Abstract
A protease-activated antimicrobial peptide (PAMP) and its inactive precursor were purified from the culture supernatant of Propionibacterium jensenii LMG 3032 and characterized at the molecular level. PAMP is a 64-amino-acid cationic peptide of 6,383 Da with physicochemical features similar to those of bacteriocins from gram-positive bacteria. This peptide displayed bactericidal activity against several propionibacteria and lactobacilli. DNA sequencing indicated that the PAMP-encoding gene (pamA) is translated as a proprotein of 198 amino acids with an N-terminal signal peptide of 27 amino acids and that PAMP constitutes the C-terminal part of this precursor. The amino acid sequence of pro-PAMP showed no similarity to those of other known proteins. By using activity tests and mass spectrometry, we showed that PAMP was formed upon protease treatment of the precursor protein. The propionibacteria produced the PAMP precursor constitutively during growth up to a level of ∼4 mg/liter, but the producing bacteria were unable to activate the precursor. The requirement for an external protease represents a novel strategy for generating antimicrobial peptides.
Antimicrobial peptides are produced by all kinds of organisms, from bacteria to mammals. In higher organisms these compounds are produced as an innate host defense mechanism to protect against pathogenic attack, whereas microorganisms presumably use these compounds as weapons in the competition for limited resources. A large number of antimicrobial peptides have been isolated from amphibians (35), fish (2, 22), insects (32), mammals (13), plants (1), and different microorganisms (12). Antimicrobial proteins and peptides from bacteria include toxins like diphtheria and cholera toxins (24, 25), bacteriolytic enzymes like lysostaphin (30) and hemolysins (8), and bacteriocins and bacteriocin-like peptides (10, 12). Numerous bacteriocins have been characterized from gram-positive bacteria, and some of them show a relatively broad spectrum of inhibition. Antimicrobial peptides produced by food-grade organisms such as lactic acid bacteria and propionibacteria have received special interest due to their potential applications in food preservation (33).
The classical propionibacteria have a long history of use in dairy fermentations, in particular the production of Swiss-type cheeses. A few antimicrobial peptides from these bacteria have been described so far (7, 9, 14, 15, 17, 21, 29), and only two bacteriocins have been characterized at the molecular level (7, 17).
Bacteria use a number of different mechanisms to regulate and produce active peptides and proteins. Most conventional bacteriocins are produced as precursor peptides, which are modified posttranslationally inside the cell or at the cell exterior during export to generate their biologically active forms (12). However, it has been shown that antimicrobial peptides from both bacteria (27, 28) and higher organisms (23, 31) can be produced from the degradation of larger proteins.
In this work we describe a novel antimicrobial peptide isolated from Propionibacterium jensenii LMG 3032. This compound is secreted from the cell as an inactive proprotein which is proteolytically activated by proteases in the environment. The mature peptide has several features in common with well-known antimicrobial peptides like class II bacteriocins and antimicrobial cationic peptides from higher organisms. This is to our knowledge the first bacteriocin-like peptide formed from an inactive extracellular protein by an external protease.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used are shown in Table 1. Propionibacteria were propagated in sodium lactate broth (SLB) (10% tryptone, 10% yeast extract, 1.2% sodium lactate, 0.25 g of K2HPO4/liter) (16) or M17 (Oxoid) (34) with glucose (5 g/liter) at 30°C. Lactobacilli were propagated in MRS (Oxoid) at 30°C.
TABLE 1.
Bacterial strains used in this study
| Indicator species | Straina | Sensitivity to purified PAMP (MIC [nM]) |
|---|---|---|
| Propionibacterium acidipropionici | ATCC 4965 | 3.6 |
| Propionibacterium acidipropionici | ATCC 4875 | 7.2 |
| Propionibacterium acidipropionici | LMG 2829 | 3.6 |
| Propionibacterium freudenreichii subsp. freudenreichii | ATCC 6207T | 7.2 |
| Propionibacterium freudenreichii subsp. shermanii | ATCC 9614T (DSM 4902) | —b |
| Propionibacterium freudenreichii | INF-P203 | 116 |
| Propionibacterium freudenreichii | INF-P205 | 58 |
| Propionibacterium freudenreichii | INF-P216 | — |
| Propionibacterium freudenreichii | LMG 2948 | — |
| Propionibacterium freudenreichii | LMG 2969 | — |
| Propionibacterium freudenreichii | LMG 3001 | — |
| Propionibacterium jensenii | ATCC 4868T | — |
| Propionibacterium jensenii | ATCC 14072 | — |
| Propionibacterium jensenii | INF-P311 | — |
| Propionibacterium jensenii | LMG 2818 | — |
| Propionibacterium jensenii | LMG 2819 | — |
| Propionibacterium jensenii | LMG 2820 | 116 |
| Propionibacterium jensenii | LMG 2821 | — |
| Propionibacterium jensenii | LMG 2823 | — |
| Propionibacterium jensenii | LMG 2942 | — |
| Propionibacterium jensenii | LMG 3032 | 116 |
| Propionibacterium thoenii | ATCC 4872 | — |
| Propionibacterium thoenii | ATCC 4874T | — |
| Propionibacterium thoenii | TL 221 | — |
| Propionibacterium thoenii | 419 | — |
| Propionibacterium thoenii | INF P409 | — |
| Propionibacterium thoenii | INF-P410 | 116 |
| Propionibacterium thoenii | INF-P414 | 0.9 |
| Propionibacterium thoenii | LMG 2792 | — |
| Lactobacillus delbrueckii subsp. lactis | ATCC 4797 | 0.5 |
| Lactobacillus delbrueckii subsp. lactis | DSM 20072T | 0.2 |
| Lactobacillus delbrueckii subsp. bulgaricus | DSM 20081T | 7.2 |
| Lactobacillus helveticus | DSM 20075T | 0.9 |
| Lactobacillus helveticus | LMG 2808 | — |
| Lactobacillus plantarum | LMG 2003 | 116 |
| Lactobacillus plantarum | LMG 2362 | — |
| Lactobacillus sakei | NCDO 2714 | 0.9 |
Abbreviations: ATCC, American Type Culture Collection (Rockville, Md.); DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen; NCDO, National Collection of Food Bacteria (Reading, United Kingdom); INF and LMG, our strain collection. Propionibacterium thoenii 419 comes from the Environmental Bacteriology Culture Collection, University of the Orange Free State, Bloemfontein, South Africa. Propionibacterium thoenii TL 221 comes from L'Institut National de la Recherche Agronomique (INRA). T, type strain.
