Abstract
Production of type IV bundle-forming pili (BFP) by enteropathogenic Escherichia coli (EPEC) requires the protein products of 12 genes of the 14-gene bfp operon. Antisera against each of these proteins were used to demonstrate that in-frame deletion of individual genes within the operon reduces the abundance of other bfp operon-encoded proteins. This result was demonstrated not to be due to downstream polar effects of the mutations but rather was taken as evidence for protein-protein interactions and their role in the stabilization of the BFP assembly complex. These data, combined with the results of cell compartment localization studies, suggest that pilus formation requires the presence of a topographically discrete assembly complex that is composed of BFP proteins in stoichiometric amounts. The assembly complex appears to consist of an inner membrane component containing three processed, pilin-like proteins, BfpI, -J, and -K, that localize with BfpE, -L, and -A (the major pilin subunit); an outer membrane, secretin-like component, BfpB and -G; and a periplasmic component composed of BfpU. Of these, only BfpL consistently localizes with both the inner and outer membranes and thus, together with BfpU, may articulate between the Bfp proteins in the inner membrane and outer membrane compartments.
The virulence of enteropathogenic Escherichia coli (EPEC) for orally challenged volunteers (3) requires genes encoded on the 69-kb EPEC adherence factor (EAF) plasmid (31) and within the chromosomal locus of enterocyte effacement (7). The EAF plasmid carries a 14-gene operon that encodes the bundle-forming pilus (BFP), a member of the widely distributed type IV family of pilin proteins (28, 29). This operon is required for the production of BFP filaments and virulence; in addition, functional studies of bfp operon mutants show that expression of the operon confers two readily assayable in vitro phenotypes. The localized adherence (LA) phenotype is characterized by circumscribed clusters of bacteria attached to the surface of cell culture monolayers (6, 16). The autoaggregation phenotype (AA) is evident when an overnight culture of dispersed EPEC is inoculated into tissue culture medium; 45 to 60 min later, the bacteria coalesce into dynamic, spherical assemblies which disaggregate after 3 to 4 h, again yielding a suspension of single cells (3).
The bfp operon, together with its transcriptional activator BfpTVW/PerABC, which is located elsewhere on the EAF plasmid, (9, 32), is sufficient for expression of BFP filaments and the LA and autoaggregation phenotypes when it is harbored in E. coli strains that normally do not express type IV pili. Thus, the operon's 14 genes appear to encode the minimal set of functions, exclusive of transcription factors and the periplasmic protein, DsbA (34), that are specifically required for BFP biogenesis and function.
Details of the environmentally responsive transcriptional regulation of the bfp operon are emerging (21); however, the interactions among the proteins expressed by the operon remain obscure. We postulate that many, if not all, of these proteins coalesce into an assembly complex necessary for the elaboration and functional attributes of BFP.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this work are described in Table 1. The growth conditions have been described previously (3). Briefly, cultures were grown at 37°C with shaking in DME (Dulbecco's modified Eagle's medium containing 4.5% glucose) starting from a 1:100 dilution of an aerated standard overnight culture (a bacterial suspension from an overnight Luria-Bertani broth culture resuspended in phosphate-buffered saline to an optical density at 600 nm of 1.8). Typically, cultures were harvested after 4 h of growth (in the mid-log to late log phase).
TABLE 1.
Bacterial strains used in this study
| Strain (abbreviation) | Relevant characteristics | Reference(s) |
|---|---|---|
| B171-8 (−8) | B171 lacking the 90-kb Tetr-Strr plasmid; retains the EAF plasmid; virulent in human challenge study | 3, 8 |
| B171-8dbfp (dbfp) | B171-8 EAF plasmid; bfp operon mutant; expresses none of the Bfp proteins | 22 |
| B171-8ΔAcm (ΔAcm) | B171-8 EAF plasmid mutant specifically lacking the bfpA gene product, BfpA; 133 of 193 amino acids removed and replaced by the Cmr gene | 3 |
| B171-8ΔG (ΔG) | B171-8 EAF plasmid mutant specifically lacking the bfpG gene product, BfpG; 94 of 133 amino acids deleted | This study |
| B171-8ΔB (ΔB) | B171-8 EAF plasmid mutant specifically lacking the bfpB gene product, BfpB; 9 of 453 amino acids deleted; two stop codons introduced into each reading frame | 22 |
| B171-8ΔC (ΔC) | B171-8 EAF plasmid mutant specifically lacking the bfpC gene product, BfpC; 371 of 402 amino acids deleted | This study |
| B171-8ΔU (ΔU) | B171-8 EAF plasmid mutant specifically lacking the bfpU gene product, BfpU; 131 of 156 amino acids deleted | This study |
| B171-8ΔD (ΔD) | B171-8 EAF plasmid mutant specifically lacking the bfpD gene product, BfpD; 400 of 534 amino acids deleted | 3 |
| B171-8ΔE (ΔE) | B171-8 EAF plasmid mutant specifically lacking the bfpE gene product, BfpE; 237 of 292 amino acids deleted | This study |
| B171-8ΔF (ΔF) | B171-8 EAF plasmid mutant specifically lacking the bfpF gene product, BfpF; 90 of 331 amino acids deleted | 3 |
| B171-8ΔP (ΔP) | B171-8 EAF plasmid mutant containing an in-frame deletion in bfpP; 113 of 249 amino acids deleted | 35; this study |
| B171-8ΔH (ΔH) | B171-8 EAF plasmid mutant containing an in-frame deletion in bfpH; 141 of 344 amino acids deleted | This study |
| B171-8ΔI (ΔI) | B171-8 EAF plasmid mutant specifically lacking the bfpI gene product, BfpI; 60 of 181 amino acids deleted | This study |
| B171-8ΔJ (ΔJ) | B171-8 EAF plasmid mutant specifically lacking the bfpJ gene product, BfpJ; 153 of 183 amino acids deleted | This study |
| B171-8ΔK (ΔK) | B171-8 EAF plasmid mutant specifically lacking the bfpK gene product, BfpK; 81 of 163 amino acids deleted | This study |
| B171-8ΔL (ΔL) | B171-8 EAF plasmid mutant specifically lacking the bfpL gene product, BfpL; 75 of 149 amino acids deleted | This study |
Recombinant DNA techniques.
