but not the degree of sialylation because the sample had been desialylated. The summed spectrum in Figure 2a shows the glycan composition for N170, which is contained in tryptic peptide T15. This glycan is fucosylated and contains 2-4 GlcNAc-Gal units as well as 1-3 sialic acids. We had previously observed 2-5 GlcNAc-Gal units at this site [13], however, in our current study, an ion corresponding to the fifth GlcNAc-Gal was not detected in any charge state. This lack of detection may be consistent with the fact that the relative abundance was also very low in our previous characterization by nanospray or that the more extensively sialylated glycopeptides have significantly reduced sensitivities compared to less highly sialylated glycopeptides in the nHPLC/ESI/MS analyses. The summed spectrum containing the fucosylated peptide T40 (N428(P)/431) is shown in Figure 2b and these data indicate the presence of 1-3 GlcNAcGals with 1-3 sialic acids. Previously, we could not determine the extent of sialylation for this site, but were able to characterize the presence of 1-7 GlcNAc-Gal units [13]. Glycopeptides which were observed previously with higher GlcNAc-Gal and SA units [13] were not observed here, presumably due to their high masses and charge.
Table 1.
Heterogeneity, Relative Abundances, and Extent of Sialylation Determined for Complex Glycans Using nHPLC/ESI/MS
| Glycopeptide Mass | |||||
|---|---|---|---|---|---|
| Site | Tryptic Peptide | aTheo. | bObs. | c(GlcNAc-Gal)x(SA)y +/− Fuc | Relative Abundance(%) |
| N99 | T6 d(K)LTPLBVTLNBTDLGK(A) | 3110.3 | 3110.4 | (GlcNAc-Gal)1 + Fuc | 100 |
| 3401.4 | 3401.8 | (GlcNAc-Gal)1(SA)1 + Fuc | 68 | ||
| 3475.5 | 3475.2 | (GlcNAc-Gal)2 + Fuc | 84 | ||
| 3766.6 | 3767.5 | (GlcNAc-Gal)2(SA)1 + Fuc | 53 | ||
| 4057.7 | 4058.2 | (GlcNAc-Gal)2(SA)2 + Fuc | 53 | ||
| 3840.6 | 3841.5 | (GlcNAc-Gal)3 + Fuc | 74 | ||
| N110 | T7 (K)ATNTNSSNWK(E) | 2745.1 | 2746.1 | (GlcNAc-Gal)2 − Fuc | 79 |
| 3036.2 | 3036.3 | (GlcNAc-Gal)2(SA)1 − Fuc | 42 | ||
| 3327.3 | 3327.1 | (GlcNAc-Gal)2(SA)2 − Fuc | 53 | ||
| 3110.3 | 3110.5 | (GlcNAc-Gal)3 − Fuc | 100 | ||
| 3401.4 | 3400.1 | (GlcNAc-Gal)3(SA)1 − Fuc | 84 | ||
| 3475.5 | 3475.2 | (GlcNAc-Gal)4 − Fuc | 65 | ||
| 3766.6 | 3767.5 | (GlcNAc-Gal)4(SA)1 − Fuc | 53 | ||
| 4057.7 | 4058.7 | (GlcNAc-Gal)4(SA)2 − Fuc | 53 | ||
| N170 | T15 (R)LIHBNR(S) | 2582.0 | 2581.2 | (GlcNAc-Gal)2 + Fuc | 48 |
| 2873.1 | 2872.8 | (GlcNAc-Gal)2(SA)1 + Fuc | 98 | ||
| 3164.2 | 3164.5 | (GlcNAc-Gal)2(SA)2 + Fuc | 100 | ||
| 2947.2 | 2945.7 | (GlcNAc-Gal)3 + Fuc | 35 | ||
| 3238.3 | 3237.2 | (GlcNAc-Gal)3(SA)1 + Fuc | 39 | ||
