Abstract
For many viruses, the final stage of assembly involves structural transitions that convert an innocuous precursor particle into an infectious agent. This process — maturation — is controlled by proteases that trigger large-scale conformational changes. In this context, protease inhibitor antiviral drugs act by blocking maturation. Recent work has succeeded in determining the folds of representative examples of the five major proteins —major capsid protein, scaffolding protein, portal, protease and accessory protein — that are typically involved in capsid assembly. These data provide a framework for detailed mechanistic investigations and elucidation of mutations that affect assembly in various ways. The nature of the conformational change has been elucidated: it entails rigid-body rotations and translations of the arrayed subunits that transfer the interactions between them to different molecular surfaces, accompanied by refolding and redeployment of local motifs. Moreover, it has been possible to visualize maturation at the submolecular level in movies based on time-resolved cryo-electron microscopy.
Introduction
In virus assembly — particularly for large and complex viruses — the sequence of events that yield progeny virions proceeds in three phases. First, a precursor particle — the provirion or procapsid — is formed. Second, the viral genome is packaged. The final phase — maturation, which overlaps with packaging — consists of programmed changes that convert the provirion into a complete, infectious, virion. The idea of first assembling an inert precursor and then activating it is not new (see http://mv.vatican.va/3_EN/pages/x-Select/20select/20select_03.html). Nevertheless, the paradigm applies well to the process of virus maturation and recent developments have yielded insight into the underlying molecular mechanisms.
Maturation is typically controlled by a viral protease that is incorporated into the provirion as a zymogen and then activated to process specific sites in the provirion interior, including itself (each other). The primary role of proteolysis is not degradative elimination of certain components but, rather, to trigger structural changes. These changes, and the altered pattern of interactions that they bring about, stabilize the fragile precursor and/or they may actuate certain downstream reactions. For enveloped viruses, maturation tends to be based on processing the glycoproteins that recognize receptors and subsequently cause the viral envelope to fuse with a host cell membrane, delivering the nucleocapsid into the cytoplasm (e.g. [1,2]).
For many viruses, both enveloped and non-enveloped, the procapsid is the key site of maturation. In effect, its surface shell is induced to undergo a solid-state phase transition that stabilizes the particle, enabling it to withstand the high pressure imposed by the densely packed genome [3] as well as other challenges; moreover, sites are generated that allow the capsid to interact with other components, completing maturation. The role of allosteric control in virus assembly has long been appreciated [4,5]. However, it is only recently that detailed information has emerged concerning the structures of the proteins, the nature of the conformational changes, and the dynamics of maturation. Acknowledging advances made with another ancient family of double-stranded DNA viruses (e.g. [6,7•]), this review will focus on maturation phenomena shared by the tailed bacteriophages, particularly HK97, and a family of animal viruses, the herpes-viruses, particularly herpes simplex virus type 1 (HSV1). In conclusion, to underscore that maturation is a widespread phenomenon, a cursory summary is given of maturation-related studies in other systems.
Assembly pathway and building blocks
A generic assembly pathway is shown in Figure 1. Formulating such schemes is an exercise in frustration as there are exceptions to every rule. Nevertheless, the emerging picture is that much the same set of proteins —with deep evolutionary roots — is used, in differing numbers and combinations, to assemble capsids of diverse sizes and shapes. Although there tends to be little or no indication of common antecedents from sequence similarity, the folds have remained true to type, while being elaborated in some cases with additional domains or through gene duplication. In the past few years, structures have been determined for examples of each of these proteins (Figure 2).
Figure 1.
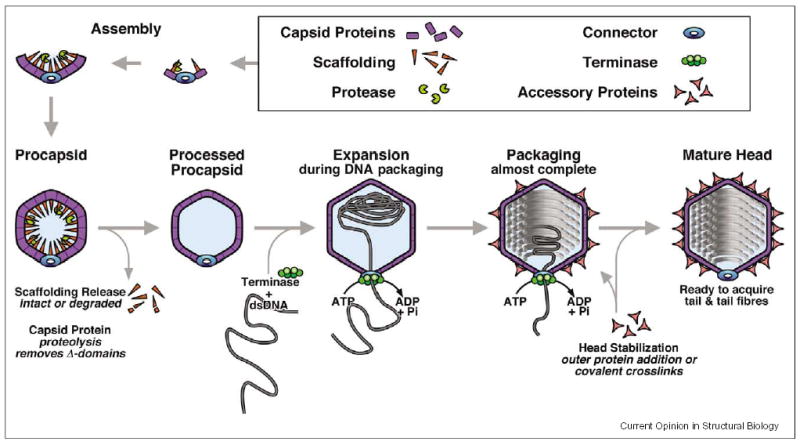
Generic assembly pathway for the heads of tailed bacteriophages.