—, no antimicrobial activity.
Colony assay for antimicrobial activity.
Strains of propionibacteria were spotted on SLB agar plates and grown for 4 to 5 days, depending on the growth rate of the bacterial strains. Five milliliters of SLB, GM17, or MRS soft agar mixed with 0.5-ml cultures of the indicator bacteria in late logarithmic growth phase was then poured over the plates. After incubation for 24 to 48 h at 30 or 37°C, depending on the indicator strain, the plates were examined for zones of growth inhibition surrounding the colonies.
Application of proteases in colony assays.
Different proteases were spotted around colonies of potential bacteriocin-producing bacteria. After an incubation period of 1 to 2 h at 30°C, soft agar with indicator strains was poured over the colonies as described above. The proteases used were protease A (20 mg/ml) from Saccharomyces cerevisiae, proteinase K (20 mg/ml) from Tritirachium album, protease P-2922 (2 U/μl) from Staphylococcus aureus, protease P-5147 (20 mg/ml) from Streptomyces griseus, protease P-5380 (20 mg/ml) from Bacillus licheniformis, α-chymotrypsin (20 mg/ml) from bovine pancreas, trypsin (20 mg/ml) from bovine pancreas, and rennet. Rennet was purchased from Christian Hansen, Copenhagen, Denmark. All other proteases were from Sigma-Aldrich.
Quantitative determination of antimicrobial activity.
Antimicrobial activity was determined by a microtiter plate assay (11). Each well of the microtiter plate contained 50 μl of a twofold serial dilution of the antimicrobial sample in SLB, GM17, or MRS and 150 μl of a 50- or 100-fold-diluted culture of the indicator strain. The plates were incubated at 30°C for 24 to 48 h, depending on the indicator used, and growth inhibition was measured spectrophotometrically at 620 nm with a microtiter plate reader. By definition, 1 unit of antimicrobial activity (AU) causes 50% growth inhibition (50% of the turbidity of a control culture without the peptide) in this microtiter plate assay with Lactobacillus sakei NCDO 2714 as the indicator. Among the indicator strains tested, L. sakei NCDO 2714 was one of the most sensitive and was therefore selected as the standard indicator. The MICs represent the concentrations of the protease-activated antimicrobial peptide (PAMP) that caused 50% growth inhibition of susceptible bacterial strains.
Purification of PAMP.
The antimicrobial peptide was purified from a 0.5-liter culture of P. jensenii LMG 3032. The bacteria were grown in SLB at 30°C until the onset of stationary phase. The culture supernatant was centrifuged at 12,000 × g for 20 min at 4°C. Proteins were precipitated from the culture supernatant by the addition of ammonium sulfate to a final concentration of 40% (wt/vol). The sample was incubated at 4°C for 1.5 h and then centrifuged at 12,000 × g for 30 min. The pellet was dissolved in 50 ml of water before proteinase K was added to a concentration of approximately 70 μg ml−1, and the sample was incubated at 30°C for 2 h. The pH was then adjusted to 3.0 with concentrated HCl, and 50 ml of water was added before the sample was applied to a 3-ml SP-Sepharose Fast-Flow cation-exchange column (Amersham Pharmacia Biotech) equilibrated with 10 mM acetic acid. The column was washed with 20 ml of 30 mM sodium phosphate buffer at pH 6.0 and 20 ml of 30 mM sodium phosphate buffer at pH 7.0 before the antimicrobial peptide was eluted in 20 ml of 0.3 M NaCl. The peptide was further purified by reverse-phase chromatography (RESOURCE RPC; Amersham Pharmacia Biotech) by using an Äkta purifier system (Amersham Pharmacia Biotech) with 2-propanol mixed with 0.1% trifluoroacetic acid in a linear gradient from 0 to 100% at a flow rate of 0.4 ml min−1. Further purification of the peptide was obtained after subjecting the active fractions to a second round of reverse-phase chromatography by using the same gradient as described above with a second column (Sephasil peptide C8, 5-μm ST 4.6/250; Amersham Pharmacia Biotech).
The eluted fractions with the highest activity were then mixed and rechromatographed on the latter reverse-phase column to obtain pure peptide.
Purification of pro-PAMP.