| 3529.4 | 3528.9 | (GlcNAc-Gal)3(SA)2 + Fuc | 55 | ||
| 3820.5 | 3819.9 | (GlcNAc-Gal)3(SA)3+ Fuc | 29 | ||
| 3312.3 | 3310.2 | (GlcNAc-Gal)4 + Fuc | 19 | ||
| 3603.4 | 3602.8 | (GlcNAc-Gal)4(SA)1 + Fuc | 29 | ||
| 3894.5 | 3893.6 | (GlcNAc-Gal)4(SA)2 + Fuc | 35 | ||
| 4185.6 | 4186.1 | (GlcNAc-Gal)4(SA)3 + Fuc | 24 | ||
| N249 | T21 (R)SDNFTNNAK(T) | 2779.1 | 2777.9 | (GlcNAc-Gal)2 + Fuc | 68 |
| 3070.2 | 3069.8 | (GlcNAc-Gal)2(SA)1 + Fuc | 100 | ||
| 3361.3 | 3362.8 | (GlcNAc-Gal)2(SA)2 + Fuc | 45 | ||
| 3144.2 | 3142.7 | (GlcNAc-Gal)3 + Fuc | 50 | ||
| 3435.3 | 3434.2 | (GlcNAc-Gal)3(SA)1 + Fuc | 64 | ||
| 2998.2 | 2997.5 | (GlcNAc-Gal)3 − Fuc | 73 | ||
| 3289.3 | 3288.2 | (GlcNAc-Gal)3(SA)1 − Fuc | 50 | ||
| 3580.4 | 3579.7 | (GlcNAc-Gal)3(SA)2 − Fuc | 64 | ||
| 3871.5 | 3872.3 | (GlcNAc-Gal)3(SA)3 − Fuc | 55 | ||
| N274 | T22 (K)TIIVQLNESVAINBTRPNNNTR(K) | 3932.3 | 3930.9 | (GlcNAc-Gal)1 + Fuc | 12 |
| 4223.4 | 4222.2 | (GlcNAc-Gal)1(SA)1 + Fuc | 16 | ||
| 4297.5 | 4296.7 | (GlcNAc-Gal)2 + Fuc | 88 | ||
| 4588.6 | 4587.5 | (GlcNAc-Gal)2(SA)1 + Fuc | 78 | ||
| 4662.6 | 4661.6 | (GlcNAc-Gal)3 + Fuc | 39 | ||
| 4953.7 | 4954.1 | (GlcNAc-Gal)3(SA)1 + Fuc | 37 | ||
| 5244.8 | 5243.8 | (GlcNAc-Gal)3(SA)2 + Fuc | 27 | ||
| 5027.7 | 5028.8 | (GlcNAc-Gal)4 + Fuc | 22 | ||
| 5318.8 | 5319.9 | (GlcNAc-Gal)4(SA)1 + Fuc | 100 | ||
| 5609.9 | 5609.5 | (GlcNAc-Gal)4(SA)2 + Fuc | 63 | ||
| N415 | T39 (K)AMYAPPIGGQISBSSNITGLLLTR(D) | 3925.5 | 3926.1 | (GlcNAc-Gal)1 + Fuc | 14 |
| 4216.5 | 4216.1 | (GlcNAc-Gal)1(SA)1 + Fuc | 13 | ||
| 4290.6 | 4291.1 | (GlcNAc-Gal)2 + Fuc | 100 | ||
| 4581.7 | 4582.4 | (GlcNAc-Gal)2(SA)1 + Fuc | 69 | ||
| 4655.7 | 4656.2 | (GlcNAc-Gal)3 + Fuc | 20 | ||
| 4946.8 | 4947.2 | (GlcNAc-Gal)3(SA)1 + Fuc | 42 | ||
| 5237.9 | 5237.6 | (GlcNAc-Gal)3(SA)2 + Fuc | 31 | ||
| N428e(P)/N431 | T40 (R)DGGTNVTNDTEVFRPGGGDMR(D) | 3598.5 | 3597.9 | (GlcNAc-Gal)1 + Fuc | 4 |
| 3889.6 | 3888.6 | (GlcNAc-Gal)1(SA)1 + Fuc | 6 | ||
| 3963.6 | 3963.5 | (GlcNAc-Gal)2 + Fuc | 5 | ||
| 4254.7 | 4254.3 | (GlcNAc-Gal)2(SA)1 + Fuc | 100 | ||
| 4545.8 | 4544.9 | (GlcNAc-Gal)2(SA)2 + Fuc | 90 | ||
| 4328.8 | 4328.2 | (GlcNAc-Gal)3 + Fuc | 2 | ||
| 4619.9 | 4618.8 | (GlcNAc-Gal)3(SA)1 + Fuc | 7 | ||
| 4910.9 | 4911.5 | (GlcNAc-Gal)3(SA)2 + Fuc | 16 | ||
| 5202.0 | 5201.1 | (GlcNAc-Gal)3(SA)3 + Fuc | 9 | ||
Theo. = theoretical
Obs. = observed
GlcNAc-Gal = N-acetylglucosamine and galactose, x = the number of GlcNAc-Gal units, SA = sialic acid, y = the number of SA units, and Fuc = fucose
B = carboxymethylcysteine
P = partially glycosylated
Figure 2.