Figure 2.
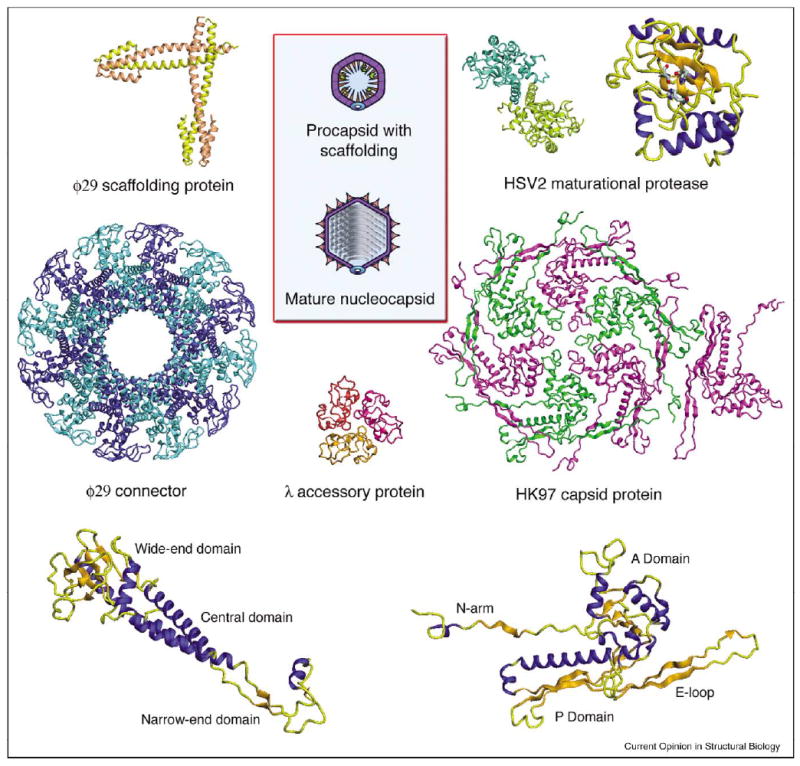
Structural genomics of capsid assembly. Diagrams of the procapsid and mature head (capsid plus internally coiled DNA) are shown at center (from Figure 1). Ribbon diagrams are shown for the φ29 scaffold (PDB code 1N04, [13••]) and portal/connector (PDB code 1IJG, [9]), the HSV2 protease (PDB code IAT3, [61]), the λ accessory protein (PDB code 1TCZ, [26]) and the HK97 MCP (PDB code 1OHG, [19]). It remains to be seen whether system-to-system variation is accomplished by embellishment of the same basic fold (e.g. by adding additional domains, as with portals, whose molecular masses vary widely [12••]) or by substituting proteins with entirely different folds. We conjecture that accessory proteins and scaffolding proteins are the likeliest candidates for variations of the latter kind.
One of the few features for which there is no documented exception (it is probably only a matter of time) is the portal/connector protein, a dodecameric ring that occupies one of the 12 vertices of the icosahedral capsid, and serves as the conduit via which DNA enters and exits the capsid [8]. It is plausible that the portal also initiates procapsid assembly and, after packing is complete, presents the template on which the tail is mounted. The structure of the φ29 connector has been determined [9,10], and its basic architecture is reproduced in SPP1 [11•] and T7 [12••].
Expression strategies: in pieces or as polyproteins?
Scaffolding proteins are employed to direct procapsid assembly with due fidelity and are subsequently expelled, either intact or in fragments. Structures have been determined for the φ29 scaffolding protein, a dimeric coiled coil [13••], and for a fragment of the P22 protein [14]. At first sight, HK97 provides the inevitable exception in having no scaffolding protein. However, its capsid protein has an unusually long propeptide — the Δ domain (102 residues) — that probably plays this role [15]. Certainly, it has a high α-helical content [16], a property of scaffolding proteins [17]. The fact that the T4 major capsid protein (MCP) also has a long propeptide while not dispensing with scaffolding proteins, of which it has two essential ones [18], does not invalidate this hypothesis; it may simply be that T4, with typical panache, has three, not two, essential scaffolding proteins. Variations in expression strategy, whereby, in one system, two or more proteins are synthesized as a polyprotein that is subsequently dissected, whereas in another system, they are expressed as separate gene products, are quite common. For example, the HSV protease enters the procapsid as a fusion with the scaffolding protein, later to release itself (see below).
The MCP of HK97 has been solved as a component of the mature capsid [19,20••]. Its fold turned out to be quite different from the β barrel of the first several capsids to be determined. The P22 MCP appears to have essentially the same fold as that of HK97, in the absence of discernible sequence similarity [21•]. Will this trend be maintained throughout this class of virus or will other MCPs exhibit different folds (in all, some half-dozen folds have been recognized to serve as building blocks for icosahedral capsids [22])? In particular, it will be interesting to know whether a version of the HK97 MCP fold is present in the floor layer of herpesvirus capsids, as seems a distinct possibility.