All DNA manipulations were performed by standard molecular and genetic techniques (23). Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.) and were used according to the manufacturer's recommendations. PCR amplifications were performed with PCR Supermix from Stratagene (San Diego, Calif.) as recommended by the manufacturer.
Construction of the Bfp mutant strains.
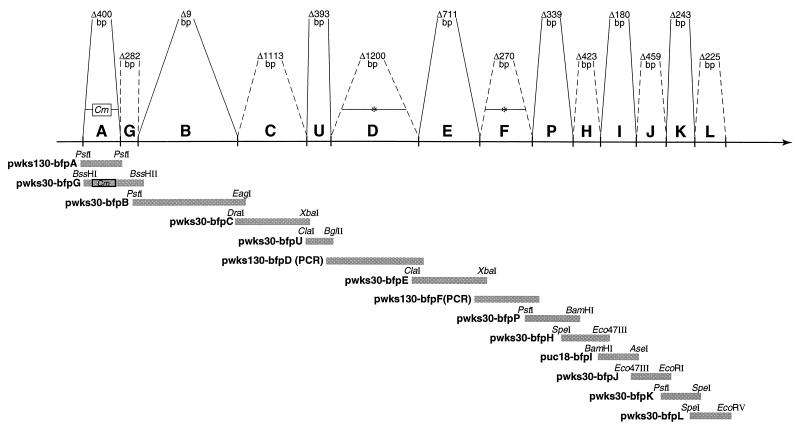
In-frame deletions were constructed as described previously (3, 22). Convenient restriction sites in each gene were used to create in-frame deletions in wild-type fragments containing the relevant bfp gene; alternatively, when no convenient restriction sites were in frame, PCR was used to generate deletions (Fig. 1). The deleted genes and approximately 400 to 900 bp of flanking sequence were cloned into a modified version of suicide plasmid pGP704 (15). Suicide vector-driven homologous recombination was used to replace the wild-type locus on the EAF plasmid with the desired in-frame deletion mutation. The resultant deletions were validated by restriction fragment analysis and, when necessary, sequence analysis across the junction sites. All restriction sites and the relative sizes of the bfp operon open reading frames (ORFs) refer to the previously published sequence of the bfp operon reported by Sohel et al. (28) (GenBank accession no. U27184).
FIG. 1.
Diagrammatic representation of the BFP operon on the EAF plasmid of EPEC strain B171-8. The horizontal solid line represents the 14-gene operon and shows the direction of transcription; the individual genes (A through L) are indicated above this line. The information above the horizontal line indicates the number of base pairs deleted in each of the individual in-frame deletions generated on the EAF plasmid; the boundaries of the genes are indicated by solid and dashed lines. Below the horizontal line the individual genes (and the plasmids which carry them) that were used to complement the deletions are indicated. The sizes of the complementing fragments (relative to the ORFs) and the restrictions sites that define the fragments are indicated.
Complementation of deletion mutations.
Wild-type fragments containing each of the bfp genes were cloned into the low-copy-number vector pWKS130 (33), except as indicated in Fig. 1. Plasmids containing individual bfp genes were electroporated into the corresponding B171-8 mutants and selected for the appropriate antibiotic resistance. The complemented bacteria were phenotypically compared with the appropriate deletion mutant, with the operon knockout, dbfp, and with wild-type B171-8 with respect to pilus fiber production (by electron microscopy) and function (by LA and autoaggregation assays).
LA and autoaggregation assays.
Analyses of the BFP-dependent LA and autoaggregation phenotypes were performed as described previously (3).
Preparation and purification of anti-peptide antibodies.
Hydrophobicity and surface accessibility predictions (5, 14) were used to identify two short regions from each Bfp protein that were predicted to be potentially antigenic (Table 2). Peptides corresponding to these regions were synthesized by using a Milligen 9050 Pep synthesizer. The synthesized peptides were conjugated to bovine gamma globulin, and the conjugates were used for production of polyclonal antisera in rabbits (Josman Laboratories, Napa, Calif.).
TABLE 2.
Synthetic peptides used to raise anti-Bfp antibodies
| Protein | Peptide position (amino acids) | Amino acid sequence of peptide |
|---|---|---|
| BfpG | 14-37 | CFTQAQESANKNEKLFSGDTTSVA |
| BfpC | 197-232 | RDKDILNEEPSPAVSEEVRNIPWEG KSKPGSLFSKC |
| BfpU | 27-50 | SGKNDSRKEENKLPDVDLEKYKDE |
| BfpD | 338-352 | TEERKKYLFSRDNEI |
| BfpE | 102-118 | DIIYMNDTKKKVKGALAG |
| BfpF | 99-114 | KLKLPVPSLDSLGFSE |
| BfpI | 76-104 | DGVISSVKSNINSGNKIKVDVKKAGK YVS |
| BfpJ | 155-162 | CDRGGYDF |
| BfpK | 44-82 | CVVATDMNSVANIIISSAVGYKTGKY DVSSITNDNDFNK |
| BfpL | 131-169 | CSINDNSGIAIIKTFNKTG |
The synthesized peptides were also conjugated to keyhole limpet hemocyanin, and the conjugates were coupled to cyanogen bromide-activated Sepharose (Amersham-Pharmacia Biotech) according to the manufacturer's instructions. The resulting affinity columns were used to affinity purify the rabbit antisera as described by Harlow and Lane (10). Purification was monitored by an enzyme-linked immunosorbent assay by using keyhole limpet hemocyanin-conjugated peptide as the antigen (10).
Immunoblotting.
Western blot analysis of protein expression was performed as described previously (22). For analysis of protein stability in the various Bfp mutants, equal masses of total protein, typically 15 μg, were loaded in the lanes. For analysis of subcellular localization, the cell equivalent for each compartment was loaded (i.e., the amounts of protein were proportional to the fractions of the total protein that the cellular compartments typically comprise; typically, 4% of the total protein is periplasmic protein [18], 9% of the total protein is inner membrane protein, 6% of the total protein is outer membrane protein [12], and the remaining 81% of the total protein is cytoplasmic protein). Affinity-purified anti-peptide rabbit antibodies specific to the Bfp proteins were used as primary antibodies. Horseradish peroxidase-labeled goat anti-rabbit secondary antibody was used at a 1:5,000 dilution, and the reaction preparation was developed by using enhanced chemiluminescence as recommended by the manufacturer (NEN Life Sciences).
Cellular fractionation protocols.