Positive ion nHPLC/ESI/MS spectra summed over the chromatographic retention window in which complex glycopeptides a) N170 and b) N428(P)/431 eluted. The inset in panel a is for the ion corresponding to [(4▪) + ▴]3+. The inset in panel b is for the ion corresponding to [(3▪) + ▴]3+. Both insets are of spectra summed over the elution window of that component. For figure clarity, the peaks corresponding to other tryptic peptides/glycopeptides are not labeled. ▪ = GlcNAc-Gal, ♦ = sialic acid, and ▴ = fucose; the number before each symbol indicates how many of each are present. The superscripted number is the charge state. In the amino acid sequence B= carboxymethylcysteine.
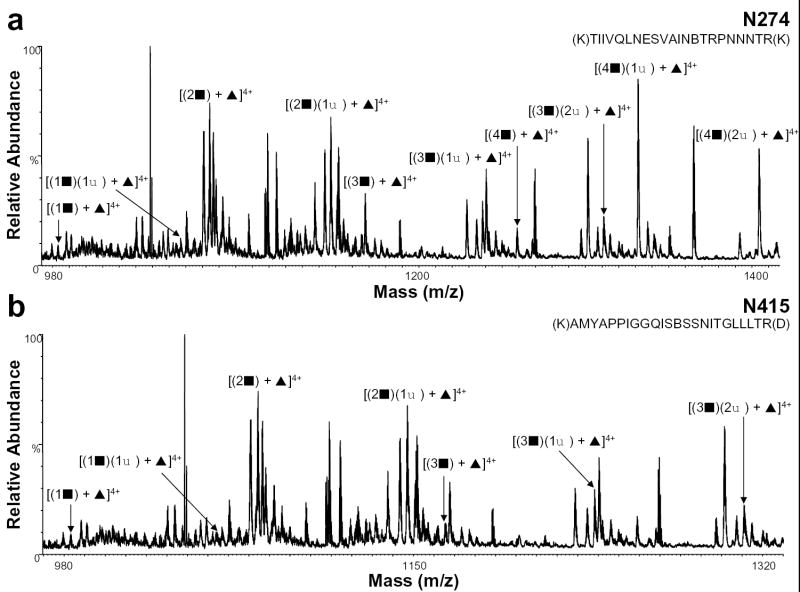
Previously, we reported that the glycan group at residue N274 was complex, but we were unable to characterize the heterogeneity at this position [13]. In addition, the carbohydrate chain at residue N415, could not be determined. Using the nHPLC/ESI/MS approach, however, we were able to determine the glycan at this residue to be complex. Figure 3 shows the positive ion nHPLC/ESI/MS spectra of these two previously uncharacterized complex glycans. From the summed mass spectrum containing peptide T22 (Figure 3a), which contains the glycosylation site at residue N274, we could determine that this glycopeptide is fucosylated and contains 1-4 GlcNAc-Gal units and 1-2 sialic acids. The summed spectrum containing the glycopeptide T39 (Figure 3b), with a consensus glycosylation site at amino acid N415, shows that the glycan structure for this peptide has 1-3 GlcNAc-Gal units and 1-2 sialic acids.
Figure 3.
Positive ion nHPLC/ESI/MS spectra summed over the chromatographic retention window in which complex glycopeptides a) N274 and b) N415 eluted. For figure clarity, the peaks corresponding to other tryptic peptides/glycopeptides are not labeled. ▪ = GlcNAc-Gal, ♦ = sialic acid, and ▴ = fucose; the number before each symbol indicates how many of each are present. The superscripted number is the charge state. In the amino acid sequence B= carboxymethylcysteine.
In an attempt to gain structural information about the glycopeptides as well as sequence information, nHPLC/ESI/MS/MS analyses were performed. Figure 4 shows a representative positive ion MS/MS spectrum obtained for m/z 1419.1 corresponding in mass to the fucosylated tryptic peptide T40, which contains glycosylation sites N428(P)/431, and has 2 GlcNAc-Gal units and 1 SA unit. As can be seen from this MS/MS spectrum, fragmentation of the sialylated complex glycopeptides is possible. No ions in this MS/MS spectrum (or any other MS/MS spectrum of a complex glycopeptide), however, could be assigned to the peptide backbone. Thus the spectrum does not provide direct information about the peptide sequence.
Figure 4.
Positive ion nHPLC/ESI/MS/MS spectrum of the peptide [(M+3H)+3 ion of m/z 1419.1] containing complex glycans at sites N428(P)/431 after transformation of all ions into the single charge state spectrum using MaxEnt. GlcNAc = N-acetylglucosamine; GlcNAc-Gal = N-acetylglucosamine and galactose; SA = sialic acid; Man=Mannose.
Based on the low relative abundance of ions for the sialylated peptides, suppression effects in the positive ion mode may be decreasing the sensitivity of these peptides. Increasing numbers of sialic acid residues on peptides may lead to an overall negative net charge, and, thus, result in a lack of observation of these peptides. Therefore, negative ion nHPLC/ESI/MS analyses were performed in order to obtain better coverage of the complex glycan sites. Under these conditions, however, no complex glycans could be identified (data not shown).