Once the procapsid has been assembled, it has to be matured. Facilitated by the action of the viral protease, this transformation is further promoted by electrostatic repulsive forces exerted by packaged DNA on the inner capsid surface [23]. These forces are outwards directed and should grow in strength as packaging proceeds.
However, P22 and T7 have no proteases and serve as examples of phages that have devised other ways to control this transition (e.g. to restrain the procapsid from starting to mature before it is complete). As yet, there are no high-resolution structures for bacteriophage proteases, but there are several for herpesvirus proteases (e.g. [24]), which are likely to be structurally similar [25••].
The principal effect of maturation is structural transformation. For most phages, this involves a substantial expansion — typically by ~20% in linear dimensions, a near doubling of enclosed volume. At the same time, the capsid changes shape, from round to polyhedral. Although the wall thins by ~50%, the expanded capsid is more robust than the precursor. In some systems (e.g. P22, T7), expansion stabilizes the capsid sufficiently to resist the pressure of encapsidated DNA. In others, further measures are taken: λ and T4 bind clamp-like accessory proteins, whose structure has been determined in the case of λ [26], whereas HK97 establishes a network of covalent cross-links [27].
Maturation of bacteriophage HK97
The HK97 system lends itself to the study of maturation. Its capsid protein, gp5, has been characterized comprehensively [15] and its structure is known [19]. Even in the absence of the portal, 60 hexamers and 12 pentamers of gp5 self-assemble to form a procapsid (Prohead I), while incorporating the correct quota (~60 copies) of protease. Interestingly, a hexamer-only structure with empty vertices, called the ‘whiffleball’, may be reassembled from mutant hexamers [28]. Maturation begins with cleavage of the Δ domains (gp5 to gp5*), producing Prohead-II, which may be expanded in vitro by acidification, then restoring to neutrality.
The structural changes between Prohead II and Head II lead to the formation of a remarkable network of intermolecular cross-links. Cryo-EM analysis combined with model building revealed that Prohead II expansion is accomplished by rigid-body rotations of ~40° and translations of up to 50 Å of the cores of gp5* subunits, accompanied by the refolding of two extended motifs — the N-arm and E-loop ([23]; Figure 2, bottom).
The controllability of HK97 maturation in vitro has allowed its dynamics to be studied in initially synchronized populations by cryo-EM and small-angle X-ray scattering (SAXS) [29,30], leading to the identification of three transition states or expansion intermediates (EIs) (Figure 3). At pH 4.15, Prohead II rapidly converts into a semi-expanded state (EI-I), then to the structurally similar EI-II and finally to a larger, thin-walled, spherical particle (EI-III). Upon neutralization, EI-III rapidly assumes the polyhedral head structure. The term Head II refers to the mature, fully cross-linked capsid. Initially, expansion was thought to be complete before cross-linking began, but closer investigation established that cross-linking starts at the EI-II stage; EI-III preparations are mixtures of structurally similar but variably cross-linked particles, of which the extreme case, EI-IV, has all cross-links in place except those donated by the penton subunits [31•].
Figure 3.
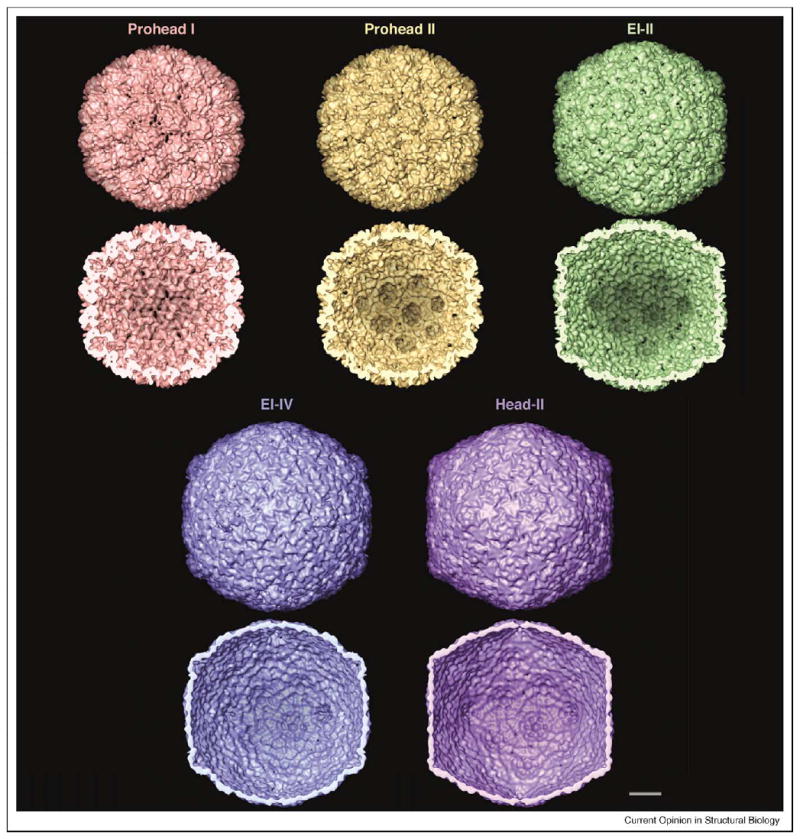