The precursor protein (pro-PAMP) was harvested and precipitated from culture supernatants as described above, with the exception that no proteinase K was added. The protein was quantified by the microtiter plate assay described above after aliquots were exposed to 20 μg of proteinase K/ml at 30°C for 1 h. The ammonium sulfate fraction was diluted 10 times with water, adjusted to pH 4.0 by the addition of concentrated HCl, and applied to a 3-ml SP-Sepharose Fast-Flow cation-exchange column (Amersham Pharmacia Biotech) equilibrated with 10 mM acetic acid. The column was washed with 20 ml of distilled water before pro-PAMP was eluted in 20 ml of 0.3 M NaCl. The pro-PAMP-containing fraction was then applied on a cation-exchange column (RESOURCE S; Amersham Pharmacia Biotech) by using an Äkta purifier system (Amersham Pharmacia Biotech). The column was equilibrated with 20 mM sodium phosphate at pH 4.0, and pro-PAMP was eluted from the column with a linear gradient of 0 to 1 M NaCl in 20 mM sodium phosphate at pH 4.0 and at a flow rate of 0.5 ml min−1. Fractions containing pro-PAMP were collected and rechromatographed (Mono-S HR 5/5; Amersham Pharmacia Biotech) by using the elution procedure described above.
Finally, pro-PAMP was purified by gel filtration on the Äkta purifier system by using a Sephadex peptide HR 10/30 column (Amersham Pharmacia Biotech) and 0.2 M NaCl as the mobile phase.
Mass spectrometry.
The molecular masses of pro-PAMP and PAMP were determined with a matrix-assisted laser desorption ionization-time of flight mass spectrometer (Voyager-RP DE; Applied Biosystems, Foster City, Calif.) in the linear positive-ion mode. The total acceleration voltage was 25 kV. The voltage on the first grid and the delay time between ion production and extraction were adapted to the masses of the different samples. One hundred single scans were accumulated for each spectrum. The matrix, α-cyano-4-hydroxycinnamic acid (C-2020; Sigma), was dissolved at a concentration of 15 mg/ml in a mixture of 1:1 acetonitrile-0.1% aqueous trifluoroacetic acid. Afterwards, 0.5 μl of sample and 1.5 μl of matrix were mixed together on the sample plate and air dried. All data were calibrated by using an external calibration standard mixture (Applied Biosystems).
Quantification of purified PAMP and pro-PAMP.
The concentrations of purified PAMP and pro-PAMP were determined spectrophotometrically at 280 nm by using the molar extinction coefficients deduced from the amino acid sequences.
Effect of proteinase K on pro-PAMP.
Fractions of purified pro-PAMP (∼100 pmol) were exposed to proteinase K at concentrations of 0 to 400 μg ml−1. The samples were incubated at 30°C for 1.5 h, and the antimicrobial activity was measured by a microtiter plate assay.
Effect of antibacterial peptide on viability of sensitive cells.
Purified PAMP was added at concentrations of 125 and 250 nM to a 50-fold-diluted 48-h culture of L. sakei NCDO 2714 grown in MRS. The optical density at 620 nm and the viable count (by dilution and plate counting) were determined at time intervals.
N-terminal amino acid sequencing.
The N-terminal amino acid sequence was determined by automated Edman degradation by using an Applied Biosystems 447A automatic sequence analyzer with an online 120A amino acid phenylthiohydantoin analyzer as described by Cornwell et al. (3).
DNA sequence analysis.
Total DNA from P. jensenii LMG 3032 was obtained by using Advamax beads (Advanced Genetic Technologies Corp., Gaithersburg, Md.) following the procedure described by the manufacturer. Restriction enzymes, Taq polymerase, and other DNA-modifying enzymes were used as recommended by the manufacturers (Promega, Madison, Wis.; New England BioLabs Inc., Hertfordshire, United Kingdom; and Advanced Biotechnologies Ltd., London, United Kingdom). PCRs were carried out in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). The reactions (reaction mixtures, 100 μl) were run with 2.5 U of Taq polymerase (Advanced Biotechnologies Ltd.) and 100 pmol of each primer. The PCR conditions included a hot start at 94°C (3 min) followed by 35 cycles of denaturation at 94°C (30 s), annealing at 60 or 62°C (30 s), and polymerization at 72°C (3 min). For small fragments (<200 bp), the polymerization time at 72°C was 5 s.
PCR fragments were isolated by agarose gel electrophoresis followed by extraction with Wizard Plus SV Minipreps columns (Promega). The isolated PCR products were sequenced with an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit and an ABI Prism 377 DNA sequencer (Applied Biosystems).
Two degenerate primers, 3032P1 (5′ MGN GCN MGN GCN CCN CAY A 3′) and 3032P2 (5′ NCC NGC NAC NGC NCC NGC D 3′), were designed from the amino acid sequence obtained by Edman degradation of the N-terminal part of the peptide and were used in PCR. New specific primers were designed from the sequence of the primary PCR product. Samples of total DNA were cut with different restriction enzymes (BamHI, EcoRI, and SpeI) and ligated to the plasmid pBluescript II SK(+) (Stratagene, La Jolla, Calif.) cut with the same restriction enzymes. These ligation mixtures were used as templates in PCRs with combinations of Propionibacterium-specific primers and the vector-specific primers T7 and KS. New primers were constructed from the sequences of the PCR products obtained, and this procedure was repeated until the sequence shown in Fig. 3 was obtained.
FIG. 3.
