Abstract
CTP synthetase (EC 6.3.4.2, UTP: ammonia ligase (ADP-forming)) is an essential enzyme in all organisms; it generates the CTP required for the synthesis of nucleic acids and membrane phospholipids. In this work we showed that the human CTP synthetase genes, CTPS1 and CTPS2, were functional in Saccharomyces cerevisiae and complemented the lethal phenotype of the ura7Δ ura8Δ mutant lacking CTP synthetase activity. The expression of the CTPS1-and CTPS2-encoded human CTP synthetase enzymes in the ura7Δ ura8Δ mutant was shown by immunoblot analysis of CTP synthetase proteins, the measurement of CTP synthetase activity, and the synthesis of CTP in vivo. Phosphoamino acid and phosphopeptide mapping analyses of human CTP synthetase 1 isolated from 32Pi-labeled cells revealed that the enzyme was phosphorylated on multiple serine residues in vivo. Activation of protein kinase A activity in yeast resulted in transient increases (2-fold) in the phosphorylation of human CTP synthetase 1 and the cellular level of CTP. Human CTP synthetase 1 was also phosphorylated by mammalian protein kinase A in vitro. Using human CTP synthetase 1 purified from Escherichia coli as a substrate, protein kinase A activity was dose- and time-dependent, and dependent on the concentrations of CTP synthetase1 and ATP. These studies showed that S. cerevisiae was useful for the analysis of human CTP synthetase phosphorylation.
CTP synthetase (EC 6.3.4.2, UTP: ammonia ligase (ADP-forming)) catalyzes the final step in the pyrimidine biosynthetic pathway (1). The end product CTP is required for the synthesis of nucleic acids and membrane phospholipids (2). Thus, CTP synthetase is an essential enzyme for the growth and metabolism of all organisms (2). In eukaryotes, CTP synthetase activity regulates the balance of nucleotide pools (3-9) and influences the pathways by which membrane phospholipids are synthesized (9-11). The importance of understanding the mode of action and regulation of CTP synthetase is further highlighted by the fact that elevated CTP synthetase activity is a common property of several cancers in humans (12-20).
CTP synthetase has been purified and characterized from bacteria (21-23), Saccharomyces cerevisiae (8, 24), and rat liver (25). In addition, crystal structures for the Escherichia coli (26) and Thermus thermophilus (27) enzymes have been solved. The enzymological properties of CTP synthetase enzymes from various sources are similar, although some differences have been identified (23). The enzyme catalyzes a complex set of reactions involving the ATP-dependent transfer of the amide nitrogen from glutamine (i.e., glutaminase reaction) to the C-4 position of UTP to generate CTP (21, 28). GTP activates the glutaminase reaction by accelerating the formation of a covalent glutaminyl enzyme intermediate (21, 29). CTP synthetase exhibits positive cooperative kinetics with respect to UTP and ATP and negative cooperative kinetics with respect to glutamine and GTP (8, 21, 23, 24, 29-33). The positive cooperative kinetics toward UTP and ATP is attributed to the nucleotide-dependent tetramerization of the enzyme (8, 21, 34, 35). Indeed, the CTP synthetase tetramer is the active form of the enzyme (8, 21, 23, 24, 29-33, 35). The enzyme may also utilize dUTP for the synthesis of dCTP.(23, 36)2
An important mode of CTP synthetase regulation is feedback inhibition by CTP (8, 21, 23-25). CTP inhibits CTP synthetase activity by increasing the positive cooperativity of the enzyme for UTP (8, 21, 24, 25). dCTP does not substitute for CTP as a feedback inhibitor of CTP synthetase activity using dUTP or UTP as a substrate (23, 36). A defect in CTP feedback inhibition results in abnormally high intracellular levels of CTP and dCTP (4, 9, 37), resistance to nucleotide analog drugs used in cancer chemotherapy (38-41), and an increased rate of spontaneous mutations (5, 39, 41).
Studies on the URA7-encoded enzyme from S. cerevisiae have revealed that CTP synthetase activity is regulated by phosphorylation. The yeast enzyme is phosphorylated on multiple serine residues in vivo (42). In vitro studies have shown that CTP synthetase is a substrate for protein kinases A (43) and C (42, 44). These phosphorylations result in the stimulation of CTP synthetase activity by a mechanism that increases catalytic turnover (42-44). In addition, phosphorylation facilitates the nucleotide-dependent tetramerization of the enzyme (35) and causes a decrease in the sensitivity of the enzyme to feedback inhibition by CTP (43, 44).
Genes encoding CTP synthetase have been isolated from a variety of bacteria (23, 45-48), yeast (6, 7), and human (49, 50). Owing to the relatively high degree of deduced amino acid sequence identity (∼ 53%) between the yeast and human enzymes, we examined the hypothesis that the human CTPS1 and CTPS2 genes are functionally expressed in S. cerevisiae. We showed in this study that the CTPS1 or CTPS2 genes complemented the lethal phenotype of an ura7Δ ura8Δ mutant lacking CTP synthetase. In addition, expression of the CTPS1-encoded human CTP synthetase in yeast revealed that the enzyme was phosphorylated and regulated by protein kinase A.
EXPERIMENTAL PROCEDURES
Materials—All chemicals were reagent grade. Growth medium supplies were purchased from Difco laboratories. Restriction endonucleases, modifying enzymes, and recombinant Vent DNA polymerase were purchased from New England Biolabs. Plasmid DNA purification and DNA gel extraction kits, and Ni2+-NTA agarose resin were purchased from Qiagen. Oligonucleotides were prepared by Genosys Biotechnologies, Inc. The Yeast Maker yeast transformation kit was purchased from Clontech. Invitrogen was the source of the DNA size ladder used for agarose gel electrophoresis. Radiochemicals were purchased from PerkinElmer Life Sciences. Nucleotides, 5FOA, phenylmethylsulfonyl fluoride, benzamidine, aprotinin, leupeptin, pepstatin, tricarboxyethylphosphine, lyticase, bovine serum albumin, and standard phosphoamino acids were purchased from Sigma. Protein assay reagents, electrophoretic reagents, and protein standards were purchased from Bio-Rad. Mouse monoclonal anti-His6 antibodies were from Cell Signaling Technology. Alkaline phosphatase-conjugated goat anti-mouse and goat anti-rabbit antibodies were purchased from Pierce. Protein kinase A catalytic subunit (bovine heart) was purchased from Promega. Hybond-P PVDF membrane and the enhanced chemifluorescence Western blotting detection kit were purchased from GE Healthcare. Ni2+-NTA column was obtained from Novagen. The Poros HQ column was purchased from Applied Biosystems. Cellulose thin layer glass plates were from EM Science. Scintillation counting supplies were purchased from National Diagnostics.
Strains and Growth Conditions—The strains used in this work are listed in Table I. Yeast were grown in SC medium containing 2% glucose at 30°C as described previously (51, 52). For selection of cells bearing plasmids, appropriate amino acids were omitted from SC medium. Cells were also grown in YEPD medium (1% yeast extract, 2% peptone, 2% glucose) or YEPA medium (1% yeast extract, 2% peptone, 2% acetate). Plasmid maintenance and amplifications were performed in E. coli strain DH5α. E. coli cells were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4) at 37 °C. Ampicillin (100 μg/ml) was added to the growth medium for E. coli carrying plasmids. Media were supplemented with either 2% (yeast) or 1.5% (E. coli) agar for growth on plates. Yeast cell numbers in liquid media were determined spectrophotometrically at A600 nm. For purification of the human CTP synthetase 1 from E. coli, BL21(DE3) cells bearing plasmid pET-28b(+)-hCTPS1 were grown in 6 liters LB medium at 37 ° of C to a cell density of A600 = 0.15. The cells were then slowly cooled to 15°C over a 2 hour period until they reached a cell density of A600= 0.3. At this point, the expression of human CTP synthetase 1 was induced by adding 0.5 mM β-isopropylthioglactoside to the culture. Maximum expression occurred after 16 h at 15 °C. Under these growth conditions, approximately 50% of human CTP synthetase 1 protein was in the soluble fraction of the cell lysate.