Maturation pathway of the HK97 capsid, visualized in surface renderings (outer surface, upper rows; inner surface, lower rows) at ~14 Å resolution. All images are from cryo-EM reconstructions, except Head II, which is a resolution-limited rendition of the crystal structure [19]. The capsid has icosahedral geometry (T=7 laevo) and is viewed along a twofold axis of symmetry. Prohead I is composed of 420 copies of gp5. In Prohead II, the N-terminal Δ domains have been removed from the inner surface. In acid-induced maturation in vitro, the first transition state, EI-I (not shown), is about 10% bigger than Prohead II [29] and very similar to EI-II [33••] (shown). The main difference between EI-I and EI-II is that EI-II has some covalent cross-links [31•]. The next structural state is a thin-walled spherical particle called the ‘balloon’. Balloons vary in their extents of cross-linking, as in EI-III (variable partial cross-linking) and EI-IV (almost complete cross-linking). The balloon structure is very similar to that of the end-state Head, except for the positions of its pentons, which move ~30 Å outwards in the final transition. Bar = 100 Å .
Determination of the structures of the successive states by cryo-EM allowed the creation of maturation movies, initially by morphing [29] and subsequently in terms of pseudo-atomic models (WR Wikoff et al., unpublished; http://mmtsb.scripps.edu/viper/MOVIES/hk97movies.html). The finding that most particles captured in cryo-micrographs of populations of maturing capsids could be used for image reconstruction based on icosahedral symmetry indicated that this symmetry is observed, to a close approximation and for most of the time, by maturing capsids. Switching from one state to the next, when it occurs, is rapid and cooperative but not synchronized over the population (i.e. the transitions are stochastically triggered). Possible pathways between these staging posts have been explored computationally [32].
As HK97 matures, covalent cross-links form between K169 at the tip of the E-loop and N356 of a neighboring subunit. This reaction is autocatalytic (i.e. it occurs without the involvement of an enzyme). The cross-linking pattern is such that each subunit in a given hexamer or pentamer connects to subunits in two (different) neighboring capsomers, creating an interlinked network or ‘chain mail’ [19,27]. The effects of cross-linking on capsid stability have been investigated by calorimetry [33••], showing that, in this system (unlike T4 or P22), expansion per se results in a slight destabilization, which is more than compensated by the emphatic stabilization that accompanies cross-linking. Given that complementation experiments showed a cross-link-incompetent mutant to be non-viable, indeed dominant negative, cross-linking is an essential function for HK97.
Capsid maturation of herpes simplex virus
Herpesviruses are a family of large DNA viruses that, despite having diversified to infect a wide range of vertebrate hosts, have maintained a fixed architecture consisting of a nucleocapsid surrounded by a lipoprotein envelope and, between them, the tegument — a compartment for delivering proteins to the host cell [34]. Although all herpesviruses have a T=16 capsid geometry that, to our knowledge, is yet to be documented among bacteriophages, they exhibit many phage-like behaviors in assembly [35]. (For a recent review of HSV assembly and DNA packaging, see [36].) Procapsid assembly is directed by an internal scaffolding protein (‘assembly protein’), which forms an inner shell, and a peripheral scaffolding protein (‘triplex’) (Figure 4a). Maturation, normally coupled with DNA packaging, is initiated by the protease clipping the terminal peptide from the assembly protein, disrupting its interaction with the outer shell. Finally, the mature nucleocapsid exits the nucleus and proceeds along a pathway on which it acquires tegument and envelope.
Figure 4.