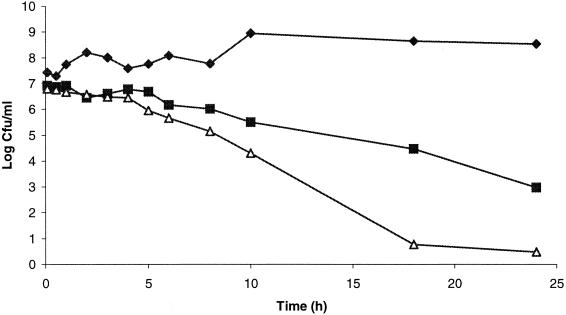
Growth and survival of L. sakei NCDO 2714 with and without purified PAMP. Symbols: ⧫, without PAMP; ▪, with 125 nM PAMP; ▵, with 250 nM PAMP.
Analyses of DNA and protein sequences were performed with OMIGA 2.0 DNA and protein sequence analysis software (Oxford Molecular, Oxford, United Kingdom). Identification of the signal peptide and cleavage site was performed with SignalP V1.1 (19).
Nucleotide sequence accession number.
The DNA sequence described here has been deposited in the GenBank database under accession number AF465935.
RESULTS
Characterization of a PAMP from P. jensenii.
A collection of dairy propionibacteria was screened for bacteriocin activity by colony assays. Common proteases, in most cases proteinase K, are used in such assays to confirm the proteinaceous nature of the antimicrobial compounds (7). Such tests were also performed during our screenings, revealing an unusual pattern of antimicrobial activity for P. jensenii LMG 3032 and P. jensenii ATCC 4868. During ordinary overlay assays, 5-day-old colonies of these two strains produced no inhibition of different propionibacteria and lactobacilli. However, clear inhibition zones were observed when proteinase K was added in the close vicinity of the colonies prior to the addition of the indicators. The addition of protease to agar plates without any bacterial colonies did not affect the growth of the indicator bacteria, indicating that the observed inhibition was not due to the protease. The results suggested that proteinase K activated a precursor of an antimicrobial compound produced by the two strains of P. jensenii. Trypsin, proteinase A, proteinase P-5147 (pronase), proteinase P-5380 (subtilisin), chymotrypsin, and rennet gave inhibitory zones of similar size as that given by proteinase K. Treatment with proteinase P-2922 (endoproteinase Glu-C) gave no inhibition zones. In all cases, the addition of more than 100 μg of protease close to the colonies eliminated the antimicrobial activity.
All Propionibacterium acidipropionici strains tested, two strains of P. freudenreichii, two strains of P. jensenii, two strains of P. thoenii, and six strains of lactobacilli were inhibited by P. jensenii LMG 3032 in overlay assays when proteinase K was used (Table 1). None of the Lactococcus, Enterococcus, Carnobacterium, or Listeria strains tested was inhibited.
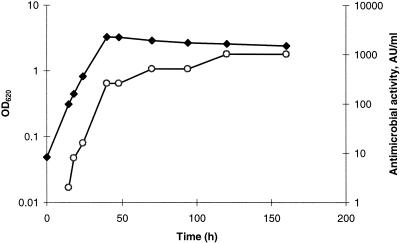
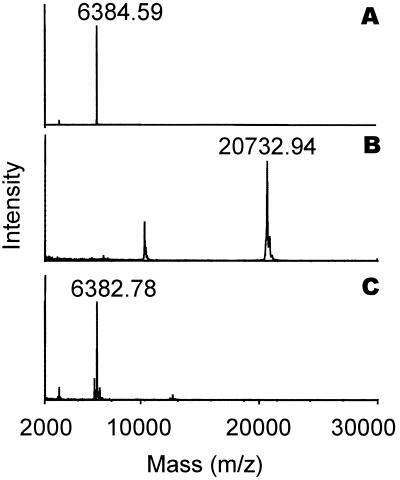
In addition, in liquid culture of P. jensenii LMG 3032, the inhibitory activity was only detected after protease treatment. The results shown in Fig. 1 indicate a constitutive production of the precursor, since antimicrobial activity could be detected in the exponential growth phase and increased in correlation with the optical density. The antimicrobial peptide was purified from P. jensenii LMG 3032 by a procedure involving ammonium sulfate precipitation, activation by proteinase K, ion exchange, and reverse-phase chromatography (Table 2). The isolated antimicrobial peptide was called PAMP. Mass spectrometry of purified PAMP resulted in one single peak with an m/z of 6,384.6 (Fig. 2A). The first 28 amino acid residues of the N-terminal amino acid sequence of PAMP were determined by Edman degradation, yielding the following sequence: RARAPHKAWYNCMTDAGISGAIAGAVAG.
FIG. 1.
Growth kinetics and bacteriocin production of P. jensenii LMG 3032. Antimicrobial activity was determined from the ammonium sulfate precipitate from cell-free culture supernatants by using L. sakei NCDO 2714 as an indicator. Symbols: ⧫, optical density at 620 nm; ○, AU/ml.
TABLE 2.
Purification of the antimicrobial peptide
| Purification step | Volume (ml) | A280 | Antimicrobial activity (AU/ml) | Specific activity (AU/ml/A280) | Fold increase in specific activity |
|---|---|---|---|---|---|
| Culture supernatant | 500 | 0.19 | 160a | 8.5 × 102 | 1 |
| Ammonium sulfate precipitate | 10 | 9.40 | 5,120a | 5.5 × 102 | 0.6 |
| Cation-exchange chromatography | 20 | 0.21 | 5,120 | 2.5 × 104 | 29 |
| Reverse-phase chromatography | 2 | 0.05 | 12,800 | 2.6 × 105 | 303 |
| Reverse-phase chromatography (C8) | 0.5 | 0.03 | 25,600 | 8.5 × 105 | 1,013 |
Antimicrobial activity measured after treatment with proteinase K.