Two protocols were used to separate bacterial cell compartments. The first generated inner membranes and outer membranes. The second provided samples of all four compartments (cytoplasm, periplasm, inner membranes, and outer membranes). These separation protocols are described below.
Membrane fractions were prepared from cells that had not been treated with lysozyme and that had been disrupted by passage through a French pressure cell rather than by sonication (17). Inner and outer membranes were separated by ultracentrifugation through a descending sucrose step gradient as described by Schmidt et al. (27). The completeness of the separation by this method was assessed by recentrifuging the separated membrane fractions in a flotation sucrose gradient as described by Possot et al. (20). Fractions from the step gradient and floatation gradient were assayed for cross contamination by using 2-keto-3-deoxyoctonate (13) and succinate dehydrogenase (19) assays, as well as immunoblotting with antisera to Bfp proteins previously shown to fractionate with either the inner or outer membrane but not with both; no cross contamination was found between the membrane fractions of these preparations.
Alternatively, complete cellular fractionation was accomplished by using a modification of the method of Hultgren et al. (11). Cultures (500 ml) were pelleted after 4 h of growth in DME, yielding approximately 1 g (wet weight) of cells. The cell pellet was resuspended in 8 ml of ice-cold 20% (wt/vol) sucrose in 20 mM Tris (pH 8.0), to which 80 μl of 0.5 M EDTA and then 80 μl of a 15-mg/ml lysozyme solution were added. The suspension was incubated on ice for 40 min. MgCl2 was added to a final concentration of 75 mM, and the suspension was centrifuged at 15,000 × g at 4°C for 20 min. The resulting supernatant was saved and stored at −20°C as the periplasmic fraction. The cell pellet was resuspended in 16 ml of ice-cold 20% sucrose-20 mM Tris (pH 8.5)-75 mM MgCl2. The resulting suspension was sonicated four times (30 s each) by using a Branson Sonifier with a microtip (setting 7) on ice and then subjected to two rounds of freezing and thawing to thoroughly lyse the spheroplasts. The lysate was centrifuged at 5,000 × g at 4°C for 10 min to pellet any unbroken cells, which were discarded, and then recentrifuged at 260,000 × g at 4°C for 2 h in a Beckman SW41 rotor. The resulting supernatant was removed and stored at −20° as the cytoplasmic fraction. The total membrane pellet was resuspended in 15 ml of ice-cold 10 mM HEPES (pH 8), and then 0.75 ml of 10% Sarkosyl was added. The suspension was incubated on a gyrating platform for 20 min at room temperature and then was centrifuged at 367,000 × g for 1 h at room temperature. The supernatant was saved at −20°C as the inner membrane fraction. The pellet was washed twice in cold 10 mM HEPES (pH 8), resuspended in 2 ml of 10 mM HEPES (pH 8), and saved at −20° as the outer membrane fraction. Fractions were validated as described above; the protein concentrations in all fractions were determined with bicinchoninic acid (Pierce).
RESULTS
Phenotype characterization of in-frame mutations of each of the 14 bfp operon ORFs.
Previous studies have reported that BfpA (8), BfpG (1, 27), BfpB (22), BfpC (1), BfpD (1), BfpE (4), and BfpP (35) are required for biogenesis of ultrastructurally normal BFP filaments and for expression of two BFP functional phenotypes, LA and autoaggregation. By contrast, BfpF mutants were found to produce filaments that appeared normal, but these mutants exhibited an aberrant autoaggregation phenotype (2, 3) and, although hyperpiliated, had reduced virulence for orally inoculated volunteers (3). A BfpH mutant was reported to be normal with respect to pilus morphology and the LA and autoaggregation phenotypes (1). To corroborate these findings and to determine if any of the five remaining, untested bfp operon-encoded proteins (BfpU, -I, -J, -K, and -L) are required for BFP filament biogenesis and phenotypes, each of the 14 ORFs of the operon was individually disrupted (Fig. 1), and the resulting mutants were examined by electron microscopy and by performing LA and autoaggregation assays. To avoid potential polar effects on the transcription or translation of downstream ORFs, in-frame mutations were generated, and any such effects were sought by determining if complementation with a wild-type copy of the mutated gene restored BFP biogenesis and function (Fig. 1). The results are presented in Table 3 and corroborate the previously published findings that BfpA, -G, -B, -C, -D, -E, and -P are required for pilus biogenesis. In-frame disruptions of five additional genes, bfpU, -I, -J, -K, and -L, also resulted in mutants that do not produce BFP or exhibit the LA and autoaggregation phenotypes. As previously reported (1), disruption of BfpH did not affect BFP production, morphology, or function.
TABLE 3.
Phenotypes altered by deleting Bfp proteins and their recovery by complementation
| Deletion | Bundles | Autoaggregation | LA | Proteins destabilizeda |
|---|---|---|---|---|
| ΔA cm | Nob,c | Nob,c | Nob,c | None |
| ΔG | Nob,d | Nob,d | Nob,e | BfpU, BfpI, BfpJ |
| ΔB | Nob,f | Noe,f | Noe,f | BfpG, BfpU, BfpI, BfpJ |
| ΔC | Nob,d | Nob,d | Nob,d | BfpG, BfpU, BfpF, BfpI, BfpJ, BfpK |
| ΔU | Nob | Nob | Nob | BfpD, BfpF, BfpI, BfpJ, BfpK |
| ΔD | Noc,e | Noc,e | Noc,e | BfpI, BfpJ, BfpK |
| ΔE | Nog,h | Noe,h | Noe,h | BfpA, all other assayed Bfp proteins |
| ΔF | Yesi,c | Aberrantc | Yesb,c | None |
| ΔP | Noi,j | Nob | Nob | BfpA, BfpI, BfpJ, BfpKk |
| ΔH | Yes | Yes | Yes | BfpU, BfpI, BfpJ, BfpK |
| ΔI | Noi | Nob | Nob | BfpJ, BfpK |
| ΔJ | Nob | Nob | Nob | BfpI, BfpK, BfpL |
| ΔK | Noe | Nob | Nob | BfpI |
| ΔL | Nob | Nob | Nob | BfpI, BfpJ, BfpK |
Boldface type indicates that the protein abundance phenotype was affected more than 10-fold. Lightface type indicates that the protein abundance phenotype was affected approximately threefold.
The functional phenotype was restored by complementation to wild-type levels.