TABLE I.
Strains used in this work
| Strain | Relevant characteristics | Source or Ref |
|---|---|---|
| E. coli | ||
| DH5α | F-, ϕ80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk- mk+), phoA, supE44, λ-thi-1, gyrA96, relA1 | (52) |
| BL21(DE3) | F- ompT hsdSB (rB-mB-) gal dcm (DE3) | Novagen |
| S. cerevisiae | ||
| SDO195 | MATa leu2-3, 112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO134] | (9) |
| GHY52 | MATa leu2-3, 112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO134] [pDO105] | This study |
| GHY53 | MATa leu2-3, 112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO134] [pDO105-hCTPS1] | This study |
| GHY54 | MATa leu2-3, 112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO134] [pDO105-hCTPS2] | This study |
| GHY55 | MATa leu2-3, 112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1] | This study |
| GHY56 | MATa leu2-3, 112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS2] | This study |
DNA Manipulations, Amplification of DNA by PCR, and DNA Sequencing—Standard methods were used to prepare genomic and plasmid DNA, to digest DNA with restriction enzymes, and to ligate DNA (52). Transformation of yeast (53, 54) and E. coli (52) was performed according to standard protocols. PCR reactions were optimized as described by Innis and Gelfand (55). DNA sequencing reactions were performed by the dideoxy method using Taq polymerase (52) and analyzed by automated DNA sequencer.
Construction of Plasmids—The plasmids used in this work are listed in Table II. To construct E. coli expression vectors for the human CTP synthetase genes, coding sequences for CTPS1 or CTPS2 were amplified by PCR using cDNA clones (Open Biosystems clones 3355881 and 5268973, respectively) as templates. The forward and reverse primers used in the amplification contained a DraI site and an XhoI site, respectively. Digestion of the PCR products (∼ 1.8 kb) with DraI and XhoI produced the CTPS1 sequence for codons 2-591 and a linker (Leu-Glu), and the CTPS2 sequence for codons 2-586 and a linker (Leu-Glu). The bacterial expression vector pET-28b(+), which contains the T7 lac promoter, was digested with NcoI, filled with T4 polymerase to provide a start codon, and digested with XhoI to provide the linker. DNA fragments of CTPS1 or CTPS2 were ligated to the linear pET-28b(+) to produce plasmids pET-28b(+)-hCTPS1 or pET-28b(+)-hCTPS2, respectively. The sequences for the human CTPS1 and CTPS2 genes in the bacterial expression vectors were confirmed by DNA sequencing. These plasmids directed the expression of full-length His6-tagged (C-terminus) versions of human CTP synthetase 1 or human CTP synthetase 2, respectively, in E. coli.
TABLE II.
Plasmids used in this work
| Plasmid | Relevant characteristics | Source or Ref |
|---|---|---|
| pET-28b(+) | E. coli expression vector with the T7 lac promoter and C-terminal His6-tag fusion | Novagen |
| pET-28b(+)-hCTPS1 | CTPS1 derivative of pET-28b(+) | This study |
| pET-28b(+)-hCTPS2 | CTPS2 derivative of pET-28b(+) | This study |
| YEpLac181 | Multicopy E. coli/yeast shuttle vector with LEU2 | (84) |
| YEpLac195 | Multicopy E. coli/yeast shuttle vector with URA3 | (84) |
| pDO105 | Derivative of YEpLac181 with the ADH1 promoter and multiple cloning sites | (9) |
| pDO134 | URA7 derivative of YEpLac195 | (9) |
| pDO105-hCTPS1 | CTPS1 derivative of pDO105 | This study |
| pDO105-hCTPS2 | CTPS2 derivative of pDO105 | This study |
To construct yeast expression vectors for the human CTPS1 or CTPS2 genes, their coding sequences were released from pET-28b(+)-hCTPS1 or pET-28b(+)-hCTPS2 by digestion with XbaI and XmnI. Two XbaI/XmnI fragments of similar size were differentiated by EcoRV treatment, which only cleaves the fragment that does not contain CTPS1 or CTPS2 coding sequences. Plasmid pDO105, a yeast expression vector with the ADH1 promoter, was digested with MluI, filled with Klenow, and digested with XbaI. The XbaI/XmnI DNA fragments (∼ 2.7 kb) containing the CTPS1 or CTPS2 coding sequences were ligated to the linear pDO105 to produce pDO105-hCTPS1 or pDO105-hCTPS2. These plasmids direct the expression of full-length His6-tagged (C-terminus) versions of human CTP synthetase 1 or human CTP synthetase 2, respectively, in S. cerevisiae.
Construction of S. cerevisiae Strains Expressing CTPS1 and CTPS2—Strain SDO195 is an ura7Δ ura8Δ mutant bearing plasmid pDO134, which contains a wild type URA7 allele (9). This strain confers uracil prototrophy because pDO134 also contains the URA3 gene (9). Strain SDO195 was transformed to leucine prototrophy with plasmid pDO105-hCTPS1 or plasmid pDO105-hCTPS2. The yeast transformants were streaked onto SCleucine plates containing 5FOA. 5FOA-resistant cells, which bear only pDO105-hCTPS1 or pDO105-hCTPS2, were further confirmed by their uracil auxotrophy.
Preparation of the Cytosolic Fraction from S. cerevisiae—All steps were performed at 4 °C. Yeast cells were suspended in 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 20 mM L-glutamine, 0.3 M sucrose, 10 mM 2-mercaptoethanol, 0.5 mM phenylmethanesulfonyl fluoride, 1 mM benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin. Cells were disrupted with glass beads (0.5 mm diameter) using a Biospec Products Mini-BeadBeater-8 as described previously (56). Unbroken cells and glass beads were removed by centrifugation at 1,500 x g for 5 min. The cytosolic fraction was obtained by centrifugation at 100,000 x g for 1 h. Protein concentration was estimated by the method of Bradford (57) using bovine serum albumin as the standard.
Purification of Human CTP Synthetase 1 from E. coli—All steps were performed at 4 °C. Cells from a 6-liter culture were harvested by centrifugation, resuspended in 100 ml of 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, 5 mM imidazole, and 0.05% sodium azide, and lysed in a Microfluidizer (Microfluidics Corp.). Unbroken cells and cell debris were removed by centrifugation and the supernatant loaded onto a 5 ml Ni2+-NTA column. The human CTP synthetase 1 enzyme was eluted (∼150 mM) from the column with a linear imidazole gradient (0-300 mM). Fractions containing the enzyme were first dialyzed against 500 mM NaCl, 10 mM EDTA, and 0.5 mM tricarboxyethylphosphine, and then dialyzed against storage buffer (20 mM Tris-HCl (pH 7.9), 0.5 mM EDTA, and 0.5 mM tricarboxyethylphosphine). The dialyzed enzyme was then applied to a 4 ml Poros HQ column that was equilibrated with storage buffer. human CTP synthetase 1 was eluted (∼ 0.3 M) from the Poros HQ column with a linear NaCl gradient (0-1 M) in Storage Buffer. The enzyme preparation was dialyzed against storage buffer and concentrated to 10-20 mg/ml using an Amicon Ultra Centrifugal filter. Analysis by SDS-PAGE indicated that the enzyme preparation was homogeneous.
Enzyme Assays—CTP synthetase activity was determined by measuring the conversion of UTP to CTP (molar extinction coefficients of 182 and 1520 M -1 cm-1, respectively) by following the increase in absorbance at 291 nm on a recording spectrophotometer (21). The standard reaction mixture contained 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM 2-mercaptoethanol, 2 mM L-glutamine, 0.1 mM GTP, 2 mM ATP, 2 mM UTP, and an appropriate dilution of enzyme protein in a total volume of 0.1 ml. Enzyme assays were performed in triplicate with an average standard deviation of ± 3 %. All assays were linear with time and protein concentration. A unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min.