FIG. 2.
Results of mass spectrometry of purified PAMP (A), purified pro-PAMP (B), and the purified precursor treated with proteinase K (C).
Stability of PAMP.
Neither exposure to 100°C for 15 min or pH 2.5 for 1 h nor storage at 4°C for 6 months or −20°C for 1 year had an effect on the antimicrobial activity of the peptide.
Effect of PAMP on viability of susceptible cells.
Generally, the PAMP-susceptible lactobacilli were inhibited by lower concentrations of the purified antimicrobial compound than were the sensitive propionibacteria (Table 1). As shown in Fig. 3, PAMP inhibited L. sakei NCDO 2714 very efficiently, leading to a substantial reduction in the viable count after 2 h of exposure to the peptide. For a culture with ∼107 cells/ml, exposure to 125 nM purified PAMP for 24 h resulted in a 4-log reduction in the viable count while exposure to 250 nM PAMP for the same period caused more than a 6-log reduction in the viable count (Fig. 3). Microscopy of PAMP-treated indicator cells showed no indication of cell lysis.
Molecular analysis of the PAMP-encoding gene in P. jensenii LMG 3032.
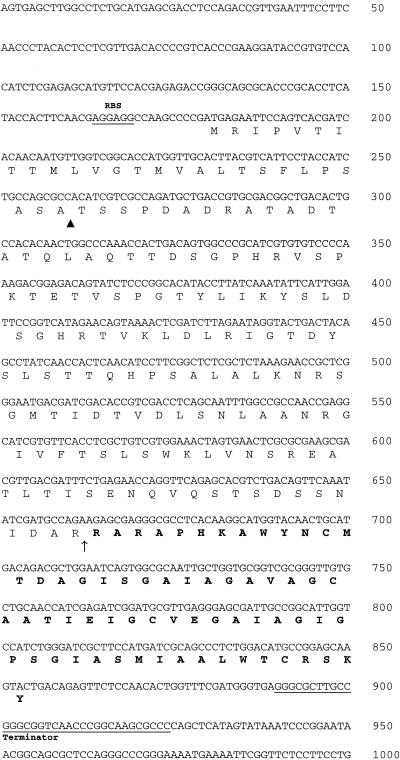
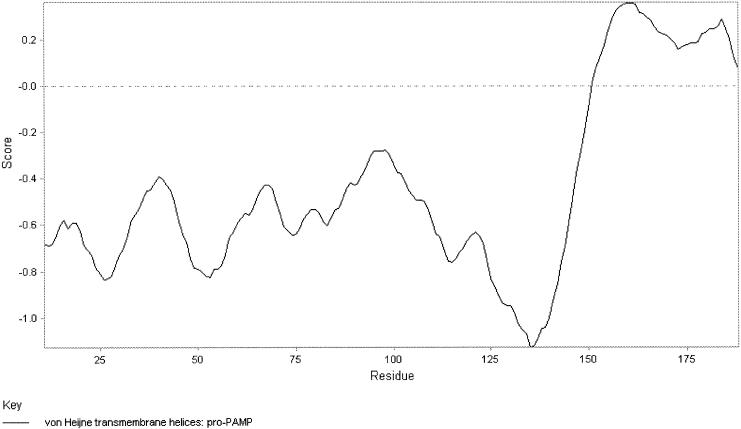
The sequence of the structural gene pamA was obtained by PCR with degenerate primers based on the N-terminal amino acid sequence followed by a primer walking strategy. Figure 4 shows the DNA sequence of 1,000 contiguous nucleotides, including the 225-amino-acid open reading frame PamA. From residue number 162, the sequence of PamA corresponds to the N-terminal sequence of PAMP. The calculated molecular mass of mature PAMP was 6,383 Da, and the pI was calculated as 8.12. This was in excellent agreement with the results of the mass spectrometry (Fig. 2A), confirming that there was no C-terminal processing. Four cysteines were found in the amino acid sequence of mature PAMP. However, the results of the mass spectrometry indicated that no disulfide bridges were present in the peptide. A putative ribosomal binding site was found 11 nucleotides upstream of the first ATG codon of pamA (Fig. 4). A putative terminator sequence consisting of two inverted repeats of 14 nucleotides was found 36 nucleotides downstream of the PAMP structural gene (Fig. 4). Extensive searches in various sequence databases revealed no significant homologies to amino acid or DNA entries. Based on the SignalP V1.1 algorithm (19), PamA is translated as a preproprotein that contains a signal peptide of 27 amino acids, typical for proteins secreted from the cell by the sec-dependent pathway (26). This discovery indicates that the pre-proprotein probably is processed in two steps. First, the signal peptide is cleaved off by the signal peptidase during export, before extracellular proteolytic processing of the proprotein gives mature PAMP of 64 amino acids (Fig. 4). Analysis of the amino acid sequence shows that PAMP can form two transmembrane helices (Fig. 5).
FIG. 4.
DNA sequence of 1,000 contiguous nucleotides, including the open reading frame pamA. A potential ribosome-binding site (RBS) and a terminator sequence are indicated. The amino acid sequence of pre-pro-PAMP is also shown, with the sequence of mature PAMP in boldface. The putative cleavage sites of the N-terminal leader sequence (▴) and the proprotein (↑) are indicated.
FIG. 5.
Protein profile analyses of pro-PAMP indicate that mature PAMP can form two transmembrane helices.
Purification of the precursor protein.