The BFP phenotype reported by Bieber et al. (3) was confirmed in this study.
The BFP phenotype reported by Anantha et al. (1) was confirmed in this study.
The functional phenotype was restored by complementation to levels that were detectable but less than wild-type levels.
The BFP phenotype reported by Ramer et al. (22) was confirmed in this study.
ND, not determined.
The BFP phenotype reported by Blank and Donnenberg (4) was confirmed in this study.
The level of the functional phenotype restored by complementation was not determined.
The BFP phenotype reported by Zhang et al. (35) was confirmed in this study.
The proteins that were destabilized were unprocessed preproteins. The phenotype reported by Bieber et al. (3) for BfpA was confirmed in this study.
Effects of in-frame disruptions of bfp operon ORFs on the abundance of other BFP assembly complex proteins.
To learn more about the possible interactions between bfp operon-encoded proteins, the effect of each ORF-specific mutation on the abundance of other operon-encoded proteins was ascertained by Western blotting. ORF-specific peptide antibodies were used to detect 10 proteins (Table 2). BfpB was detected with affinity-purified antibodies elicited by a BfpB fusion protein (22), and BfpA was detected with an antiserum to the intact pilin (8). Antibodies to two unique BfpH synthetic peptides failed to detect the corresponding protein by Western analysis; no attempt was made to prepare antibodies to BfpP, the prepilin peptidase (35). Only proteins whose abundance was reduced by ≥75% in each of at least three assays were considered to have been significantly affected by a particular mutation (Table 3).
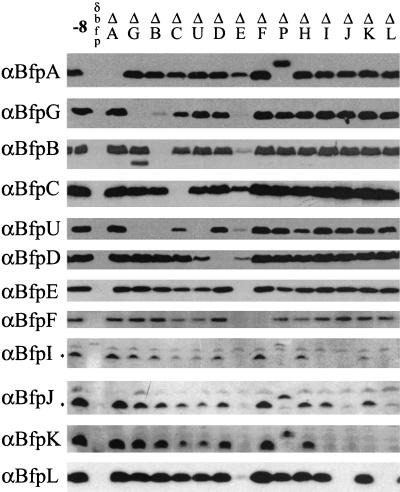
We reasoned that protein-protein interactions within an assembly complex might contribute to the stability of one or more of the proteins that compose that complex. If this is true, then deletion of one protein species might be expected to destabilize other proteins with which it physically interacts in the complex, possibly by exposing them to cellular proteases or otherwise causing them to be degraded by mechanisms related to their normal turnover. A necessary precondition for this experimental approach is proof that disruption of a particular bfp operon gene does not alter the expression of a downstream, nonmutated gene owing to a polar effect. This possibility is refuted by the results shown in Fig. 2, which shows that wild-type levels of proteins encoded by genes downstream of a mutation were seen for most of the deletions (Fig. 1 shows the gene order in the operon). The exceptions are reductions in the levels of proteins for the following bfp deletions: ΔG, reduced levels of BfpU, -I, and -J; ΔB, reduced levels of BfpG, -U, -I, and -J; ΔC, reduced levels of BfpG, -U, -F, -I, -J, and -K; ΔU, reduced levels of BfpD, -F, -I, -J, and -K; ΔD, reduced levels of BfpI, -J, and -K; ΔE, reduced levels of all proteins except BfpA and -C; ΔH, reduced levels of BfpU, -I, -J, and -K; ΔI, reduced levels of BfpJ and -K; ΔJ, reduced levels of BfpI, -K, and -L; ΔK, reduced level of BfpI; and ΔL, reduced levels of BfpI, -J, and -K. However, of these, only ΔJ could be construed to have a true polar effect on BfpK and BfpL (i.e., all of the genes immediately downstream). In all other cases, an intervening gene showed normal levels of expression. Moreover, for each of the deletions mentioned above, including the BfpJ deletion, wild-type levels of proteins (and phenotypes) were restored when the mutation was complemented with a single, specific fragment covering only the deleted gene (data not shown). Finally, for seven of the mutations (ΔB, -C, -E, -H, -J, -K, and -L), the protein(s) showing altered abundance was encoded by a gene upstream of the mutated gene. Thus, its level of transcription and translation would not have been reduced by a polar effect. Taken together, these findings provide compelling evidence that downstream polar effects cannot account for the observed differences in Bfp protein abundance that are described below.
FIG. 2.
Bfp protein abundance as affected by in-frame deletions throughout the BFP operon. Equal amounts of whole-cell lysates from equally dense cultures of the B171-8 strains listed at the top were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes, prepared in parallel, were immunoblotted with the anti-Bfp sera indicated on the left. −8 is wild-type strain B171-8; δdbfp is the operon knockout B171-8dbfp; the other designations refer to the specific in-frame deletions of ORFs in the operon. Asterisks indicate the positions of the processed BfpI- and BfpJ-specific bands.
As demonstrated in Fig. 2 and summarized in Table 3, none of the mutations significantly affected the abundance of BfpA, which encodes pilin, the stoichiometrically major component of the BFP filament. By contrast, the bfpE deletion reduced, by at least 10-fold, the levels of all of the proteins in the operon except BfpA and BfpC, whose levels were decreased only 2- to 3-fold. The ΔB mutation significantly affected the stability of BfpG, as we reported previously (27). The bfpG deletion, while not affecting the abundance of BfpB, was associated with limited cleavage of BfpB, as demonstrated by the appearance of a BfpB antibody-reacting band about 5 kDa smaller than the mature BfpB protein. The results obtained with BfpB and -G corroborate the idea that changes in the abundance or stability of a protein can predict physical associations between proteins because BfpB and BfpG were shown to physically interact by chemical cross-linking and coimmunoprecipitation studies (27). Deletion of the bfpG, -B, or -C gene eliminated or dramatically reduced the abundance of BfpU. Disruption of bfpU decreased the abundance of BfpD. Deletion of bfpC or bfpU significantly destabilized BfpI, -J, and -K. Deletion of bfpG, -B, or -U promoted significant accumulation of lower-molecular-weight BfpC bands, as described below. Taken together, these observations point to several patterns of Bfp protein-protein interactions within the Bfp assembly complex. For example, a pattern of mutual dependence was demonstrated by mutations in genes encoding the BfpJ and BfpL proteins; disruption of bfpJ was associated with the absence of BfpL, and conversely, disruption of bfpL was associated with the absence of BfpJ. Similarly, a reciprocal but less substantial pattern of dependence was observed between BfpI and -K; in ΔI BfpK was less abundant than BfpI was in ΔK. Taken together, these results indicate that there is a possible interaction among BfpI, -J, -K, and -L.