SDS-PAGE and Immunoblot Analysis—SDS-PAGE (58) and immunoblotting (59) using PVDF membrane were performed as described previously. For His6-tagged human CTP synthetase 1 and human CTP synthetase 2, mouse monoclonal anti-His6 antibodies were used at a dilution of 1:1,000. For yeast CTPS, rabbit anti-yeast CTPS antibodies were used at a dilution of 1: 5,000. The anti-yeast CTPS antibodies were raised against the peptide sequence QDVIEGKYDLEAGENKFNF (residues 561-579 at the C-terminal end of the deduced protein sequence of the URA7 gene) in New Zealand White rabbits by standard procedures (60) at Bio-Synthesis, Inc. Goat anti-mouse and goat anti-rabbit IgG-alkaline phosphatase conjugates were used as secondary antibodies at a dilution of 1:5,000. His6-tagged human CTP synthetase 1 and human CTP synthetase 2 proteins and yeast CTPS were detected on immunoblots using the enhanced chemifluorescence Western blotting detection kit as described by the manufacturer. Images were acquired by FluorImaging analysis. Immunoblotting signals were in the linear range of detectability.
In Vivo Labeling—The S. cerevisiae ura7Δ ura8Δ mutant bearing the human CTPS1 gene was used to examine the phosphorylation of the human CTP synthetase 1 enzyme. Cultures (50 ml) in SC medium were grown to the exponential phase of growth. Cells were harvested and resuspended in 5 ml of fresh medium containing 32Pi (0.25 mCi/ml) and incubated for 3 h. The labeled cells were harvested by centrifugation and disrupted with glass beads in 50 mM Tris-HCl (pH 7.4) containing protease (0.5 mM phenylmethylsulfonyl fluoride, 1 mM benzamide, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin) and phosphatase (10 mM NaF, 5 mM β-glycerophosphate, 1 mM sodium vanadate) inhibitors. The His6-tagged human CTP synthetase 1 was isolated from the cell extracts with Ni2+-NTA resin as described by the manufacturer. SDS-PAGE treatment buffer was added to the Ni2+-NTA resin-bound enzyme followed by SDS-PAGE and transfer of proteins to PVDF membrane. Images of the 32P-labeled proteins were acquired by phosphorimaging analysis.
To examine the effect of the Ras-cAMP pathway on the phosphorylation of human CTP synthetase 1, exponential phase cells (50 ml) grown in YEPD medium were washed and incubated overnight in YEPA medium (100 ml) to deplete internal stores of glucose. Cells were harvested and resuspended in low phosphate (61) YEPA medium (2 ml) and labeled with 32Pi(0.5 mCi/ml) for 3 h. Glucose was added to a final concentration of 5 % to activate the Ras-cAMP pathway and protein kinase A activity (62). At the indicated time intervals, cells (0.5 ml) were treated with 20 mM NaN3 and harvested by centrifugation. The cells were suspended in 0.25 ml of spheroplast buffer (25 mM Tris-HCl (pH 7.4), 1 M sorbitol, 5 mM DTT, 10 mM NaF, 10 mM NaN3, and 30 units of lyticase) and incubated for 30 min at 30 °C to prepare spheroplasts. The spheroplasts were lysed by boiling for 3 min in 2% SDS. His6-tagged human CTP synthetase 1 was isolated from the lysates with Ni2+-NTA resin as described above followed by SDS-PAGE and transfer of proteins to PVDF membrane. The membrane was used for phosphorimaging analysis of 32P-labeled human CTP synthetase 1 and for immunoblot analysis using anti-His6 antibodies. ImageQuant software was used to quantify the relative amounts of the phosphorylated human CTP synthetase 1 enzyme.
Phosphoamino Acid Analysis and Phosphopeptide Peptide Mapping—32P-labeled human CTP synthetase 1 was extracted from PVDF membrane slices and subjected to acid hydrolysis with 6 N HCl as described previously (43). Hydrolysates were dried in vacuo and applied to 0.1 mm cellulose thin-layer chromatography plates with standard phosphoamino acids (2.5 μg phosphoserine, 2.5 μg phosphothreonine, and 5 μg phosphotyrosine). Phosphoamino acids were separated by two-dimensional electrophoresis (63). Following electrophoresis, the plates were dried and subjected to phosphorimaging analysis. Standard phosphoamino acids were visualized by spraying the plate with 0.25% ninhydrin in acetone. PVDF membrane slices containing 32P-labeled human CTP synthetase 1 were subjected to digestion with L-1-tosylamido-2-phenylethyl chloromethyl ketonetrypsin and two-dimensional peptide mapping analysis (64). Electrophoresis (1% ammonium bicarbonate buffer at 1000 volts for 45 min) and ascending chromatography (n-butyl alcohol/glacial acetic acid/pyridine/water, 10:3:12:15 for 8 h) were performed on cellulose thin-layer glass plates. Dried plates were then subjected to phosphorimaging analysis.
Phosphorylation of Human CTP Synthetase 1 with Protein Kinase A—Phosphorylation reactions were measured for 10 min at 30 °C in a total volume of 40 μl. Samples of pure human CTP synthetase 1 were incubated with 50 mM Tris-HCl (pH 8.0), 60 mM dithiothreitol, 50 μM [γ-32P]ATP (5 μCi/nmol), 10 mM MgCl2, and the indicated concentrations of the bovine heart protein kinase A catalytic subunit (43). At the end of the phosphorylation reactions, samples were treated with 2x Laemmli's sample buffer (58), followed by SDS-PAGE. SDS-polyacrylamide gels were dried and the phosphorylated proteins were subjected to phosphorimaging analysis. The relative amounts of phosphate incorporated into human CTP synthetase 1 were quantified using ImageQuant software.
Extraction and Mass Analysis of CTP—CTP were extracted from yeast cells with 1 M formic acid-10% 1-butanol (v/v) as described by OzierKalogeropoulos et al. (6). The extract was analyzed for CTP by high-performance liquid chromatography using a Partisil 10 SAX column as described by Pappas et al. (36).
Analyses of Data—Statistical analyses were performed with SigmaPlot software.
RESULTS
Human CTPS1 and CTPS2 Genes Complement the CTP Synthetase Defect of the ura7Δ ura8Δ Mutations in S. cerevisiae—To determine if the human CTP synthetase genes (CTPS1 and CTPS2) were functional in S. cerevisiae, we examined whether these genes could rescue the lethal phenotype of the ura7Δ ura8Δ double mutant by plasmid shuffling (65). The ura7Δ ura8Δ mutant bearing the URA7 gene on an URA3-based plasmid was transformed with either the CTPS1 or CTPS2 gene on a LEU2-based plasmid (Table I). When these transformants were subjected to 5FOA selection, cells that were resistant to the drug were produced (Fig. 2). 5FOA selects against the cells carrying the URA3 gene, and this result indicated that the 5FOA-resistant cells did not carry the URA3-based plasmid, and thus the URA7 gene. In addition, the viability of 5FOA-reistant cells in the absence of the URA7 gene indicated that the CTPS1 and CTPS2 genes were functional and complemented the CTP synthetase defect of the ura7Δ ura8Δ mutant. As expected, transformants bearing the plasmid that did not contain the human CTP synthetase gene were sensitive to 5FOA because the cells required the URA7 gene and could not lose the URA3-based plasmid. The lack of the URA3 gene in the 5FOA-reistant cells was further confirmed by the uracil auxotrophy (Fig. 2). The 5FOA-resistant cells, which are ura7Δ ura8Δ mutant cells expressing the human CTPS1 or CTPS2 gene, showed growth that was indistinguishable from that of the mutant cells expressing the yeast URA7 gene.
Fig. 2.