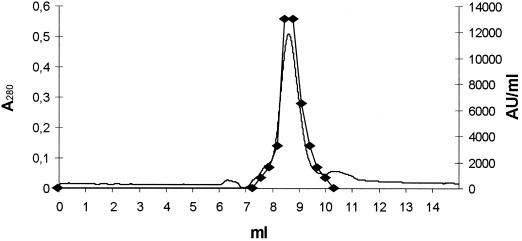
Based on the DNA sequence, the molecular weight of the precursor after removal of the signal peptide was calculated as 20,734. The proprotein (pro-PAMP) was isolated and purified by a procedure involving ammonium sulfate precipitation, ion-exchange chromatography, and gel filtration (Fig. 6). Mass spectrometry of purified pro-PAMP revealed a single peak with an m/z of 20,733 (Fig. 2B). This result was consistent with the calculated mass of the proprotein based on the DNA sequence. Treatment of purified pro-PAMP with proteinase K (40 μg ml−1) resulted in one dominant peptide with the same molecular mass as purified PAMP (Fig. 2C). This result confirmed that processing of the precursor with proteinase K results in the generation of one specific antimicrobial peptide. We obtained ∼0.4 mg of pure pro-PAMP from a 0.5-liter culture supernatant (Fig. 6). Based on the results presented in Fig. 1, we calculated the maximal production of the PAMP precursor in culture supernatant as 4 mg/liter.
FIG. 6.
Chromatogram (optical density at 280 nm) and antimicrobial activity of the fractions (AU per milliliter) obtained after gel filtration of pro-PAMP. Symbol: ⧫, AU/ml.
The effect of proteinase K on the purified PAMP precursor.
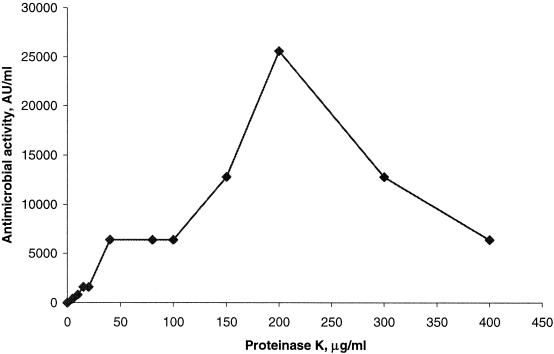
Purified pro-PAMP was exposed to different concentrations of proteinase K for 1.5 h and subsequently tested for activity against L. sakei NCDO 2714 in a microtiter plate assay. As shown in Fig. 7, pro-PAMP had no inhibitory activity, but a concentration of 5 μg of proteinase K ml−1 resulted in almost 2% processing of the precursor, and a dose-dependent increase in activity was observed up to 200 μg ml−1. Exposure of pro-PAMP to concentrations above 200 μg ml−1 resulted in a loss of antimicrobial activity. This result indicates that PAMP is susceptible to degradation by proteinase K, which was further supported by the finding that partly purified PAMP was completely inactivated after exposure to 400 μg of proteinase K ml−1 for 1 h at 37°C (data not shown).
FIG. 7.
Antimicrobial activity obtained after treatment of purified pro-PAMP with various amounts of proteinase K.
DISCUSSION
In this paper we describe an antimicrobial peptide with an unusual mode of activation. The formation of PAMP differs from that of other bacteriocins isolated from gram-positive bacteria, since it is secreted as an inactive proprotein and has to be proteolytically activated outside the cell. Ratnam et al. observed similar results when they found that inhibitory zones from some strains of propionibacteria became larger upon protease treatment (29). They suggested that an antagonist produced by the propionibacteria was enhanced by the protease.
The formation of antimicrobial peptides from larger peptides or proteins with other defined functions, such as paracin I, which is an antimicrobial peptide derived from the histone H2A in catfish (23), and the strongly antimicrobial peptide lactoferricin, derived from pepsin-hydrolyzed lactoferrin (31), has been reported. Among bacteria, it has been shown that the degradation of ribosomal proteins may result in antimicrobial peptides like the cecropin-like antimicrobial peptide made by Helicobacter pylori from ribosomal protein L1 (27, 28). The PAMP precursor protein showed no significant sequence similarity to any known protein. Thus, we can suggest no other purpose for pro-PAMP than that of being a bacteriocin precursor, although an additional function cannot be excluded.
To our knowledge this is the first description of a bacteriocin-like peptide formed from a bacterial extracellular protein by an external protease. However, PAMP has many properties in common with bacteriocins from both lactic acid bacteria and propionibacteria. These properties include size, stability, hydrophobicity, and cationic nature. Our results show that PAMP has a bactericidal mode of action, killing sensitive bacteria at nanomolar concentrations of the peptide, which is typical for many bacteriocins from gram-positive bacteria (Table 1) (5, 11, 20).
The activity of PAMP is probably restricted to specific strains, killing members among all species of classical propionibacteria and several lactobacilli. While most bacteriocins show inhibitory spectra limited to species closely related to the producer, the inhibitory spectrum of PAMP does not have such restrictions. Interestingly, lactobacilli were found to be among the most sensitive strains. Similar inhibition spectra have also been found for other bacteriocins from propionibacteria (9, 15, 29). Propionibacteria and lactobacilli are often found in the same habitat, and it may be tempting to speculate that these antimicrobial compounds evolved to promote competition for limited resources between organisms sharing the same environment.