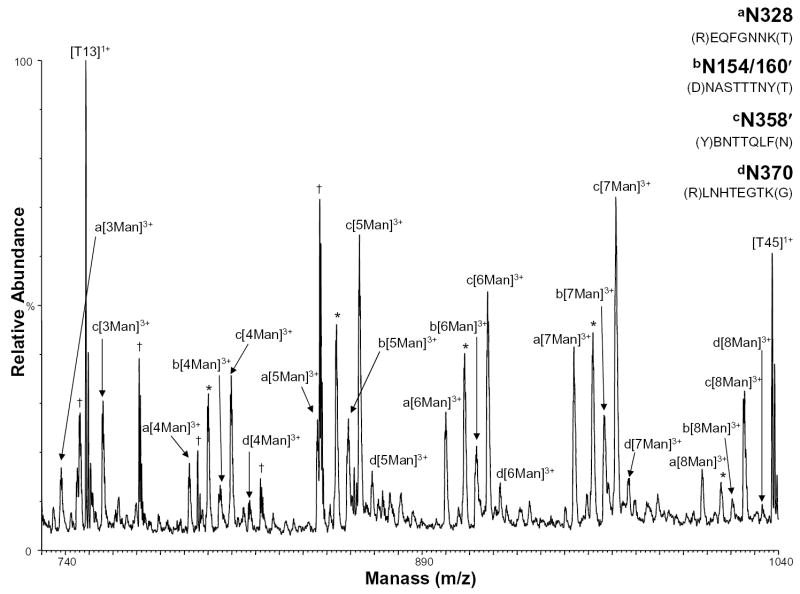
Because one of the glycosylation sites was found to contain high mannose glycans in this study, but was previously characterized to contain complex glycans [13], the high mannose glycosylation sites were further investigated using the nHPLC/ESI/MS approach. Figure 5 shows the positive ion nHPLC/ESI/MS spectra of two glycopeptides in which the glycan structure was previously determined to be high mannose, but the number of mannoses had not been determined and two glycosylated peptides found in partial peptides, which arise from anomalous cleavages. The mass spectrum of tryptic peptide T32 (Figure 5, labeled as ion series a) shows that the glycan at N328 has 3-8 mannoses present, where previously the number of mannoses could not be determined. The mass spectrum of the partial peptide T14′ (Figure 5, labeled as ion series b), residues 154-161 (unconfirmed by MS/MS due to the lack of peptide sequence ions), which contains glycosylation sites N154/160′, shows 4-8 mannoses present. The site where this glycan chain in the peptide sequence was located could not be distinguished between the N154 and N160 sites. However, based on theoretical calculations, it is likely that only one site is occupied and not both because the observed mass is less than the theoretical mass of the glycopeptide with the core glycosylation at both sites. The full-length T14 peptide (amino acids 146 to 165) was also observed in these nHPLC/ESI/MS analyses as having 0-6 mannoses present. Using MALDI/MS, this same peptide was previously characterized by our group to only contain complex glycans [13]. In Figure 5, the ions labeled as series c show the anomalously cleaved partial peptide T34′ (which contains residues 357-363 and the consensus glycosylation site N358′) as having 3-8 mannoses present. The corresponding full-length peptide (which contains the consensus site N358 as well as site N364) was characterized previously as having 11-18 mannoses [13]. In Figure 5, the ions labeled as series d show the mass spectrum of peptide T35 with site N370 shows 4-8 mannoses in the glycan chain, which had not been determined previously.
Figure 5.
Positive ion nHPLC/ESI/MS spectra of a peptide with high mannoses and the anomalous cleavage partial peptides containing high mannose consensus sites a) N328, b) N154/160′, c) N358′, and d) N370. The ′ indicates a partial peptide. For figure clarity, the peaks corresponding to high mannose consensus site N304 are labeled with an *. Other tryptic peptides are labeled. Peaks that were not identified are labeled with a †. The mannoses are labeled depending on the number present, where Man=mannose. The superscripted number is the charge state.
Table 2 shows the summary of glycosylation sites where high mannose (HM) glycans were found in these experiments. The corresponding tryptic peptides containing these glycosylation sites, the masses of the glycopeptides, the number of mannoses observed, as well as their corresponding relative abundances are also shown. The relative abundances were determined in the same way as for Table 1. It is not known why higher numbers of mannoses cannot be seen that were present in our previous work for some of the peptides. One possibility is that the gp120 being used is from a different lot. The observation of an abundant nine mannose glycan at higher m/z (T6, N99) argues against a dramatic decrease in sensitivity for nine mannose glycosylated peptides, and the observation of relatively abundant eight mannose glycopeptides make suppression effects relatively unlikely.