Complementation of the CTP synthetase defect of the S. cerevisiae ura7Δ ura8Δ mutant by the human CTPS1 or CTPS2 gene. Strain SD195, a ura7Δ ura8Δ mutant carrying the URA7 gene on a URA3-based plasmid (pDO134), was transformed to leucine prototrophy with the empty LEU2-based plasmid (pDO105), or with the same plasmid containing the human CTP synthetase genes (pDO105-hCTPS1 or pDO105-hCTPS2). The transformants were streaked onto SC-leucine plates containing 5FOA and incubated for 3 days at 30 °C. The 5FOA-resistant colonies were produced only from the transformants carrying pDO105-hCTPS1 or pDO105-hCTPS2. Four independent colonies from 5FOA-resistant cells were patched onto SC-leucine plates and grown for 2 days at 30 °C. Strain SD195 carrying pDO105 was included in this analysis as a negative control. The patches were replica plated onto SC-leucine, SC-leucine + 5FOA, and SC-uracil plates. The plates were incubated for 3 days at 30 °C.
Expression of the Human CTP Synthetase Enzymes in S. cerevisiae—The CTPS1 and CTPS2 genes, which were functional in the ura7Δ ura8Δ mutant, directed the expression of CTP synthetase enzymes containing a His6 tag at the C-terminus of the proteins. Immunoblot analysis showed that anti-His6 antibodies specifically recognized the His6-tagged human CTP synthetase 1 and CTP synthetase 2 proteins from the extracts of ura7Δ ura8Δ mutant cells expressing the CTPS1 or CTPS2 gene (Fig. 3, left). Anti-Ura7p antibodies, which were directed against a peptide sequence at the C-terminal end of the URA7-encoded CTP synthetase, did not show cross-reactivity to the cell extracts containing the human enzymes (Fig. 3, middle). There is no sequence homology between the peptide sequence and either of the human CTP synthetase enzymes. Thus, this result also confirmed that the 5FOA-resistant cells (i.e., ura7Δ ura8Δ mutant cells expressing the CTPS1 or CTPS2 gene) did not express the yeast CTP synthetase enzyme. The His6-tagged human CTP synthetase 1 and CTP synthetase 2 proteins showed migration on SDS-polyacrylamide gel with molecular masses of 75 kDa and 73 kDa, respectively, which is in close agreement with the deduced sizes of the proteins (Fig. 3, left). The human CTP synthetase enzymes expressed in yeast showed different protein levels. From about equal amounts of cell extracts (Fig. 3, right), the level of human CTP synthetase 1 protein was about 3 fold higher than that of human CTP synthetase 2 protein (Fig. 3, left).
Fig. 3.
Expression of human CTP synthetase 1 and human CTP synthetase 2 proteins in S. cerevisiae. The ura7Δ ura8Δ mutant bearing plasmids with the yeast URA7 gene (strain GHY52) or the human CTPS1 (strain GHY55) or CTPS2 (strain GHY56) genes were grown to the exponential phase of growth. Cell extracts were prepared and samples (20 μg of protein) were subjected to SDS-PAGE, followed by transfer to PVDF membrane. The membrane was probed with anti-His6 antibodies to detect the His6-tagged human CTP synthetase 1 (hCTPS1) and human CTP synthetase 2 (hCTPS2) proteins (left). The membrane was stripped of the anti-His6 antibodies and reprobed with anti-Ura7p antibodies to detect the yeast URA7-encoded CTP synthetase (yCTPS) protein (middle). Prior to probing with the antibodies, the PVDF membrane was stained with Ponceau S to detect the total amount of protein on the membrane (right). The data shown were representative of two independent experiments.
Next, we examined the CTP synthetase activity of human CTP synthetase 1 and CTP synthetase 2 proteins in vitro from the cytosolic fractions of ura7Δ ura8Δ mutant cells expressing the CTPS1 or CTPS2 gene. The human CTP synthetase 1 and CTP synthetase 2 proteins exhibited CTP synthetase activities that were in the same range as that of the CTP synthetase activity encoded by the URA7 gene product (Fig. 4A). The CTP synthetase activity of human CTP synthetase 1 was about 2 fold higher than the activity of human CTP synthetase 2. The difference in the CTP synthetase activities correlated with the difference in the amounts of human CTP synthetase 1 and CTP synthetase 2 proteins detected from cell extracts by immunoblot analysis. We also examined the CTP synthetase activity of human CTP synthetase 1 and human CTP synthetase 2 in vivo by analyzing the levels of CTP from ura7Δ ura8Δ mutant cells expressing the CTPS1 or CTPS2 gene. High performance liquid chromatography analysis of CTP extracted from the cells at the exponential phase showed that the human enzymes did indeed synthesize CTP in vivo (Fig. 4B). The amounts of CTP found in the yeast cells expressing the human CTP synthetase enzymes were similar, but slightly lower than that found in cells expressing the yeast enzyme.
Fig. 4.
CTP synthetase activities and the synthesis of CTP in S. cerevisiae expressing human CTP synthetase 1 and human CTP synthetase 2. The ura7Δ ura8Δ mutant bearing plasmids with the yeast URA7 gene (strain GHY52) or the human CTPS1 (strain GHY55) or CTPS2 (strain GHY56) genes were grown to the exponential phase of growth. Panel A, The cytosolic fraction was isolated from the cells and assayed for CTP synthetase activity from the indicated yeast (yCTPS) or human CTP synthetase 1 (hCTPS1) and CTP synthetase 2 (hCTPS2) enzymes. Panel B, CTP was extracted from the cells and analyzed by high performance liquid chromatography. Each data point represents the average of duplicate determinations from two independent experiments ± S.D.
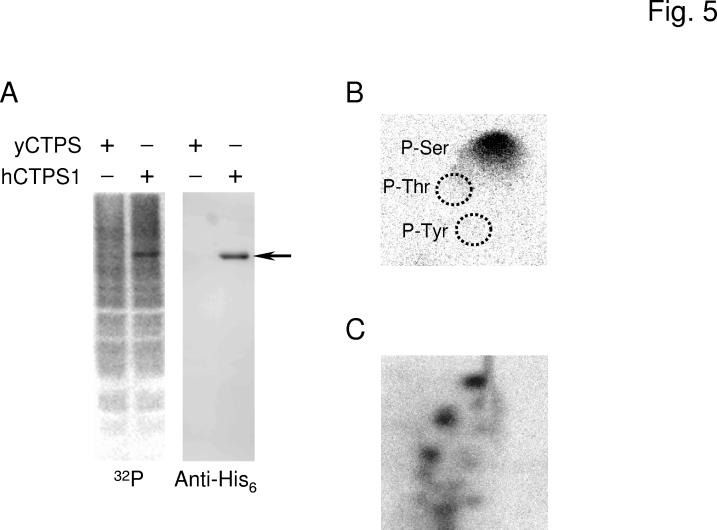
Phosphorylation of Human CTP Synthetase 1 in S. cerevisiae—Analysis of the human CTP synthetase 1 and CTP synthetase 2 sequences using the NetPhosK 1.0 server (http://www.cbs.dtu.dk/services/NetPhosK/) (66) indicated potential phosphorylation sites for several protein kinases (e.g., protein kinase A, protein kinase C, protein kinase G, calmodulin-dependent protein kinase, casein kinase I, casein kinase II, and glycogen synthase 3). Accordingly, we questioned whether the human CTP synthetase enzymes are phosphorylated when expressed in S. cerevisiae. To examine the in vivo phosphorylation of the human CTP synthetase enzymes, ura7Δ ura8Δ mutant cells expressing the His6-tagged human CTP synthetase 1 or CTP synthetase 2 enzymes were labeled with 32Pi. The human enzymes were then isolated from cell extracts with Ni2+-NTA resin and subjected to SDS-PAGE, followed by transfer to PVDF membrane. Phosphorimaging analysis showed that protein phosphorylation was detected at the expected molecular mass of the His6-tagged human CTP synthetase 1 (Fig. 5A). The protein phosphorylation was specific to the human CTP synthetase 1, and it was not detected from cells expressing the URA7-encoded CTP synthetase enzyme. Immunoblot analysis using anti-His6 antibodies confirmed that the protein phosphorylated in vivo corresponded to the His6-tagged human CTP synthetase 1 (Fig. 5A). Phosphoamino acid analysis of 32P-labeled human CTP synthetase 1 showed that the human enzyme was phosphorylated on serine residues (Fig. 5B). Digestion of the 32P-labeled enzyme with trypsin followed by phosphopeptide mapping analysis showed multiple phosphopeptides (Fig. 5C). Taken together, these results indicated that human CTP synthetase 1 was phosphorylated at multiple serine residues when expressed in yeast. In contrast to human CTP synthetase 1, it was difficult to determine unambiguously whether human CTP synthetase 2 was phosphorylated in S. cerevisiae. This may be due to the abundance and/or extent of phosphorylation of CTP synthetase 2 when compared with CTP synthetase 1.