The main difference between PAMP and conventional bacteriocins is the generation of the antimicrobial peptide, and it might be a matter of discussion as to whether PAMP, based on its biochemical and antimicrobial properties, might be called a bacteriocin or not. Bacteriocins from gram-positive bacteria are usually produced as precursors with N-terminal leader peptides (12). The leader peptides serve as recognition signals for the export of the bacteriocins out of the cells, and they also protect the producers against their own bacteriocin by making them biologically inactive inside the cell. The PAMP precursor has an N-terminal leader peptide typical of proteins and peptides exported by the general sec-dependent pathway (26). Both of the previously characterized bacteriocins from propionibacteria, propionicin SM1 and propionicin T1 (7, 17), are produced as precursors with similar sec-dependent signal peptides, but unlike these bacteriocins, active PAMP is not formed concomitantly with export through the cell membrane.
Biologically inactive protein precursors are quite common in nature, and several peptides and proteins are activated by proteolytic cleavage. This is often the case with enzymes and hormones whose biological activity must be temporarily suppressed. Although extracellular activation of bacteriocins is unusual, a small number of antimicrobial peptides and proteins from bacteria are produced as precursors that are activated subsequent to export from the cell. Cytolysin is a two-peptide lantibiotic with bactericidal and hemolytic activity (10). The cytolysin subunits are produced as prepeptides that are posttranslationally modified before both peptides are activated outside the cell by the extracellular protease CylA. The gene encoding CylA is located together with the structural cytolysin genes, and mutants unable to produce this protease exhibit no cytolysin activity (10). Lysostaphin (peptidoglycan hydrolase) and hexosaminidase (endo-β-N-acetylglucosaminidase) from Staphylococcus simulans are both secreted as proproteins that are activated concomitantly with processing by an extracellular sulfhydryl protease (18). The export of these bacteriolytic enzymes as inactive precursors may be important in providing protection against the action of the compound and may therefore be considered a means of immunity.
No open reading frames were found in the vicinity of pamA. Genes encoding immunity factors are often located downstream next to the bacteriocin structural gene (6, 12). The PAMP structural gene is followed by a putative terminator sequence, indicating that no other open reading frames downstream of pamA are cotranscribed with this gene. Our results showed that the producer strain itself was sensitive to purified PAMP although the MIC was relatively high (Table 1). This producer sensitivity and the lack of any obvious immunity factor suggest that the secretion of the inactive pro-PAMP is a self-protecting mechanism.
The amino acid sequence of pro-PAMP revealed numerous possible proteolytic cleavage sites for proteinase K, and one might therefore expect to find several differently sized variants of the peptide. Mass spectrometry analyses revealed only one dominant product after proteolysis of purified pro-PAMP (Fig. 2C). The proteolytic cleavage generating PAMP occurs at one specific site between two arginines, which is a typical cleavage site for serine proteases like proteinase K (4). These findings might indicate that this particular site is more susceptible for proteolytic cleavage than other sites in its vicinity. However, mass spectrometry showed no other major peptides, indicating that the N-terminal part of pro-PAMP is extensively degraded by proteinase K (Fig. 2C). Thus, PAMP appears to be particularly recalcitrant to proteolytic attack. It is possible that proteases with other cleavage specificities, like chymotrypsin, may produce active peptides from the proprotein that differ in length from PAMP by one or more amino acids.
The variation in protease sensitivity between PAMP and the rest of the precursor molecule may reflect structural differences. Overall, structure predictions showed that the PAMP part was more hydrophobic than the rest of the molecule (data not shown).
The prediction that PAMP can form two transmembrane helices (Fig. 5) suggests that PAMP acts by pore formation. This is a feature similar to those of many characterized antimicrobial peptides from gram-positive bacteria which have been shown to permeabilize the membranes of susceptible microorganisms via pores (6, 12).
Even though several proteolytic enzymes were able to initiate antimicrobial activity in colony assays, no evidence of endogenous protease activity of the producer strain leading to the activation of PAMP was detected in our investigations. The antimicrobial activity performed by PAMP is thus most probably dependent on protease activity in the surroundings. It is possible that proteases secreted from other microorganisms in the same environment as the producing propionibacteria activate pro-PAMP. The secretion of an inactive precursor might thus give the producer a selective advantage and could represent a novel variant of niche exclusion strategy to compete with other bacteria. Besides its interesting biological aspects, the production of PAMP may be of practical importance in the manufacturing of dairy products, especially cheese, as rennet and proteases produced by other bacteria may affect the ripening process. Further work will clarify whether endogenous proteases from propionibacteria, lactobacilli, or other microorganisms are capable of activating pro-PAMP in situ.
Acknowledgments
We are indebted to D. Mantzilas for performing all of the mass spectrometry analysis. We also thank K. Sletten for performing the amino acid sequencing.
T. Faye was funded by a grant from the Norwegian Research Council. D. A. Brede was funded by The Nordic Industrial Fund grant P98089.
REFERENCES
- 1.Cammue, B. P., M. F. De Bolle, F. R. Terras, P. Proost, J. Van Damme, S. B. Rees, J. Vanderleyden, and W. F. Broekaert. 1992. Isolation and characterization of a novel class of plant antimicrobial peptides from Mirabilis jalapa L. seeds. J. Biol. Chem. 267:2228-2233. [PubMed] [Google Scholar]
- 2.Cole, A. M., P. Weis, and G. Diamond. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008-12013. [DOI] [PubMed] [Google Scholar]
- 3.Cornwell, G. G., K. Sletten, B. Johansson, and P. Westermark. 1988. Evidence that the amyloid fibril protein in senile systemic amyloidosis is derived from normal prealbumin. Biochem. Biophys. Res. Commun. 154:648-653. [DOI] [PubMed] [Google Scholar]
- 4.Creighton, T. E. 1993. Proteins: structures and molecular properties, 2nd ed. W. H. Freeman, New York, N.Y.