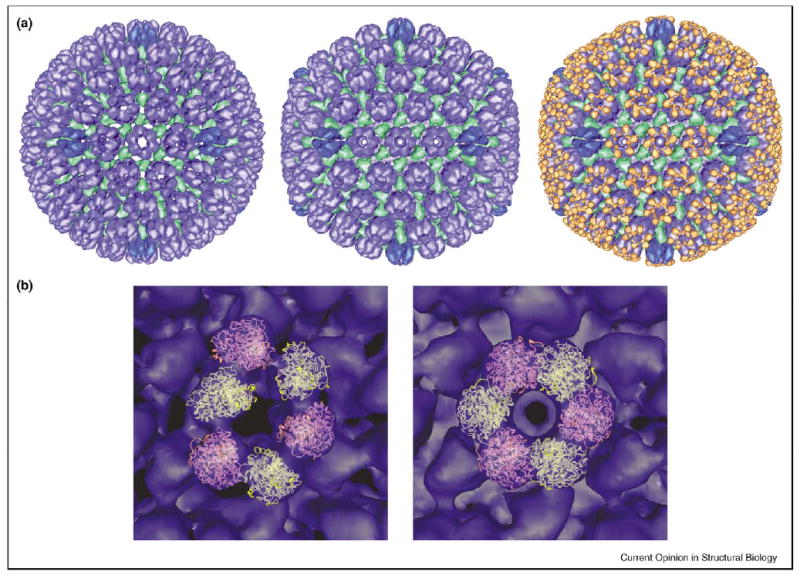
Movements of and VP26 binding by the protrusion domains of HSV1. (a) Maturation of the 1250 Å diameter HSV1 procapsid (left), whose shell contains VP5 (the MCP, blue) and the triplex proteins, VP19c and VP23 (green). If the portal protein, UL6, is available, it occupies one vertex. The scaffolding protein, preUL26.5, and the protease–scaffold fusion, preUL26, form an inner shell that is not shown. The matured capsid (middle) exposes binding sites around the tips of the VP5 hexamers that bind six copies of VP26 (orange) — right. VP26 does not bind to penton VP5. The differing conformations of penton and hexon VP5 are denoted by differing shades of blue in all three panels. The binding site for VP26 on hexons involves two adjacent subunits of VP5: inappropriate juxtaposition of these subunits offers an explanation for the failure of VP26 to bind to procapsid hexons or pentons (either state). The left and middle images were adapted from [44••], and the right from [62]. (b) The crystal structure of the protrusion domain (PDB code 1NO7, [40••]) has been fitted into the P-hexons (i.e. the peripentonal hexons) of the procapsid (left) and the mature capsid (right). The initial fitting was done by hand and then refined with an automated program (JB Heymann, unpublished). In the precursor state, there is essentially no contact between adjacent protrusion domains. In maturation, they swivel about hinges at the top of the underlying floor domain to make extensive nearest-neighbor contacts in a now highly sixfold symmetric hexon protrusion. The cryo-EM envelope of the capsid is blue. Ribbon diagrams of alternating protrusion domains around a hexon are pink and yellow.
As noted above, the structures of several herpesvirus proteases have been determined (Figure 2). They are dimers, like the scaffolding protein to which they are initially attached. HSV1 has a portal protein [37] that resembles phage portals in symmetry and structure [38•]. The triplex is a heterotrimer that coordinates capsomers at threefold lattice sites. Unlike assembly protein, it is not discarded during maturation but remains in situ, switching its role from morphogenesis (scaffold) to structural reinforcement (accessory protein). The enormous HSV1 capsid (1250 Å diameter) has been visualized at 8.5 Å resolution in its mature state [39]. Its MCP has a very high mass for a capsid protein, 150 kDa, about half of which is in the ‘protrusion domain’, which extends 80 Å outwards in an elaboration reminiscent of the excrescences on adenovirus hexon [6]. A crystal structure has been determined for the protrusion domain, revealing a highly α-helical fold [40••].
The procapsid, which is short lived, was first detected in in vitro assembly experiments with recombinant baculovirus-expressed HSV1 proteins [41]. It was then realized that the capsids that accumulate in cells infected with a virus carrying a temperature-sensitive mutation in the protease are, in fact, procapsids. When cells infected with this mutant are switched to the permissive temperature, the proteases in assembled procapsids are activated and maturation ensues. This remarkable property has enabled study of downstream events in synchronized populations [42]. The in vitro assembled procapsids were validated when they were shown to be indistinguishable from particles extracted from mutant-infected cells [43]. In HSV1 procapsids, maturation results in enhanced interactions in the hexameric rings of protrusion domains as they convert from highly asymmetric to symmetric configurations, and the establishment of strong interactions between the floor domains of adjacent capsomers (Figure 4b). These movements involve substantial rigid-body rotations, pointing to a mechanism similar to that of HK97.