Table 2.
Heterogeneity and Relative Abundances Determined for High Mannose Glycans Using nHPLC/ESI/MS
| Glycopeptide Mass | |||||
|---|---|---|---|---|---|
| Site | Tryptic Peptide | aTheo. | bObs. | cMan | Relative Abundance(%) |
| N99 | T6 d(K)LTPLBVTLNBTDLGK(A) | 3570.5 | 3569.5 | 6 Man | 36 |
| 3732.5 | 3731.6 | 7 Man | 33 | ||
| 3894.6 | 3893.5 | 8 Man | 64 | ||
| 4056.6 | 4055.5 | 9 Man | 37 | ||
| 4218.7 | 4217.8 | 9 Man + Glc | 91 | ||
| 4380.7 | 4379.9 | 9 Man + 2Glc | 100 | ||
| eN154/160′ | T14′ (D)NASTTTNY(T) | 2411.4 | 2410.4 | 4 Man | 55 |
| 2573.4 | 2572.2 | 5 Man | 93 | ||
| 2735.5 | 2734.1 | 6 Man | 79 | ||
| 2897.5 | 2896.8 | 7 Man | 100 | ||
| 3059.6 | 3058.7 | 8 Man | 34 | ||
| N154/160 | T14 (R)NLDVVPIDNASTTTNYTNYR(L) | 3163.8 | 3164.9 | Peptide + Core | 4 |
| 3325.8 | 3326.0 | 1 Man | 33 | ||
| 3487.9 | 3488.1 | 2 Man | 54 | ||
| 3649.9 | 3650.1 | 3 Man | 100 | ||
| 3812.0 | 3812.1 | 4 Man | 32 | ||
| 3974.0 | 3974.1 | 5 Man | 47 | ||
| 4136.1 | 4136.3 | 6 Man | 58 | ||
| N170 | T15 (R)LIHBNR(S) | 2839.1 | 2840.5 | 7 Man | 45 |
| 3001.1 | 3002.4 | 8 Man | 80 | ||
| 3163.2 | 3164.1 | 9 Man | 100 | ||
| N203 | T18 (K)BNNK(T) | 2075.7 | 2074.8 | 4 Man | 28 |
| 2237.8 | 2236.6 | 5 Man | 75 | ||
| 2399.8 | 2398.4 | 6 Man | 70 | ||
| 2561.9 | 2561.2 | 7 Man | 100 | ||
| 2723.9 | 2723.0 | 8 Man | 83 | ||
| 2886.0 | 2885.7 | 9 Man | 95 | ||
| N214/235 | T20 (K)GPBTNVSTVQBTHGIRPIVSTQLLLNGSLAEEEVVIR(S) | 4943.9 | 4944.4 | Peptide + Core | 22 |
| 5106.0 | 5107.2 | 1 Man | 24 | ||
| 5268.0 | 5269.2 | 2 Man | 28 | ||
| 5430.1 | 5431.1 | 3 Man | 28 | ||
| 5592.2 | 5593.1 | 4 Man | 50 | ||
| 5754.2 | 5755.3 | 5 Man | 75 | ||
| 5916.3 | 5917.1 | 6 Man | 100 | ||
| N304 | T28 (K)AHBNISR(A) | 2397.9 | 2398.5 | 4 Man | 70 |
| 2559.9 | 2559.3 | 5 Man | 100 | ||
| 2722.0 | 2722.1 | 6 Man | 85 | ||
| 2884.1 | 2883.8 | 7 Man | 95 | ||
| 3046.1 | 3044.5 | 8 Man | 25 | ||
| N328 | T32 (R)EQFGNNK(T) | 2213.9 | 2212.7 | 3 Man | 40 |
| 2375.9 | 2374.5 | 4 Man | 46 | ||
| 2537.9 | 2536.3 | 5 Man | 67 | ||
| 2700.0 | 2699.1 | 6 Man | 65 | ||
| 2862.0 | 2860.9 | 7 Man | 100 | ||