Fig. 5.
Phosphorylation of human CTP synthetase 1 in S. cerevisiae. Panel A, the ura7Δ ura8Δ mutant bearing plasmids with the yeast URA7 gene (strain GHY52) or the human CTPS1 gene (strain GHY55) was grown in 50 ml of growth medium to the exponential phase of growth. Cells were harvested, resuspended in 5 ml of fresh medium containing 32Pi (0.25 mCi/ml), and incubated for 3 h. Cell extracts were prepared from the labeled cells and incubated with Ni2+-NTA resin to bind the His6-tagged human CTP synthetase 1. Proteins bound to the Ni2+-NTA resin were eluted with SDS-PAGE treatment buffer, followed by electrophoresis and transfer to PVDF membrane. The membrane was subjected to phosphorimaging (left) followed by immunoblot analysis using anti-His6 antibodies (right). The arrow indicates the position of human CTP synthetase 1 (hCTPS1). Panel B, a PVDF membrane slice containing 32P-labeled human CTP synthetase 1 was hydrolyzed with 6N HCl for 90 min at 110 °C, and the hydrolysate was separated by 2-dimensional electrophoresis. The positions of the standard phosphoamino acids phosphoserine (P-Ser), phosphothreonine (P-Thr), and phosphotyrosine (P-Tyr) are indicated in the figure. Panel C, a PVDF membrane slice containing 32P-labeled human CTP synthetase 1 was digested with trypsin. The resulting peptides were separated on cellulose thin layer plates by electrophoresis (from left to right) in the first dimension and by chromatography (from bottom to top) in the second dimension. The data shown in the three panels were representative of two independent experiments.
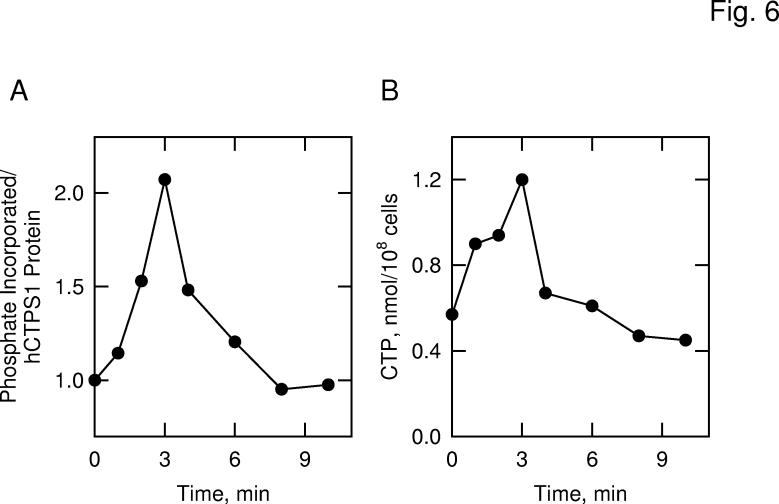
Effect of Protein Kinase A Activation on the Phosphorylation and Stimulation of Human CTP Synthetase 1 in S. cerevisiae—The human CTP synthetase 1 enzyme contains potential phosphorylation sites for protein kinase A. To determine whether the phosphorylation of the enzyme in S. cerevisiae was mediated by protein kinase A, we examined the extent of the protein phosphorylation in cells under the condition where protein kinase A activity is stimulated (67). In S. cerevisiae, protein kinase A activity is stimulated by the addition of glucose to nonfermenting cells (62, 67). Glucose (a fermentable carbon source) triggers a rapid and transient increase in cAMP levels due to the activation of adenylate cyclase activity via the Ras-cAMP pathway (67, 68). In turn, cAMP stimulates protein kinase A activity by causing the dissociation of the regulatory subunits from the catalytic subunits of the enzyme (67, 68). Cells expressing the His6-tagged human CTP synthetase 1 were grown in medium containing acetate (a non-fermentable carbon source) to attenuate the Ras-cAMP pathway (62). The cells were labeled with 32Pi for 3 h, followed by the addition of 5% glucose to activate the Ras-cAMP pathway (62). The human CTP synthetase 1 enzyme was then isolated with Ni2+-NTA resin, and the extent of the enzyme phosphorylation was determined. The activation of the Ras-cAMP pathway resulted in a time-dependent and transient increase (maximum 2-fold) in the phosphorylation of human CTP synthetase 1 (Fig. 6A). To assess the physiological consequences of human CTP synthetase 1 phosphorylation by protein kinase A, we examined the cellular levels of CTP. The transient increase in human CTP synthetase 1 phosphorylation correlated with a time-dependent and transient increase (maximum of 2-fold) in the cellular concentration of CTP (Fig. 6B).
Fig. 6.
Effects of protein kinase A activation on the phosphorylation of human CTP synthetase 1 and on the cellular concentration of CTP in S. cerevisiae. Panel A, cells expressing human CTP synthetase 1 (strain GHY55) were labeled with 32Pi for 3 h in low phosphate YEPA medium. Following the labeling period, glucose was added to a final concentration of 5% to activate the Ras-cAMP pathway and protein kinase A activity. Human CTP synthetase 1 was precipitated from lysates with Ni2+-NTA resin, followed by SDS-PAGE and transfer of proteins to PVDF membrane. The membrane was subjected to phosphorimaging and immunoblot analyses followed by the quantification of the signals using ImageQuant software. Relative phosphorylation was calculated by dividing the signal intensity of 32P-labeled human CTP synthetase 1 by that of enzyme protein. The extent of phosphorylation of human CTP synthetase 1 before glucose addition was set at 1. Panel B, in a separate experiment, the cellular concentration of CTP was measured from unlabeled cells that were activated in the Ras-cAMP pathway. The data are representative of two independent experiments.
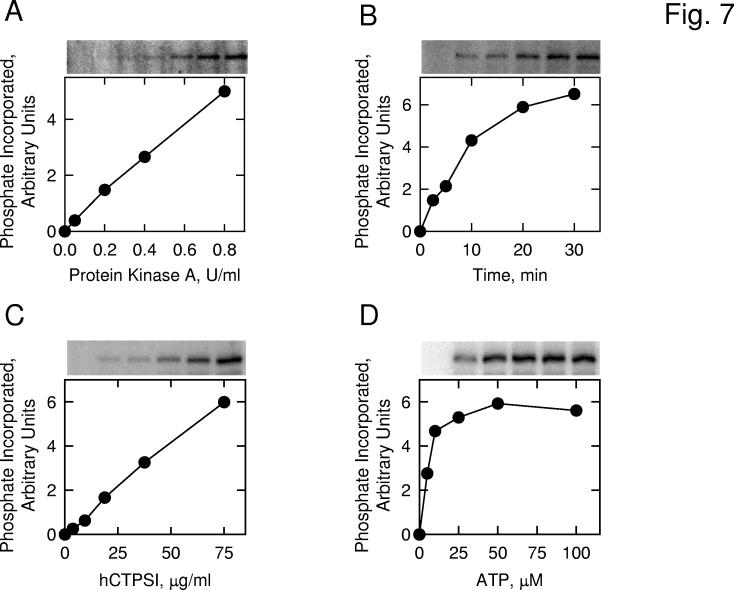
Phosphorylation of Human CTP Synthetase 1 by Protein Kinase A In Vitro— To examine the hypothesis that human CTP synthetase 1 is a substrate for mammalian protein kinase A, we utilized a homogeneous preparation of the protein that was expressed in E. coli, and the protein kinase A that was purified from bovine heart. The human CTP synthetase 1 protein was incubated with the protein kinase A in the presence of [γ-32P]ATP, and the phosphorylation of human CTP synthetase 1 was monitored by the incorporation of the radioactive γ phosphate into the protein. SDS-polyacrylamide gel electrophoresis followed by phosphorimaging analysis showed that human CTP synthetase 1 was phosphorylated by mammalian protein kinase A in vitro (Fig. 7). Using human CTP synthetase 1 as a substrate, protein kinase A activity was dependent on the concentration of the kinase (Fig. 7A) and on the time of the reaction (Fig. 7B). In addition, protein kinase A activity was dependent on the concentration of human CTP synthetase 1 (Fig. 7C) and on the concentration of ATP (Fig. 7D). Phosphoaminoacid and phosphopeptide mapping analyses of 32P-labeled human CTP synthetase 1 showed that the enzyme was phosphorylated on multiple serine residues (data not shown). The stoichiometry of the phosphorylation of human CTP synthetase 1 by protein kinase A was determined by calculating the amount of radioactive phosphate incorporated into enzyme protein after the kinase reaction was carried out to completion. At the point of maximum phosphorylation, protein kinase A catalyzed the incorporation of 0.12 mol of phosphate per mol of human CTP synthetase 1. This stoichiometry was based on the tetrameric form of the enzyme.