Procapsids mature spontaneously, albeit slowly, under ambient conditions, even without proteolysis. This property facilitated the analysis of its dynamics by time-resolved cryo-EM, whereby 17 states could be reconstructed, temporally ordered, and assembled into movies [44••]. The complexity of this transition is generated, in part, by the staggering of the same events taking place at different quasi-equivalent sites. In this way, the integrity of the capsid is maintained as it progresses through incremental changes that ultimately transform the pattern of intermolecular interactions. One by-product is the creation of binding sites on the outer surface for a small protein, VP26; this protein binds to hexamers of the MCP but not to the pentamers (Figure 4a, right). VP26 binding appears to involve the interfaces between adjacent protrusion domains and thus may require these domains to be appropriately juxtaposed. The role of VP26 remains somewhat mysterious. It does not confer additional stability [45] and it is dispensable, although its absence somehow decreases infectious virion production [46]. Another consequence of HSV1 procapsid maturation is weakening of the interaction between the inner (scaffold) shell and the MCP. On the normal pathway, the scaffold is expelled, but in capsids that mature without packaging DNA, it is retained in a shrunken form in particles called B-capsids.
Maturation of other viruses
In this section, we touch on some of the other systems in which capsid maturation has been detected. Early studies on tailed phages have been reviewed [47]. Studies of the P22 system have continued in considerable depth (e.g. [48]) and this Salmonella phage shares many aspects of the paradigm to which HK97 subscribes.
The double-stranded RNA viruses have several notable properties, including that they must select and package one copy each of multiple (up to 11) linear segments; their capsids serve not only as a repository for nucleic acid but also as a functional compartment where transcription and replication take place. It is envisaged that, in the three-segment phage φ6 system, packaging proceeds sequentially and each successful packaging event elicits a change in the maturing procapsid, exposing a binding site that serves as a receptor for the next segment [49]. This pathway may emulate the succession of EIs in the HK97 system.
Tetraviruses are small (T=4) ssRNA viruses of insects whose assembly and maturation have been investigated for NωV. Recombinant expression of the MCP led to the identification of a procapsid that is ~25% larger than the mature capsid — the inverse of the phage paradigm [50]. Its maturation (a condensation process) has been characterized [51]. Autocatalytic cleavage of the MCP is required to render maturation irreversible, whereas the transition may be induced, reversibly, by pH change in the absence of cleavage.
Papillomaviruses are non-enveloped DNA viruses that propagate in mammalian epithelia and consist of a T=7 all-pentamer capsid [52] containing the chromatinized circular dsDNA genome. Several have been implicated in cervical carcinoma, and encouraging progress has been achieved with vaccines of recombinant capsids or virus-like particles (VLPs) [53]. Recently, a considerably larger provirion has been identified that, in maturing, undergoes compaction correlated with the formation of disulfide bonds [54••].
Lentiviruses, including HIV, follow elaborate assembly/ maturation pathways that include the envelopment (budding) of a spheroidal precursor shell of Gag and Gag-Pol polyproteins [55•], whose principal components, once separated, reorganize to form the conical capsid (CA) [56] and a thin-walled surface shell or matrix (MA) [57]. Maturation also involves processing of the glycoprotein precursor (by some other protease), activating it to a fusion-competent state.
The ability to probe precursor-product relationships depends on having in hand both the procapsid and the mature form. In vivo, procapsids mature rapidly and therefore do not accumulate: accordingly, their existence may not be suspected unless the system is perturbed so as to impede maturation. Two such situations are mutations in the maturational protease(s) and the expression of combinations of capsid protein genes in heterologous systems. It is likely that precursors exist, and maturation plays a comparably important role, in systems in which they have not yet been discovered.
Finally, we note that comparable structural transitions are undergone by the capsids of some viruses (e.g. picornaviruses) during cell entry [58•,59•]. In such cases, the mature infectious virion — although endowed with long-term stability — does not represent a global minimum free-energy state but, rather, a local minimum, from which it shifts upon interaction with the receptor [60].
Conclusions
Maturation, the crucial final phase of virus assembly, involves large cooperative conformational changes that either directly render the virion infectious or enable it to bind additional components needed for infectivity. For the class of viruses that encompasses the tailed bacteriophages and the herpesviruses, high-resolution structures have now been determined for examples of each of the five proteins that are typically involved in capsid assembly, either as building blocks or as morphogenic factors. Maturation is usually triggered by a protease whose activity triggers a structural transformation of the precursor procapsid by rigid-body rotations of the capsid protein subunits that transfer the intersubunit interactions to different, mutually complementary, regions of molecular surface. Refolding of local motifs also takes place in at least some cases. In addition to providing enhanced stability, these changes abolish binding sites that are no longer needed and create new binding sites required for downstream reactions. As a physical process, virus maturation appears similar to and may cast light on other multi-protein assemblies that also undergo structural changes in their reaction cycles. For bacteriophage HK97 and HSV, it has been possible to capture metastable intermediates of the maturation transformation in cryo-electron micrographs and, consequently, to visualize maturation as a dynamic event.