Localization of proteins encoded by the bfp operon.
Four of the Bfp proteins have previously been localized to bacterial cell compartments; BfpB and -G fractionate exclusively with the outer membrane (22, 27), and BfpA and -E are associated mainly with the inner membrane (4), although some BfpA signal is found in outer membrane fractions (1, 22). ORF-specific peptide antibodies to BfpC, -U, -I, -J, -K, and -L were used to localize these six additional proteins to bacterial cellular compartments by Western blotting (Fig. 3a). Localization data for the remaining four proteins are not reported here; BfpD and -F are the subjects of a future report, BfpP has previously been inferred to be located in or on the cytoplasmic membrane (35), and two peptide antisera to BfpH failed to identify the corresponding protein.
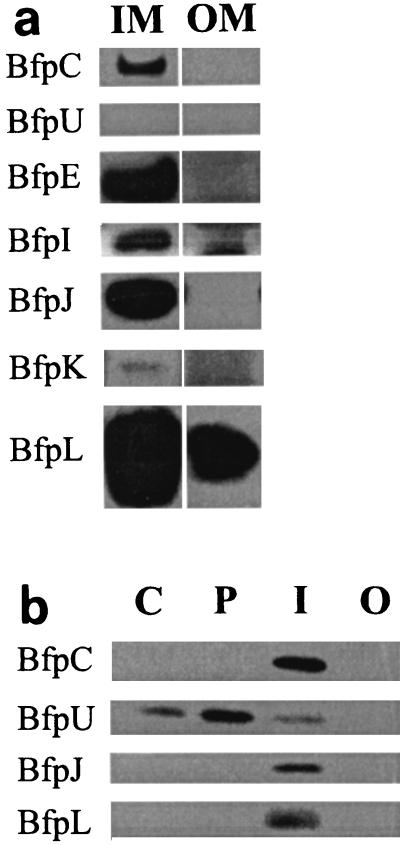
FIG. 3.
Localization of Bfp proteins in bacterial cell compartments. (a) Bfp proteins associated with the inner and outer bacterial membranes. Equal amounts of B171-8 membrane proteins isolated from sucrose step gradients (27) were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes. The membranes were immunoblotted with the anti-Bfp sera indicated on the left. IM, inner membrane fraction; OM, outer membrane fraction. (b) Bfp proteins fractionating with bacterial cell compartments. A single culture of B171-8 was fractionated into cytoplasmic (C), periplasmic (P), inner membrane (I), and outer membrane (O) fractions by the protocol of Hultgren et al. (11). An amount of protein proportional to a cell equivalent (see Materials and Methods) was loaded into each lane. The proteins were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The membranes were immunoblotted with the anti-Bfp sera indicated on the left.
Inner and outer membrane fractions, prepared from French pressure cell-disrupted bacteria, were separated from each other by descending sucrose step gradient centrifugation (27). Equal masses of inner and outer membrane proteins were assayed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting. The results of the fractionation experiments (Fig. 3a) show that BfpC, -I, -J, and -K fractionate with the bacterial inner membrane. BfpL fractionates primarily with the inner membrane, but reproducibly 2 to 10% of BfpL fractionates with the outer membrane. BfpU was not found in either of these membrane fractions.
To assess the purity of the inner and outer membrane fractions, three different assays were performed. First, the French pressure cell-derived fractions were subjected to ascending linear sucrose gradient centrifugation, and the resulting fractions were analyzed by Western blotting. The results (data not shown) corroborated the localization patterns shown in Fig. 3a. Second, 2-keto-3-deoxyoctonate assays were conducted with inner and outer membrane fractions, and they demonstrated that there was no contamination of the isolated inner membranes with outer membranes. This finding was supported by the absence of BfpB (an outer membrane protein) in the inner membrane fraction. Third, BfpE, which was convincingly demonstrated by Blank and Donnenberg (4) to be an integral inner membrane protein, was not found by Western blotting in the outer membrane fractions, demonstrating that the outer membrane fractions were not contaminated by inner membranes. Finally, neither BfpB nor BfpE was found to be associated with the periplasmic and cytoplasmic fractions, demonstrating that these fractions were not contaminated with outer and inner membranes, respectively.
To determine if BfpC, -U, -J, or -L is present in the cytoplasm or periplasm, we employed the lysozyme-detergent fractionation method of Hultgren et al. (11) to isolate outer membranes, inner membranes, periplasm, and cytoplasm. The results of this alternative fractionation protocol, in which we used lysozyme to promote hypotonic lysis and Sarkosyl extraction of the resulting cell envelope (membrane) fraction, demonstrated that BfpC and BfpJ still localized to the inner membrane fractions, with no significant amounts in the cytoplasm, periplasm, and outer membrane fractions (Fig. 3b). However, consistent with its hydrophilic amino acid composition, BfpU was found to localize primarily to the periplasmic fraction, with smaller amounts of the protein fractionating with the cytoplasm and inner membrane fractions. Interestingly, BfpL was not detected in outer membrane fractions prepared by this procedure. This suggests that in contrast to outer membranes prepared by using the French pressure cell and sucrose gradient centrifugation, either lysozyme digestion of the peptidoglycan or Sarkosyl extraction of the outer membrane disrupts the association of BfpL with these fractions (see below).
Anti-BfpC recognizes three protein species.
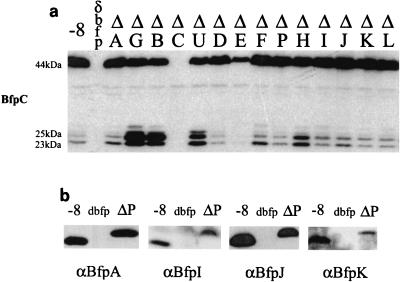
During the course of these experiments, Western blot studies performed with an anti-peptide antibody disclosed that BfpC produced by the wild-type strain migrated as three bands: a quantitatively major ∼44-kDa band that corresponds closely to the 45.4-kDa molecular mass predicted from the bfpC ORF and two quantitatively minor bands at molecular masses of ∼25 and ∼23 kDa (Fig. 4a). This wild-type BfpC pattern was also evident in 13 of the14 bfp operon knockouts; it varied only in the intensities of the lower-molcular-mass bands, which were more abundant in ΔG, ΔB, ΔU, and ΔH and less abundant in ΔD and ΔE (Fig. 4a). We hypothesized that each of the three proteins is encoded by the bfpC ORF because each band was detected by an antibody elicited to an internal BfpC peptide sequence (Table 2) and because each of the three protein species was absent from the BfpC deletion.