Fig. 7.
Phosphorylation of E. coli-expressed human CTP synthetase 1 by protein kinase A. Panel A, pure human CTP synthetase 1 (1 μg) was incubated with [γ-32P]ATP (50 μM) and the indicated amounts of protein kinase A for 10 min. Panel B, pure human CTP synthetase 1 (1 μg) was incubated with protein kinase A (1 U/ml) and [γ-32P]ATP (50 μM) for the indicated time intervals. Panel C, protein kinase A (1 U/ml) and [γ-32P]ATP (50 μM) were incubated with the indicated concentrations of pure human CTP synthetase 1 for 10 min. Panel D, protein kinase A (1 U/ml) and pure human CTP synthetase 1 (1 μg) were incubated with the indicated concentrations of [γ-32P]ATP for 10 min. Following the phosphorylation reactions, samples were subjected to SDS-PAGE. The SDS polyacrylamide gels were dried and the phosphorylated proteins were subjected to phosphorimaging analysis. The relative amounts of phosphate incorporated into human CTP synthetase 1 were quantified using ImageQuant software. Portions of the images with the phosphorylated human CTP synthetase 1 are shown above each graph. The data are representative of two independent experiments.
DISCUSSION
CTP synthetase is an important regulatory enzyme that is essential to life because it catalyzes the formation of the CTP required for the synthesis of nucleic acids and membrane phospholipids (2). The importance of understanding the regulation of CTP synthetase is emphasized by the fact that unregulated levels of its activity is a common property of leukemia cells (12-14) and rapidly growing tumors of liver (15), colon (16), and lung (17). Notwithstanding the importance of the human CTP synthetase enzymes, they have not been well characterized.
In this study, we initiated the characterization of the human CTP synthetase enzymes by expressing the genes encoding the enzymes in yeast. The yeast S. cerevisiae is a convenient model organism to study the structure/function and regulation of human gene products (69-76). We showed that the human CTP synthetase genes, CTPS1 and CTPS2, were functional in S. cerevisiae and could rescue the lethal phenotype of the yeast ura7Δ ura8Δ double mutant that lacked CTP synthetase activity. In previous studies, CTPS1 and CTPS2 were identified as genes encoding human CTP synthetase enzymes by genetic complementation of mutant (mouse or E. coli) cells defective in CTP synthetase activity (49, 50). However, the direct gene-enzyme relationship for the human CTP synthetases had not been established. In this report, we demonstrated that the genetic complementation of the yeast ura7Δ ura8Δ mutant by CTPS1 and CTPS2 was the consequence of enzymatically active CTP synthetase proteins. Although CTPS1 and CTPS2 genes were expressed in yeast from the same ADH1 promoter, the relative abundance of these proteins in cells differed; CTP synthetase 1 was more abundant when compared with CTP synthetase 2. The reason for this difference was unclear, but may be due to differential forms of regulation at the posttranslational level. Additional studies are required to address this question.
Phosphorylation is a major mechanism by which enzymes are regulated (77, 78), and indeed, the URA7-encoded CTP synthetase of yeast is phosphorylated and regulated by phosphorylation (42-44). As demonstrated in this study, human CTP synthetase 1 expressed in S. cerevisiae was also phosphorylated. Phosphorylation of human CTP synthetase 1 in human cells had been indicated by the identification of a phosphopeptide derived from the human enzyme in a large-scale characterization of HeLa cell nuclear phosphoproteins (27). These findings support the hypothesis that human CTP synthetase 1 is a bona fide phosphoprotein in human cells.
The yeast URA7-encoded CTP synthetase is phosphorylated and regulated by protein kinase A (43). Ser424 is the sole protein kinase A phosphorylation site in the yeast enzyme (79). The phosphorylation of CTP synthetase at Ser424 results in the stimulation of activity by a mechanism that includes an increase in catalytic turnover and a decrease in sensitivity to feedback inhibition by CTP (79). In addition, this phosphorylation stimulates the synthesis of the phospholipid phosphatidylcholine via the CDP-choline pathway (80). The protein kinase A phosphorylation site in yeast CTP synthetase is not conserved in human CTP synthetase 1. However, multiple protein kinase A sites are predicted in the human enzyme. Given the fact that the yeast protein kinase A catalytic subunit is structurally and functionally identical to the mammalian protein kinase A catalytic subunit (81), we predicted that protein kinase A would phosphorylate and regulate human CTP synthetase 1 in yeast. In this work, we showed that the phosphorylation state of human CTP synthetase 1 increased (2-fold) in yeast cells activated for the Ras-cAMP pathway. The response of human CTP synthetase 1 phosphorylation to the activation of the Ras-cAMP pathway was similar to that previously shown for the URA7-encoded CTP synthetase (43) and other enzymes known to be phosphorylated by protein kinase A in yeast (62, 67, 82). Moreover, the protein kinase A-mediated increase in human CTP synthetase 1 phosphorylation correlated with the stimulation of CTP synthesis in vivo. Taken together, these results indicated that protein kinase A mediated the phosphorylation and regulation of human CTP synthetase 1 in vivo.
Human CTP synthetase 1, expressed and purified from E. coli, was also a substrate for mammalian protein kinase A in vitro. Characterization studies showed that mammalian protein kinase A activity was both dose- and time-dependent, and dependent on the concentrations of human CTP synthetase 1 and ATP. That the enzyme was phosphorylated on multiple serine residues by protein kinase A was shown by phosphopeptide mapping analysis. This supports the notion that human CTP synthetase 1 would be phosphorylated by protein kinase A at multiple sites in human cells. Although human CTP synthetase 2 was not unambiguously identified as a phosphoprotein in yeast cells, preliminary studies indicated that CTP synthetase 2 purified from E. coli was also phosphorylated by protein kinase A (data not shown).
The phosphorylation of the E. coli-expressed human CTP synthetase enzymes with protein kinase A did not stimulate their activities. In fact, the human CTP synthetase enzymes expressed and purified from E. coli did not exhibit detectable CTP synthetase activity. Likewise, when the yeast URA7-encoded enzyme was expressed and purified from E. coli (8), it did not exhibit detectable CTP synthetase activity (our unpublished data). Although the human and yeast CTP synthetase genes can complement a CTP synthetase defect in E. coli (6, 50), these eukaryotic enzymes expressed in this organism did not show significant activity when measured in vitro. One possible explanation is that the E. coli-expressed enzymes might not be subject to all of the posttranslational modifications required for maximal expression of their activities.