Acknowledgments
We are grateful to many outstanding collaborators, and particularly thank Bob Duda, Roger Hendrix, Phil Ross, Jack Johnson, Bill Wikoff, Kelly Lee, Lu Gan, Jay Brown, William Newcomb and Fred Homa.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3 :13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 3.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 4.Kellenberger E. Control mechanisms in the morphogenesis of bacteriophage heads. Biosystems. 1980;12:201–223. doi: 10.1016/0303-2647(80)90018-0. [DOI] [PubMed] [Google Scholar]
- 5.Caspar DL. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 7.•.Abrescia NG, Cockburn JJ, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martin C, Burnett RM, Stuart DI, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. The authors report a crystal structure at ~4 Å resolution for the 700 Å diameter virion of bacteriophage PRD1. The virion exhibits many points of resemblance to the animal virus adenovirus, including capsid geometry (a pseudo T=25 icosahedron) and the fold of the MCP (a double β barrel). [DOI] [PubMed] [Google Scholar]
- 8.Valpuesta JM, Carrascosa JL. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- 9.Simpson AA, Tao YZ, Leiman PG, Badasso MO, He YN, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL. Structure of the bacteriophage phi 29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 11.•.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. The 12-fold symmetric ring complex analyzed was isolated from SPP1 viral particles. It consists of the portal protein surmounting two other rings that are proposed to serve a valve function controlling the passage of DNA through the axial channel. Densities around this channel matched α helices present in the smaller φ29 portal [9,10], implying that they are conserved elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.••.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL: Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA-translocating machinery J Mol Biol 2005, in press.Portal proteins are also called connectors because they connect the phage head to its tail. This paper reports an 8 Å cryo-EM reconstruction of the T7 connector, a dodecameric ring whose subunit is approximately twice the size of the φ29 portal subunit previously solved as a crystal structure [9,10]. Analysis reveals conserved elements of secondary structure around the DNA channel, and indicates the placement of additional domains that are grafted onto a conserved nucleus in larger and more complex connector/portal molecules.
- 13.••.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. This paper reports the first high-resolution structure for an intact scaffolding protein, both for an assembly-naïve molecule (expressed in E. coli) at 2.2 Å resolution and an assembly-experienced molecule (extracted from procapsids) at 2.8 Å resolution. Both reveal a 70 Å long dimeric coiled coil, with the N-terminal helices back-folded to give a segment of four-helix bundle. The last 15 residues were not seen. A packing of dimers inside the procapsid is proposed, based on a cryo-EM density map at 27 Å resolution, whereby they line the inner surface of the capsid with their long axes parallel to its surface. [DOI] [PubMed] [Google Scholar]
- 14.Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PE, Jr, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J Mol Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- 15.Hendrix RW, Duda RL. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- 16.Benevides JM, Bondre P, Duda RL, Hendrix RW, Thomas GJ., Jr Domain structures and roles in bacteriophage HK97 capsid assembly and maturation. Biochemistry. 2004;43:5428–5436. doi: 10.1021/bi0302494. [DOI] [PubMed] [Google Scholar]
- 17.Fane BA, Prevelige PE., Jr Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 18.Black LW, Showe MK, Steven AC: Morphogenesis of the T4 head In Molecular Biology of Bacteriophage T4 Edited by Karam JD. American Society of Microbiology; 1994:218–258.
- 19.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked rings of covalently joined protein subunits form the dsDNA bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 20.••.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 Å resolution. J Mol Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. A refined structure is reported for the fold of the HK97 MCP [19], as manifested in the mature T=7 capsid, Head II, which contains seven quasi-equivalent conformers. The structure clarifies many details and substantiates the observation that the quasi-equivalent variations are confined to the extended N terminus, which invades neighboring subunits in Head II; swiveling of the E-loop, a β hairpin whose tip engages in cross-linking; and a loop at the tip of the A-domain. [DOI] [PubMed] [Google Scholar]
- 21.•.Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. Cryo-EM reconstructions of the P22 procapsid and capsid revealed that the P22 subunit fold resembles that of HK97 [19,20••] and that the procapsid-to-capsid transition involves rigid-body rotations of the arrayed subunits plus some local refolding, as in HK97 [23]. [DOI] [PubMed] [Google Scholar]
- 22.Chapman MS, Liljas L. Structural folds of viral proteins. Adv Protein Chem. 2003;64:125–196. doi: 10.1016/s0065-3233(03)01004-0. [DOI] [PubMed] [Google Scholar]
- 23.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 24.Waxman L, Darke PL. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir Chem Chemother. 2000;11:1–22. doi: 10.1177/095632020001100101. [DOI] [PubMed] [Google Scholar]