FIG. 4.
Processing of Bfp proteins. (a) BfpC proteins distributed in the in-frame deletions throughout the BFP operon. The molecular mass of BfpC predicted from the sequence of the bfpC ORF is 45.5 kDa; a band at about 44 kDa corresponds closely to the predicted size and is presumed to be the mature form of the full-length protein. Two additional proteins with molecular masses of approximately 25 and 23 kDa have the same distribution pattern in the full series of deletions. (b) BfpA, -I, -J, and -K all encode sequences predicted to be recognized by the prepilin peptidase, BfpP. The sizes of these proteins in B171-8 and B171-8ΔP indicate that all four are preproteins in ΔP and are processed to the mature, smaller form in the wild type (−8). None of these proteins was seen in the operon knockout mutant, dbfp.
The BfpC migration pattern shown in Fig. 4a could be due to any of the following effects: an internal translation start site; a degradation artifact arising from exposure of susceptible cleavage sites to cellular proteases during sample preparation; or a processing event that is required for biogenesis of the BFP assembly complex. To assess the possibility that the bfpC ORF contains an internal start site, the first ATG in the bfpC ORF was deleted on the complementing plasmid (Fig. 1). Expressing this plasmid in either the operon knockout, designated dbfp (Table 1), or the ΔC mutant failed to result in the 25- and 23-kDa bands of BfpC, suggesting that the smaller bands are not due to internal start sites (data not shown).
To determine if procedure-dependent degradation could account for the BfpC SDS-PAGE pattern, wild-type bacterial cells were collected and prepared at 4°C (until they were boiled in SDS-containing sample buffer) in buffer either containing or lacking protease inhibitors, which were included at the time of the centrifugation used to collect the bacteria and during resuspension and sonication. To accentuate any proteolytic event, aliquots of the lysates either containing or lacking protease inhibitors were incubated at 37°C for 30 min. Western blot analysis of these lysates revealed no difference in the cleavage patterns or abundance of the three BfpC-specific bands (data not shown). These results suggest that the cleavage pattern is not due to procedure-dependent proteolytic degradation. To determine if this pattern depends on EPEC-specific proteins other than BfpC, bfpC was expressed in the non-EPEC strain DH5α. The wild-type pattern of ∼44-, ∼25-, and ∼23-kDa bands was observed, indicating either that BfpC is autocatalytic or that the protein is cleaved by proteases found in a laboratory strain of E. coli.
Our attempt to deduce the site of proteolytic cleavage described above was inconclusive. A 36-amino-acid peptide (Table 2) corresponding to residues 197 to 232 of the 402-amino-acid full-length protein predicted from the bfpC ORF was used to elicit the BfpC antiserum employed in the Western blot experiments. The location of this peptide within the intact BfpC protein and the size of the two BfpC fragments led to two possible explanations for the observed cleavage pattern. First, the length and amino acid sequence of the immunizing peptide raise the possibility that the peptide encompasses two or more immunogenic epitopes. If it does, each epitope would evoke its own epitope-specific antibody. If these epitopes were located on each side of the BfpC proteolytic cleavage site, then an N-terminal fragment consisting of ∼215 residues (∼25 kDa) and a C-terminal fragment consisting of ∼187 residues (∼23 kDa) would be produced. The sizes of these predicted products conform closely to the estimated masses of the two lower-molecular-mass BfpC-specific bands. Alternatively, incomplete cleavage of BfpC at two or more sites could have produced overlapping peptide fragments corresponding to the two smaller species shown in Fig. 4a.
BfpP is required for processing of BfpA, -I, -J, and -K.
Four bfp operon-encoded proteins contain N-terminal domains predicted to be processed by the prepilin peptidase encoded by bfpP. Processing of BfpA by BfpP has been convincingly demonstrated by Zhang et al. (35). However, the role of BfpP in the processing of three pilin-like proteins, BfpI, -J, and -K, has not been investigated. To examine this issue, the wild-type parent and the bfpP mutant were studied by Western blotting using peptide antibodies to BfpA, -I, -J, and -K. Mutation of bfpP was associated with loss of the mature forms of four proteins, BfpA, -I, -J, and -K, and the appearance of higher-molecular-weight species whose sizes corresponded to the sizes of the four unprocessed proteins (Fig. 4b).
Time course of protein stability.
BFP are transiently expressed on EPEC during mid- to late-logarithmic-phase growth, as shown by electron microscopy, the autoaggregation assay, and gene expression studies (21). To test if Bfp proteins other than BfpB (27) stably accumulate during the transition from the logarithmic phase to the stationary phase and in the stationary phase of growth, aliquots of a wild-type EPEC culture grown in DME were removed after 1.5, 3, 5, 9, and 12 h. Equal amounts of total protein from the whole-cell lysates for all the time points were analyzed by Western blotting by using the 12 anti-Bfp antibodies described above. BfpG, -C, -U, -D, -F, -I, -J, -K, and -L (BfpP and BfpH were not tested) showed a consistent pattern: maximal accumulation at 3 to 5 h (mid- to late log phase), followed by a steady decline in abundance at 9 h and little signal present at 12 h (stationary phase). BfpA, BfpB, BfpE, and processed BfpC were exceptions; they maintained at least 80% of their maximal abundance at 12 h, although BfpE was the least stable of the group (data not shown). These results demonstrate that subcomponents of the assembly complex exhibit both topographical (i.e., localization) differences and temporal (i.e., growth phase) differences.