In addition to protein kinase A, the human CTP synthetases are likely phosphorylated by other protein kinases. Clearly, the phosphorylation of human CTP synthetase is complex and the analysis of these phosphorylations in human cells would be a daunting task. The studies reported here indicated that human CTP synthetase enzymes were expressed and analyzed in yeast in the absence of competing CTP synthetase activities. Moreover, we showed that human CTP synthetase 1 was phosphorylated and regulated by protein kinase A in yeast. The yeast expression system, like bacterial systems, facilitates the isolation of large quantities of protein for structure-function studies. However, the yeast expression system has the advantage over bacterial systems in that the effects of posttranslational modifications (e.g., phosphorylation) on human enzymes can be evaluated. Thus, expression of mutant human CTP synthetase enzymes in yeast should facilitate the identification of phosphorylation sites by protein kinase A, as well as by other protein kinases, to assess the biochemical and physiological consequences of specific phosphorylation site mutations.
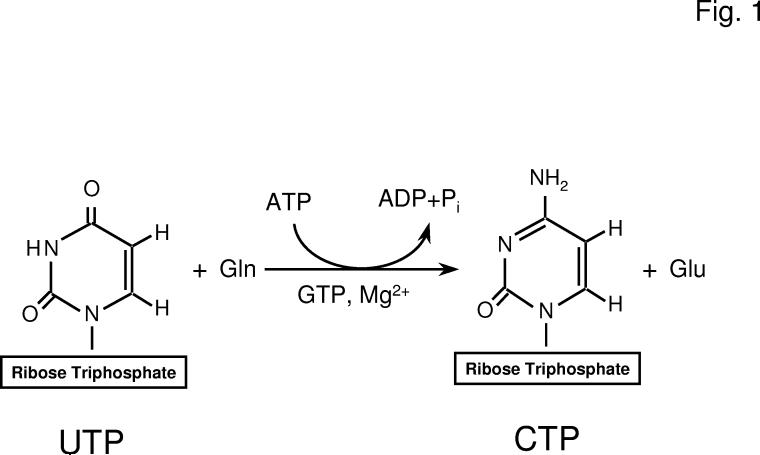
Fig. 1.
Reaction catalyzed by CTP synthetase. The figure shows the structures of UTP and CTP and the reaction catalyzed by CTP synthetase.
Footnotes
This work was supported in part by United States Public Health Service, National Institutes of Health Grants GM-50679 (to G.M.C.) and GM-63109 (to E.P.B).
The abbreviations used are: protein kinase A, cAMP-dependent protein kinase; SC, synthetic complete; 5FOA, 5-fluoroorotic acid; PVDF, polyvinylidene difluoride.
The major pathway for dCTP synthesis is via the reaction sequence CTP → CDP → dCDP → dCTP (83).
REFERENCES
- 1.Evans DR, Guy HI. J Biol.Chem. 2004;279:33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- 2.Stryer L. Biochemistry. Fourth W.H. Freeman and Company; New York: 1995. [Google Scholar]
- 3.Aronow B, Ullman B. J.Biol.Chem. 1987;262:5106–5112. [PubMed] [Google Scholar]
- 4.Robert de Saint Vincent B, Buttin G. Biochim.Biophys.Acta. 1980;610:352–359. [Google Scholar]
- 5.Meuth M, L'Heureux-Huard N, Trudel M. Proc.Nat.Acad.Sci.USA. 1979;76:6505–6509. doi: 10.1073/pnas.76.12.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozier-Kalogeropoulos O, Fasiolo F, Adeline M-T, Collin J, Lacroute F. Mol.Gen.Genet. 1991;231:7–16. doi: 10.1007/BF00293815. [DOI] [PubMed] [Google Scholar]
- 7.Ozier-Kalogeropoulos O, Adeline M-T, Yang W-L, Carman GM, Lacroute F. Mol.Gen.Genet. 1994;242:431–439. doi: 10.1007/BF00281793. [DOI] [PubMed] [Google Scholar]
- 8.Yang W-L, McDonough VM, Ozier-Kalogeropoulos O, Adeline M-T, Flocco MT, Carman GM. Biochemistry. 1994;33:10785–10793. doi: 10.1021/bi00201a028. [DOI] [PubMed] [Google Scholar]
- 9.Ostrander DB, O'Brien DJ, Gorman JA, Carman GM. J.Biol.Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 10.Hatch GM, McClarty G. J.Biol.Chem. 1996;271:25810–25816. doi: 10.1074/jbc.271.42.25810. [DOI] [PubMed] [Google Scholar]
- 11.McDonough VM, Buxeda RJ, Bruno MEC, Ozier-Kalogeropoulos O, Adeline M-T, McMaster CR, Bell RM, Carman GM. J.Biol.Chem. 1995;270:18774–18780. doi: 10.1074/jbc.270.32.18774. [DOI] [PubMed] [Google Scholar]
- 12.van den Berg AA, van Lenthe H, Busch S, de Korte D, Roos D, van Kuilenburg ABP, van Gennip AH. Eur.J.Biochem. 1993;216:161–167. doi: 10.1111/j.1432-1033.1993.tb18128.x. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg AA, van Lenthe H, Kipp JB, de Korte D, Van Kuilenburg AB, van Gennip AH. Eur J Cancer. 1995;31A:108–112. doi: 10.1016/0959-8049(94)00442-8. [DOI] [PubMed] [Google Scholar]
- 14.Verschuur AC, van Gennip AH, Muller EJ, Voute PA, Van Kuilenburg AB. Adv Exp Med Biol. 1998;431:667–671. doi: 10.1007/978-1-4615-5381-6_129. [DOI] [PubMed] [Google Scholar]
- 15.Kizaki H, Williams JC, Morris HP, Weber G. Cancer Res. 1980;40:3921–3927. [PubMed] [Google Scholar]
- 16.Weber G, Lui MS, Takeda E, Denton JE. Life Sci. 1980;27:793–799. doi: 10.1016/0024-3205(80)90333-1. [DOI] [PubMed] [Google Scholar]
- 17.Weber G, Olah E, Lui MS, Tzeng D. Adv.Enzyme Regul. 1979;17:1–21. doi: 10.1016/0065-2571(79)90005-0. [DOI] [PubMed] [Google Scholar]
- 18.Verschuur AC, van Gennip AH, Brinkman J, Voute PA, Van Kuilenburg AB. Adv.Exp.Med.Biol. 2000;486:319–325. doi: 10.1007/0-306-46843-3_62. [DOI] [PubMed] [Google Scholar]
- 19.Verschuur AC, Brinkman J, van Gennip AH, Leen R, Vet RJ, Evers LM, Voute PA, Van Kuilenburg AB. Leuk.Res. 2001;25:891–900. doi: 10.1016/s0145-2126(01)00047-9. [DOI] [PubMed] [Google Scholar]
- 20.Williams JC, Kizaki H, Weber G, Morris HP. Nature. 1978;271:71–72. doi: 10.1038/271071a0. [DOI] [PubMed] [Google Scholar]
- 21.Long CW, Pardee AB. J.Biol.Chem. 1967;242:4715–4721. [PubMed] [Google Scholar]
- 22.Anderson PM. Biochemistry. 1983;22:3285–3292. doi: 10.1021/bi00282a038. [DOI] [PubMed] [Google Scholar]
- 23.Wadskov-Hansen SL, Willemoes M, Martinussen J, Hammer K, Neuhard J, Larsen S. J.Biol.Chem. 2001;276:38002–38009. doi: 10.1074/jbc.M100531200. [DOI] [PubMed] [Google Scholar]
- 24.Nadkarni AK, McDonough VM, Yang W-L, Stukey JE, Ozier-Kalogeropoulos O, Carman GM. J.Biol.Chem. 1995;270:24982–24988. doi: 10.1074/jbc.270.42.24982. [DOI] [PubMed] [Google Scholar]
- 25.Thomas PE, Lamb BJ, Chu EHY. Biochim.Biophys.Acta. 1988;953:334–344. doi: 10.1016/0167-4838(88)90042-8. [DOI] [PubMed] [Google Scholar]
- 26.Endrizzi JA, Kim H, Anderson PM, Baldwin EP. Biochemistry. 2004;43:6447–6463. doi: 10.1021/bi0496945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto M, Omi R, Nakagawa N, Miyahara I, Hirotsu K. Structure.(Camb.) 2004;12:1413–1423. doi: 10.1016/j.str.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Liberman I. J.Biol.Chem. 1956;222:765–775. [PubMed] [Google Scholar]
- 29.Levitzki A, Koshland DE. Biochemistry. 1972;11:241–246. doi: 10.1021/bi00752a015. [DOI] [PubMed] [Google Scholar]
- 30.Levitzki A, Koshland DE. Proc.Nat.Acad.Sci.USA. 1969;62:1121–1128. doi: 10.1073/pnas.62.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levitzki A, Koshland DE. Biochemistry. 1971;10:3365–3371. doi: 10.1021/bi00794a008. [DOI] [PubMed] [Google Scholar]