| 3024.1 | 3022.6 | 8 Man | 38 | ||
| N334 | T33 (K)TIVFNQSSGGDPEIVMHSFNBR(G) | 3551.1 | 3550.2 | 1 Man | 25 |
| 3713.2 | 3712.4 | 2 Man | 28 | ||
| 3875.2 | 3874.5 | 3 Man | 56 | ||
| 4037.3 | 4036.5 | 4 Man | 55 | ||
| 4199.4 | 4198.4 | 5 Man | 100 | ||
| 4361.4 | 4360.4 | 6 Man | 66 | ||
| N358′ | T34′ (Y)BNTTQLF(N) | 2262.5 | 2263.5 | 3 Man | 45 |
| 2424.5 | 2425.4 | 4 Man | 53 | ||
| 2586.6 | 2587.2 | 5 Man | 88 | ||
| 2748.6 | 2748.9 | 6 Man | 69 | ||
| 2910.7 | 2910.7 | 7 Man | 100 | ||
| 3072.7 | 3072.7 | 8 Man | 39 | ||
| N370 | T35 (R)LNHTEGTK(G) | 2439.0 | 2439.3 | 4 Man | 74 |
| 2601.0 | 2601.6 | 5 Man | 96 | ||
| 2763.1 | 2763.8 | 6 Man | 87 | ||
| 2925.2 | 2925.7 | 7 Man | 100 | ||
| 3087.2 | 3087.6 | 8 Man | 80 | ||
| N378 | T36 (K)GNDTIILPBR(I) | 2699.1 | 2698.1 | 4 Man | 65 |
| 2861.2 | 2861.8 | 5 Man | 100 | ||
| 3023.2 | 3021.6 | 6 Man | 38 | ||
| 3185.3 | 3184.2 | 7 Man | 17 | ||
Theo. = theorectical
Obs. = observed
Man = mannose
B = carboxymethylcysteine
′= Partial peptide
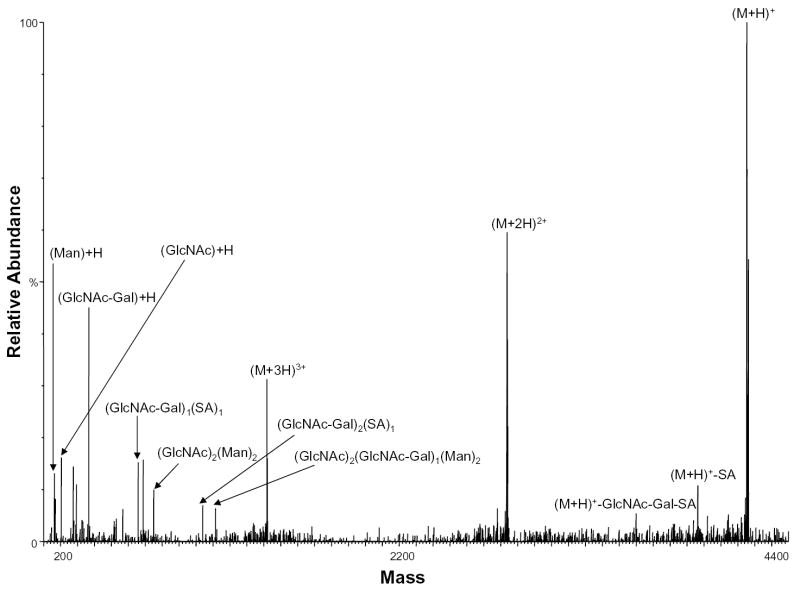
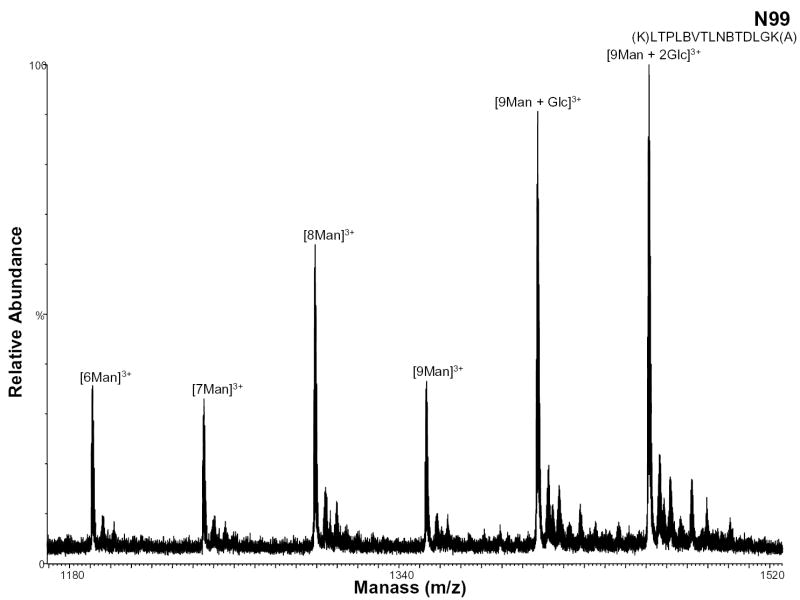
During protein high mannose glycosylation, a precursor oligosaccharide composed of N-acetylglucosamine (GlcNAc), mannose (Man), and glucose (Glc) is transferred to proteins in the endoplasmic reticulum (ER). The diversity of these N-linked oligosaccharide structures on mature glycoproteins results from extensive modification of the precursor structure. While still in the ER, the glucose residues are quickly removed from the oligosaccharides of most glycoproteins. This process known as oligosaccharide “trimming” continues in the Golgi apparatus, and thus the glucose is not seen on the glycoprotein [25]. Figure 6 shows the negative ion nHPLC/ESI/MS spectrum of a high mannose glycopeptide, T6, which was not fully processed. These data show that high mannose glycosylation of this peptide occurs (most likely at residue N99 with 9 mannoses present). The ions of m/z 1406.74 and 1460.75 correspond to the tryptic peptide with nine mannoses plus one and two Glcs, respectively, indicating that complete “trimming” did not occur.