DISCUSSION
Type IV pili are important virulence factors for many human, animal, and plant pathogens. Although the function of these structures in pathogenesis is an active area of research, little is known about their biogenesis. Like pyelonephritis-associate pili (26) and the type II (for a recent review see reference 24) and III secretion systems (30) of gram-negative organisms, type IV pilus biogenesis and function likely require a multicomponent assembly complex. Such a complex is postulated to consist of components that (i) physically interact, (ii) confer a common function that requires each of the composing elements, and (iii) localize to the same compartment or to contiguous compartments of the cell (e.g., some outer membrane components might articulate with components in the periplasm). In the study reported here we began to address these predictions by (i) localizing several of the BFP assembly proteins to specific cellular compartments, (ii) demonstrating, through the use of a comprehensive suite of in-frame deletions, that 12 of the 14 bfp operon ORFs are required for pilus biogenesis, and (iii) obtaining evidence, based on protein stability patterns, that subsets of these proteins are likely to physically interact.
Direct biochemical evidence that two or more proteins physically interact in the mature BFP assembly complex, as demonstrated by chemical cross-linking and coimmunoprecipitation results, has been obtained only for BfpB and BfpG (27). Evidence that BfpP, -A, -I, -J, and -K interact has also been obtained. Disruption of bfpP, which encodes the prepilin peptidase, was shown to prevent maturation of BfpA (35), as well as BfpI, -J, and -K (Fig. 4b), indicating that this enzyme (BfpP) and its substrates (BfpA and the pilin-like proteins BfpI, -J, and -K) physically interact during biogenesis of the assembly complex. By contrast, the data shown in Fig. 2 and Table 3 and the implications of these data for the structure of the BFP assembly complex are based on a different kind of evidence: the premise that protein stability patterns can predict protein-protein interactions within a hetero-oligomeric complex. The plausibility of this premise rests on the idea that the assembly complex resides in a proteolytic environment and that the proper deployment and arrangement of assembly proteins, including their juxtaposition with their nearest normal neighbor, prevent degradation. Evidence that this approach can accurately predict protein-protein interactions comes from the study of BfpB and BfpG; deletion of BfpB causes BfpG to degrade, and deletion of BfpG prevents multimerization of the BfpB secretin complex (27). Similar evidence was obtained by Sandkvist et al. (25) for the interaction of EpsL and EpsM within the type II extracellular protein secretion complex of Vibrio cholerae. However, interpretation of protein degradation data, by itself, is inherently ambiguous because any deletion that prevents normal docking, processing, or transport of a protein can cause it to be improperly folded or to accumulate in the wrong compartment of the cell. Such a protein is normally targeted for degradation. In such circumstances, it would not be possible to draw valid conclusions about protein-protein interactions from the observed degradation patterns. In the discussion of our data below, we attempt to use protein degradation patterns as indicative of protein-protein interactions by applying the following conservative criteria: (i) the deleted and degraded protein should reside in the same compartment or in contiguous compartments of the cell; (ii) the degradation patterns should exhibit reciprocity; and (iii) the effect of a deletion should not be pleiotropic (i.e., associated with degradation of most or all BFP assembly complex proteins). Two or more proteins that fulfill these requirements are operationally defined as interacting within the assembly complex. By contrast, other less stringent criteria suggest some kind of dependence, ranging from the possibility of a direct interaction to the kind of pleiotropic indirect effect discussed above. However, it must be borne in mind that any such conjectures about the physical association of two protein species need to be proven by other methods that provide direct evidence for the physical interaction, including chemical cross-linking and coimmunoprecipitation studies.
We deduced from our survey that the stability of most of the proteins encoded by the bfp operon depends on the presence of one or more of the other Bfp proteins. Four dependence patterns are evident, not including the effect of BfpP on the four pilin-like proteins. The first pattern, a pattern of reciprocal dependence, was demonstrated for BfpJ and BfpL or BfpI and BfpK. By contrast, a pattern of pleiotropic dependence is evident from the deletion of BfpE, which destabilizes 12 of the 14 Bfp proteins, an effect which suggests that BfpE plays a central role in the stability or distribution of the proteins. The third dependence pattern, characterized by unequal or unidirectional stability, is seen for BfpG, -B, -C, -U, -D, -I, -J, -K, and -L. This pattern is exemplified by the deletion of BfpC, which dramatically affects the stability of BfpI, -J, and -K, whereas deletion of any of the latter three proteins does not affect the stability of BfpC. Similarly, deletion of BfpB destabilizes BfpU, but deletion of BfpU has no effect on BfpB. The last or null dependence pattern is exemplified by BfpA and BfpF; deletion of these proteins does not affect the stability of any other Bfp protein.
The reciprocal and unidirectional dependence patterns described above correlate with the subcellular localization of the corresponding proteins; deletion of a protein localized to a specific compartment most significantly affects the stability of other proteins in that compartment and/or in an adjoining compartment. This permits us to postulate that there are four subassembly protein ensembles, based on localization of proteins to one compartment or contiguous compartments and the patterns of stability dependence. (i) BfpB, -G, and -U compose an outer membrane subassembly complex. We have previously shown that deleting BfpB affects the stability of BfpG (27). The results shown in Fig. 2 support this finding but also demonstrate that in ΔB, BfpU is absent; also, in ΔG, although BfpB is present, BfpU is also absent. This establishes that there is an interaction between the outer membrane-bound secretin-like structure of the BfpB-BfpG complex and a periplasmic protein, BfpU. (ii) BfpU defines a periplasmic subassembly complex, which, based on the stability dependence results and protein localization data, interacts with both the outer membrane subassembly complex, as discussed above, and the inner membrane subassembly complex. Thus, although BfpU is located mainly in the periplasm, deletion of BfpU affects the stability of the inner membrane proteins BfpI, -J, and -K. (iii) BfpC, -U, -I, -J, -K, and -L define a subassembly complex that we believe resides on the periplasmic face of the inner membrane. Subsets of these inner membrane-localized proteins (BfpI plus BfpK and BfpJ plus BfpL) exhibit reciprocal stability dependence patterns. In addition, deletion of either BfpJ or BfpL destabilizes both BfpI and BfpK. By contrast, deletion of BfpK has no noticeable effect on the abundance of either BfpJ or BfpL. These components of the inner membrane subassembly complex are stabilized by BfpU, as discussed above, and by BfpC, which also localizes to the inner membrane. (iv) BfpE, an integral inner membrane protein (4), comprises a distinct entity based on its localization pattern and pleiotropic effects on protein stability. We propose that BfpE serves two functions: it forms the inner membrane scaffold of the assembly complex, and it is required for movement of assembly proteins across the inner membrane into the developing assembly complex.