- 32.Lewis DA, Villafranca JJ. Biochemistry. 1989;28:8454–8459. doi: 10.1021/bi00447a027. [DOI] [PubMed] [Google Scholar]
- 33.von der Saal W, Anderson PM, Villafranca JJ. J.Biol.Chem. 1985;260:14993–14997. [PubMed] [Google Scholar]
- 34.Levitzki A, Koshland DE. Biochemistry. 1972;11:247–252. doi: 10.1021/bi00752a016. [DOI] [PubMed] [Google Scholar]
- 35.Pappas A, Yang W-L, Park T-S, Carman GM. J.Biol.Chem. 1998;273:15954–15960. doi: 10.1074/jbc.273.26.15954. [DOI] [PubMed] [Google Scholar]
- 36.Pappas A, Park T-S, Carman GM. Biochemistry. 1999;38:16671–16677. doi: 10.1021/bi9920127. [DOI] [PubMed] [Google Scholar]
- 37.Trudel M, van Genechten T, Meuth M. J.Biol.Chem. 1984;259:2355–2359. [PubMed] [Google Scholar]
- 38.Meuth M, Goncalves O, Thom P. Somat.Cell Genet. 1982;8:423–432. doi: 10.1007/BF01538705. [DOI] [PubMed] [Google Scholar]
- 39.Aronow B, Watts T, Lassetter J, Washtien W, Ullman B. J.Biol.Chem. 1984;259:9035–9043. [PubMed] [Google Scholar]
- 40.Kaufman ER. Muta.Res. 1986;161:19–27. doi: 10.1016/0027-5107(86)90096-5. [DOI] [PubMed] [Google Scholar]
- 41.Chu EHY, McLaren JD, Li I-C, Lamb B. Biochem.Genet. 1984;22:701–715. doi: 10.1007/BF00485854. [DOI] [PubMed] [Google Scholar]
- 42.Yang W-L, Carman GM. J.Biol.Chem. 1995;270:14983–14988. doi: 10.1074/jbc.270.25.14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W-L, Carman GM. J.Biol.Chem. 1996;271:28777–28783. doi: 10.1074/jbc.271.46.28777. [DOI] [PubMed] [Google Scholar]
- 44.Yang W-L, Bruno MEC, Carman GM. J.Biol.Chem. 1996;271:11113–11119. doi: 10.1074/jbc.271.19.11113. [DOI] [PubMed] [Google Scholar]
- 45.Weng M, Makaroff CA, Zalkin H. J.Biol.Chem. 1986;261:5568–5574. [PubMed] [Google Scholar]
- 46.Trach K, Chapman JW, Piggot P, Lecoq D, Hoch JA. J.Bacteriol. 1988;170:4194–4208. doi: 10.1128/jb.170.9.4194-4208.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tipples G, McClarty G. J.Biol.Chem. 1995;270:7908–7914. doi: 10.1074/jbc.270.14.7908. [DOI] [PubMed] [Google Scholar]
- 48.Kim JH, Kim JK, Choe YK. Biochem.Mol.Biol.Int. 1997;43:925–933. doi: 10.1080/15216549700204741. [DOI] [PubMed] [Google Scholar]
- 49.Yamauchi M, Yamauchi N, Meuth M. EMBO J. 1990;9:2095–2099. doi: 10.1002/j.1460-2075.1990.tb07377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Biochim.Biophys.Acta. 2000;1492:548–552. doi: 10.1016/s0167-4781(00)00141-x. [DOI] [PubMed] [Google Scholar]
- 51.Rose MD, Winston F, Heiter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1990. [Google Scholar]
- 52.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1989. [Google Scholar]
- 53.Ito H, Yasuki F, Murata K, Kimura A. J.Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiestl RH, Gietz RD. Curr.Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 55.Innis MA, Gelfand DH. In: PCR Protocols. A Guide to Methods and Applications. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Academic Press, Inc.; San Diego: 1990. pp. 3–12. [Google Scholar]
- 56.Klig LS, Homann MJ, Carman GM, Henry SA. J.Bacteriol. 1985;162:1135–1141. doi: 10.1128/jb.162.3.1135-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradford MM. Anal.Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 58.Laemmli UK. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 59.Haid A, Suissa M. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 60.Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 61.Warner JR. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 62.Thevelein JM, Beullens M. J.Gen.Microbiol. 1985;131:3199–3209. doi: 10.1099/00221287-131-12-3199. [DOI] [PubMed] [Google Scholar]
- 63.Boyle WJ, Van der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 64.MacDonald JIS, Kent C. J.Biol.Chem. 1994;269:10529–10537. [PubMed] [Google Scholar]
- 65.Sikorski RS, Boeke JD. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 66.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 67.Thevelein JM. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- 68.Broach JR, Deschenes RJ. Adv.Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 69.Kruger WD, Cox DR. Proc.Natl.Acad.Sci U.S.A. 1994;91:6614–6618. doi: 10.1073/pnas.91.14.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee RT, Peterson CL, Calman AF, Herskowitz I, O'Donnell JJ. Proc.Natl.Acad.Sci U.S.A. 1992;89:10887–10891. doi: 10.1073/pnas.89.22.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fridovich-Keil JL, Jinks-Robertson S. Proc.Natl.Acad.Sci U.S.A. 1993;90:398–402. doi: 10.1073/pnas.90.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shan X, Wang L, Hoffmaster R, Kruger WD. J Biol.Chem. 1999;274:32613–32618. doi: 10.1074/jbc.274.46.32613. [DOI] [PubMed] [Google Scholar]
- 73.Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW. Nat.Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 74.Outeiro TF, Lindquist S. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kokkola T, Watson MA, White J, Dowell S, Foord SM, Laitinen JT. Biochem Biophys.Res.Commun. 1998;249:531–536. doi: 10.1006/bbrc.1998.9182. [DOI] [PubMed] [Google Scholar]
- 76.Shan X, Kruger WD. Nat.Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- 77.Chock PB, Rhee SG, Stadtman ER. Annu.Rev.Biochem. 1980;49:813–843. doi: 10.1146/annurev.bi.49.070180.004121. [DOI] [PubMed] [Google Scholar]
- 78.Krebs EG, Beavo JA. Ann.Rev.Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- 79.Park T-S, Ostrander DB, Pappas A, Carman GM. Biochemistry. 1999;38:8839–8848. doi: 10.1021/bi990784x. [DOI] [PubMed] [Google Scholar]
- 80.Choi MG, Park TS, Carman GM. J.Biol.Chem. 2003;278:23610–23616. doi: 10.1074/jbc.M303337200. [DOI] [PubMed] [Google Scholar]
- 81.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 82.Kim K-H, Carman GM. J.Biol.Chem. 1999;274:9531–9538. doi: 10.1074/jbc.274.14.9531. [DOI] [PubMed] [Google Scholar]
- 83.Traut TW. Crit.Rev.Biochem. 1988;23:121–169. doi: 10.3109/10409238809088318. [DOI] [PubMed] [Google Scholar]
- 84.Gietz RD, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]