Figure 6.
Negative ion nHPLC/ESI/MS spectrum of the glycopeptide containing high mannose glycans at site N99. This carbohydrate was not fully processed and still contains 2 glucoses. The mannoses are labeled depending on the number present, where Man=mannose and Glc=glucose. In the amino acid sequence B= carboxymethylcysteine.
Conclusions
nHPLC/ESI/MS was used to characterize the glycosylation and extent of sialylation of HIV-ISF2 gp120. Specifically, we have shown that both sialylated complex glycans and high mannose glycans are present. We were able to determine the sialylation profile of eight of the fifteen sites that have been shown to contain complex glycans. The glycosylation sites for complex glycans which could be determined are N99, N110, N170, N249, N274, N415 and N428(P)/431. The other consensus complex sites that could not be characterized are N57, N124/128, N154/160, N235, and N378. A number of factors may be involved in our inability to profile the extent of sialylation of the remaining complex glycopeptides. A combination of all these factors which may have contributed include high mass tryptic peptides (some of which have m/z greater than 5000), multiple glycosylation sites within one peptide, the large number of negatively charged groups present, and possible incomplete digestion.
Since a previously determined complex glycan site was observed to be high mannose as well in these analyses, we investigated the high mannose glycans and were able to determine the number of mannoses present on thirteen of the nineteen sites containing high mannose glycans. These consensus sites are N99, N154/160, N170, N203, N214/235, N304, N328, N334, N358, N370, and N378. Glycosylation sites N154/160 and N358 were observed in two peptides with anomalous cleavage sites and had not been previously observed or characterized. Since most glycans on mature glycoproteins are first processed, it is interesting that glycosylation site N99 was observed as still possessing one and two glucose molecules, thus, indicating this glycan was not fully processed.
Due to the complexity of the gp120 tryptic digest mixture, it is possible that suppression or low protonation efficiency of the negatively ionized sialic acid residues still occurs. This appears to be leading to the suppression of the higher mass glycopeptides by the presence of the abundant lower mass peptides. Therefore, the sensitivity of the sialylated peptides is decreased. The maximum number of SA residues observed was three. An increase in the number of SA residues on a peptide may lead to a net negative charge of the peptide, thereby, resulting in these peptides being not observed in the positive ion MS spectra. No sialylated peptides, however were identified in the negative ion nHPLC/ESI/MS analyses of the digest. Future work will focus on alternative proteolysis steps and/or affinity purification of the glycopeptides. As demonstrated here, nHPLC/ESI/MS can be used as a method to characterize the type of glycan present in a glycopeptide, as well as to determine the extent of sialylation (within the experimental limits). These analyses in combination with previous efforts [13] have now allowed the characterization of all 26 consensus glycosylation sites on HIV-ISF2 gp120.
Acknowledgments
We would like to thank Dr. Christine Hager-Braun for all of her assistance with this project.
References
- 1.Luciw, P. A., Human immunodeficiency viruses and their replication. In Fundamental Virology, 3rd ed., Fields, B.N.; Chanock, R.M.; Knipe, D.M.; Howley, P.M., Eds.; Lippincott-Raven Publishers: Philadelphia, PA, 1996; p 845–897.