How might these putative subassembly complexes interact during pilus biogenesis? In contrast to the Pap pilus assembly system (26), where the pilus is assembled at the outer membrane and the pilins and accessory proteins are conveyed to the assembly site by chaperones, we believe that the BFP assembly complex spans the periplasmic space. Consistent with this idea is the observation that three proteins either localize with both inner and outer membrane fractions or show protein stability dependence with proteins in each of these subassembly complexes. BfpU is one such protein, and its localization and stability dependence characteristics are discussed above. BfpL, although mainly an inner membrane-associated protein, is also well represented in the outer membrane (Fig. 3a). However, its interaction with the outer membrane can evidently be disrupted by Sarkosyl (Fig. 3b), and deletion of this protein has no effect on the stability of the outer membrane BfpB-BfpG secretin complex. BfpL is too small (∼24 kDa) to span a normally configured periplamic space, nor was it detected in the periplasm (Fig. 3b). It is possible that it is deployed as a homopolymeric structure linking the inner and outer membrane subassembly complexes. BfpC has been assigned to the inner membrane subassembly complex based on the localization and protein stability data discussed above. However, its cleavage pattern is dramatically affected by deletions of the outer membrane subassembly complex proteins, BfpB, -G, and -U (Fig. 4a). Thus, it too may coassociate with the inner and outer membrane subassembly complexes. Evidence that supports this conclusion comes from the observation that treatment of intact bacteria with chemical cross-linking reagents cross-links BfpC with proteins in the inner and outer membrane subassembly complexes (Hwang, unpublished observations).
The amounts of different Bfp proteins seem to be temporally regulated according to the growth phase. BfpA, BfpB, BfpE, and the processed BfpC moieties are present throughout the exponential and stationary phases. The persistence of BfpA, -B, -E, and -C suggests that these proteins comprise a durable preassembly consortium that prepares the organism for the next period of active pilus production. Curiously, each of these proteins seems to represent a critical subassembly complex; BfpA is thought to compose an unassembled pool in the inner membrane, BfpB is a crucial component of the outer membrane secretin complex, BfpE is the probable inner membrane gatekeeper and scaffold for most of the other components of the assembly complex, and BfpC is hypothesized to articulate between the inner and outer membrane subassembly complexes. Preliminary evidence indicates that full-length BfpC can be cross-linked to the outer membrane BfpB-BfpG complex, either directly or indirectly (Hwang, unpublished observations) and that BfpC (both full-length and processed fragments) mainly fractionates with the inner membrane (Bieber, unpublished observations). By contrast, the remaining Bfp proteins are present only during the exponential phase of growth and thus seem to be present when the complex needs to assemble pili.
In conclusion, several lines of investigation lead to an emerging picture of the assembly complex required for type IV pilus biogenesis. Most of the protein members of this complex are translated during the exponential phase of growth when pili need to be elaborated. Once assembled, this complex consists of proteins spanning the inner membrane, bridging the periplasmic space, and protruding through the outer membrane as extracellular filamentous organelles.
Acknowledgments
This work was supported by grant AI39521 from the National Institutes of Health. S.A.S. was supported in part by a grant from the Stanford Medical Scholars Program.
We thank Rosemary Fernandez for synthesizing the Bfp peptides.
REFERENCES
- 1.Anantha, R. P., K. D. Stone, and M. S. Donnenberg. 2000. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J. Bacteriol. 182:2498-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anantha, R. P., K. D. Stone, and M. S. Donnenberg. 1998. Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect. Immun. 66:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 4.Blank, T. E., and M. S. Donnenberg. 2001. Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli. J. Bacteriol. 183:4435-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, P. Y., and G. D. Fasman. 1974. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from protein. Biochemistry 13:211.. [DOI] [PubMed] [Google Scholar]
- 6.Cravioto, A., R. Gross, S. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 7.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giron, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Hultgren, S. J., F. Lindberg, G. Magnuson, J. Kihlberg, J. M. Tennent, and S. Normark. 1989. The PapG adhesin of uropathogenic Escherichia coli contains separate regions for receptor binding and the incorporation into the pilus. Proc. Natl. Acad. Sci. USA 86:4357-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadner, R. J. 1996. Cytoplasmic membrane, p. 58-87. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 13.Keleti, G. W. L. 1973. Handbook of micromethods for the biological sciences. Van Nostrand Reinhold, New York, N.Y.
- 14.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 15.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nataro, J. P., I. Scaletsky, J. Kaper, M. Levine, and L. Trabulsi. 1985. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect. Immun. 48:378-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido, H. 1994. Isolation of outer membranes, p. 225-344. In V. L. Clark and P. M. Bavoil (ed.), Bacterial pathogenesis, part A. Identification and regulation of virulence factors, vol. 235. Academic Press, Inc, San Diego, Calif.
- 18.Nossal, N. G., and L. A. Heppel. 1966. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J. Biol. Chem. 241:3055-3062. [PubMed] [Google Scholar]
- 19.Osborn, M. J., and R. Munson. 1974. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 31:642-650. [DOI] [PubMed] [Google Scholar]
- 20.Possot, O. M., M. Gerard-Vincent, and A. P. Pugsley. 1999. Membrane association and multimerization of secreton component pulC. J. Bacteriol. 181:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 22.Ramer, S. W., D. Bieber, and G. K. Schoolnik. 1996. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J. Bacteriol. 178:6555-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol 40:271-283. [DOI] [PubMed] [Google Scholar]
- 25.Sandkvist, M., L. P. Hough, M. M. Bagdasarian, and M. Bagdasarian. 1999. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J. Bacteriol. 181:3129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer, F. G., M. Barnhart, D. Choudhury, S. D. Knight, G. Waksman, and S. J. Hultgren. 2000. Chaperone-assisted pilus assembly and bacterial attachment. Curr. Opin. Struct. Biol. 10:548-556. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:4848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C.-Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone, K. D., H.-Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol 20:325-337. [DOI] [PubMed] [Google Scholar]
- 30.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galan. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobe, T., T. Hayashi, C. G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol 21:963-975. [DOI] [PubMed] [Google Scholar]
- 33.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 34.Zhang, H. Z., and M. S. Donnenberg. 1996. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 21:787-797. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, H. Z., S. Lory, and M. S. Donnenberg. 1994. A plasmid-encoded prepilin peptidase gene from enteropathogenic Escherichia coli. J. Bacteriol. 176:6885-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]