- 5.Eijsink, V. G., M. Skeie, P. H. Middelhoven, M. B. Brurberg, and I. F. Nes. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 7.Faye, T., T. Langsrud, I. F. Nes, and H. Holo. 2000. Biochemical and genetic characterization of propionicin T1, a new bacteriocin from Propionibacterium thoenii. Appl. Environ. Microbiol. 66:4230-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilmore, M. S. 1991. Enterococcus faecalis hemolysin/bacteriocin, p. 206-213. In G. M. Dunny, P. P. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, D.C.
- 9.Grinstead, D. A., and S. F. Barefoot. 1992. Jenseniin G, a heat-stable bacteriocin produced by Propionibacterium jensenii P126. Appl. Environ. Microbiol. 58:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas, W., and M. S. Gilmore. 1999. Molecular nature of a novel bacterial toxin: the cytolysin of Enterococcus faecalis. Med. Microbiol. Immunol. 187:183-190. [DOI] [PubMed] [Google Scholar]
- 11.Holo, H., Ø. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehrer, R. I., A. K. Lichtenstein, and T. Ganz. 1993. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 11:105-128. [DOI] [PubMed] [Google Scholar]
- 14.Lyon, W. J., and B. A. Glatz. 1993. Isolation and purification of propionicin PLG-1, a bacteriocin produced by a strain of Propionibacterium thoenii. Appl. Environ. Microbiol. 59:83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon, W. J., and B. A. Glatz. 1991. Partial purification and characterization of a bacteriocin produced by Propionibacterium thoenii. Appl. Environ. Microbiol. 57:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik, A. C., G. W. Reinbold, and E. R. Vedamuthu. 1968. An evaluation of the taxonomy of Propionibacterium. Can. J. Microbiol. 14:1185-1191. [DOI] [PubMed] [Google Scholar]
- 17.Miescher, S., M. P. Stierli, M. Teuber, and L. Meile. 2000. Propionicin SM1, a bacteriocin from Propionibacterium jensenii DF1: isolation and characterization of the protein and its gene. Syst. Appl. Microbiol. 23:174-184. [DOI] [PubMed] [Google Scholar]
- 18.Neumann, V. C., H. E. Heath, P. A. LeBlanc, and G. L. Sloan. 1993. Extracellular proteolytic activation of bacteriolytic peptidoglycan hydrolases of Staphylococcus simulans biovar staphylolyticus. FEMS Microbiol. Lett. 110:205-211. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 20.Nissen-Meyer, J., H. Holo, L. S. Håvarstein, K. Sletten, and I. F. Nes. 1992. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J. Bacteriol. 174:5686-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paik, H.-D., and B. A. Glatz. 1995. Purification and partial amino acid sequence of propionicin PLG-1, a bacteriocin produced by Propionibacterium thoenii P127. Lait 75:367-377. [Google Scholar]
- 22.Park, C. B., J. H. Lee, I. Y. Park, M. S. Kim, and S. C. Kim. 1997. A novel antimicrobial peptide from the loach, Misgurnus anguillicaudatus. FEBS Lett. 411:173-178. [DOI] [PubMed] [Google Scholar]
- 23.Park, I. Y., C. B. Park, M. S. Kim, and S. C. Kim. 1998. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 437:258-262. [DOI] [PubMed] [Google Scholar]
- 24.Parker, M. W., A. D. Tucker, D. Tsernoglou, and F. Pattus. 1990. Insights into membrane insertion based on studies of colicins. Trends Biochem. Sci. 15:126-129. [DOI] [PubMed] [Google Scholar]
- 25.Pore, R. S. 1978. Microbial toxins, their functional role and phylogenetic validity. BioSystems 10:189-198. [DOI] [PubMed] [Google Scholar]
- 26.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pütsep, K., C. I. Bränden, H. G. Boman, and S. Normark. 1999. Antibacterial peptide from H. pylori. Nature 398:671-672. [DOI] [PubMed] [Google Scholar]
- 28.Pütsep, K., S. Normark, and H. G. Boman. 1999. The origin of cecropins; implications from synthetic peptides derived from ribosomal protein L1. FEBS Lett. 451:249-252. [DOI] [PubMed] [Google Scholar]
- 29.Ratnam, P., S. F. Barefoot, L. D. Prince, A. B. Bodine, and L. H. McCaskill. 1999. Partial purification of the bacteriocin produced by Propionibacterium jensenii B1264. Lait 79:125-136. [Google Scholar]
- 30.Schindler, C. A., and V. T. Schuhardt. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA 51:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin, K., K. Yamauchi, S. Teraguchi, H. Hayasawa, M. Tomita, Y. Otsuka, and S. Yamazaki. 1998. Antibacterial activity of bovine lactoferrin and its peptides against enterohaemorrhagic Escherichia coli O157:H7. Lett. Appl. Microbiol. 26:407-411. [DOI] [PubMed] [Google Scholar]
- 32.Steiner, H., D. Hultmark, Å. Engstrom, H. Bennich, and H. G. Boman. 1981. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292:246-248.7019715 [Google Scholar]
- 33.Stiles, M. E. 1996. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek 70:331-345. [DOI] [PubMed] [Google Scholar]
- 34.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zasloff, M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 84:5449-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]