- 2.Polzer S, Dittmar MT, Schmitz H, Meyer B, Müller H, Kräusslich HG, Schreiber M. Loss of N-linked glycans in the V3-loop region of gp120 is correlated to an enhanced infectivity of HIV-1. Glycobiology. 2001;11:11–19. doi: 10.1093/glycob/11.1.11. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Rey-Cuille MA, Hu SL. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res Hum Retroviruses. 2001;17:1473–1479. doi: 10.1089/08892220152644179. [DOI] [PubMed] [Google Scholar]
- 4.Polzer S, Dittmar MT, Schmitz H, Schreiber M. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology. 2002;304:70–80. doi: 10.1006/viro.2002.1760. [DOI] [PubMed] [Google Scholar]
- 5.Wyatt R, Moore J, Accola M, Desjardins E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeyarajah S, Parker CE, Sumner MT, Tomer K. Matrix-assisted laser desorption ionization/mass spectrometry mapping of human immunodeficiency virus-gp120 epitopes recognized by a limited polyclonal antibody. J Am Soc Mass Spectrom. 1998;9:157–165. doi: 10.1016/s1044-0305(97)00247-x. [DOI] [PubMed] [Google Scholar]
- 8.Luckow VA, Summers MD. Trend in the development of baculovirus expression vectors. Bio-technol. 1988;6:47–55. [Google Scholar]
- 9.Biller M, Bolmstedt A, Hemming A, Olofsson S. Simplified procedure for fractionation and structural characterization of complex mixtures of N-linked glycans, released from HIV-1 gp120 and other highly glycosylated viral proteins. J Virol Methods. 1998;76:87–100. doi: 10.1016/s0166-0934(98)00127-x. [DOI] [PubMed] [Google Scholar]
- 10.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuochi T, Matthews TJ, Kato M, Hamako J, Titani K, Soloman J, Feizi T. Diversity of oligosaccharide structures on the envelope glycoprotein gp120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J Biol Chem. 1990;265:8519–8524. [PubMed] [Google Scholar]
- 12.Mizuochi T, Spellman MW, Larkin M, Solomon J, Basa LJ, Feizi T. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem J. 1988;254:599–603. doi: 10.1042/bj2540599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;9:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 14.Ezkowitz RAB, Kuhlman M, Groopman JE, Byrn RA. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J Exp Med. 1989;169:185–196. doi: 10.1084/jem.169.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart ML, Saifuddin M, Uemura K, Bremer EG, Hooker B, Kawasaki T, Spear GT. High mannose glycans and sialic acid on gp120 regulate binding of mannose-binding lectin (MBL) to HIV type 1. AIDS Res Hum Retroviruses. 2002;18:1311–1317. doi: 10.1089/088922202320886352. [DOI] [PubMed] [Google Scholar]
- 16.Haurum JS, Thiel S, Jones IM, Fischer PB, Laursen SB, Jensenius JC. Complement activation upon binding of mannan-binding protein to HIV envelope glycoproteins. AIDS. 1993;7:1307–1313. doi: 10.1097/00002030-199310000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Saifuddin M, Hart ML, Gewurz H, Zhang Y, Spear GT. Interaction of mannose-binding lectin with primary isolates of human immunodeficiency virus type 1. J Gen Virol. 2000;81:949–955. doi: 10.1099/0022-1317-81-4-949. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, Shioda T, Moriya C, Xin X, Hasan MK, Miyake K, Shimada T, Nagai Y. Infectivities of human and other primate lentiviruses are activated by desialylation of the virion surface. J Virol. 1996;70:7462–7470. doi: 10.1128/jvi.70.11.7462-7470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RE, Talhouk JW, Brown EE, Edgar SE. The significance of hypersialylation of dipeptidyl peptidase IV (CD26) in the inhibition of its activity by tat and other cationic peptides. AIDS Res Hum Retroviruses. 1998;14:851–868. doi: 10.1089/aid.1998.14.851. [DOI] [PubMed] [Google Scholar]
- 20.Bolmstedt A, Biller M, Hansen JES, Moore JP, Olofsson S. Demonstration of peripheral fucose units in N-linked glycans of human immunodeficiency virus type 1 gp120: effects on glycoprotein conformation. Arch Virol. 1997;142:2465–2481. doi: 10.1007/s007050050255. [DOI] [PubMed] [Google Scholar]
- 21.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nature Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 22.Küster B, Wheeler SF, Hunter AP, Dwek RA, Harvey DJ. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high performance liquid chromatography. Anal Biochem. 1997;250:82–101. doi: 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- 23.Küster B, Krogh TN, Mørtz E, Harvey DJ. Glycosylation analysis of gel-separated proteins. Proteomics. 2001;1:350–361. doi: 10.1002/1615-9861(200102)1:2<350::AID-PROT350>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 24.Bozue JA, Tullius MV, Wang J, Gibson BW, Munson S., Jr Haemophilus ducreyi produces a novel sialyltransferase. J Biol Chem. 1999;274:4106–4114. doi: 10.1074/jbc.274.7.4106. [DOI] [PubMed] [Google Scholar]
- 25.Alberts, B.; Bray, D.; Lewis, J.; Raff, M.; Roberts, K.; Watson, J.D. Intracellular sorting and the maintenance of cellular compartments. In Molecular Biology of the Cell, 2nd ed., Robertson, M., Ed.; Garland Publishing, Inc.: New York, NY, 1989; p 405–479.