- 25.••.Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. Using automated sequence comparison techniques, the authors sifted faint traces of sequence similarity to unify two previously identified classes of bacteriophage proteases and herpesvirus proteases, and propose that all members of this superfamily have similar folds. These folds and their oligomeric status (dimer) have already been determined for at least five herpesviruses but, as yet, no bacteriophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol. 2000;7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 27.Duda RL. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Conway JF, Cheng N, Steven AC, Hendrix RW, Duda RL: Control of virus assembly: HK97 ‘‘whiffleball’’ mutant capsids without pentons J Mol Biol 2005, in press. [DOI] [PubMed]
- 29.Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–263. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- 30.Lee KK, Gan L, Tsuruta H, Hendrix RW, Duda RL, Johnson JE. Evidence that a local refolding event triggers maturation of HK97 bacteriophage capsid. J Mol Biol. 2004;340:419–433. doi: 10.1016/j.jmb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 31.•.Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, Johnson JE, Duda RL. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol Cell. 2004;14:559–569. doi: 10.1016/j.molcel.2004.05.015. Detailed analysis of HK97 capsid maturation, as induced by acidification, revealed that cross-link formation, which is controlled by changes in the quaternary structure of the capsid, commences as early as the EI-II stage and may be almost complete by the penultimate ‘balloon’ stage. [DOI] [PubMed] [Google Scholar]
- 32.Kim MK, Jernigan RL, Chirikjian GS. An elastic network model of HK97 capsid maturation. J Struct Biol. 2003;143:107–117. doi: 10.1016/s1047-8477(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 33.••.Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, Steven AC: Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization EMBO J 2005, in press.As HK97 matures, a network of covalent cross-links is formed that sew the capsid together. Here, differential scanning calorimetry was used in conjunction with cryo-EM to gauge the relative contributions of maturation-related structural changes and cross-linking to capsid stabilization. Stabilization was found to stem entirely from cross-linking. By genetic complementation, it was shown that cross-linking is an essential function for HK97. Cross-linking starts at EI-II stage and proceeds progressively as a Brownian ratchet, as E-loops are captured in cross-link-compatible positions. [DOI] [PMC free article] [PubMed]
- 34.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 35.Steven AC, Spear PG: Herpesvirus capsid assembly and envelopment In Structural Biology of Viruses. Edited by Chiu W, Burnett RM, Garcea RL. Oxford University Press; 1997:312–351.
- 36.Baines JD, Weller SK: Cleavage and packaging of herpes simplex virus 1 DNA In Viral Genome Packaging Machines Edited by Catalano C. Kluwer Academic/Plenum Publishers; 2005:in press.
- 37.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75 :10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.•.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. This study followed a paper [37] that answered the long-standing question of whether the HSV1 capsid, with its many phage-like characteristics, also has a portal, presenting evidence that UL6 plays this role. EM and image analysis [38•] demonstrated that baculovirus-expressed UL6 is polymorphic, ranging from 11-fold to 14-fold rings. A three-dimensional structure is presented for the 12-mer, probably the physiologically relevant oligomer, which exhibits the same architecture as phage portals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 40.••.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22:757–765. doi: 10.1093/emboj/cdg086. Recombinant expressed VP5, the 150 kDa MCP of HSV1, was reduced by trypsin digestion to a 65 kDa fragment of contiguous polypeptide chain (residues 451–1054 crystallized) and solved, revealing a novel α-helix-rich fold. This fragment fitted well into a previously published cryo-EM map at 8.5 Å resolution of the mature capsid [39], validating the structure, and also largely confirmed the assignment of α helices made from that map. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newcomb WW, Homa FL, Booy FP, Thomsen DR, Trus BL, Steven AC, Spencer JV, Brown JC. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 42.Church GA, Wilson DW. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.••.Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. HSV procapsids isolated from infected cells undergo maturation in vitro, even in the absence of proteolysis. Cryo-EM captured 17 distinct conformational states of these maturing capsids and used them as a basis for molecular movies. The underlying mechanism was observed to be the relative rotation of domains of the MCPs about hinged joints, with the same twist being staggered between different quasi-equivalent sites. [DOI] [PubMed] [Google Scholar]
- 45.Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 46.Desai P, DeLuca NA, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 47.King J, Chiu W: The procapsid-to-capsid transition in double-stranded DNA bacteriophages In Structural Biology of Viruses. Edited by Chiu W, Burnett RM, Garcea R. Oxford University Press; 1997:288–311.
- 48.Tuma R, Tsuruta H, Benevides JM, Prevelige PE, Jr, Thomas GJ., Jr Characterization of subunit structural changes accompanying assembly of the bacteriophage P22 procapsid. Biochemistry. 2001;40:665–674. doi: 10.1021/bi001965y. [DOI] [PubMed] [Google Scholar]
- 49.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canady MA, Tihova M, Hanzlik TN, Johnson JE, Yeager M. Large conformational changes in the maturation of a simple RNA virus, nudaurelia capensis omega virus (NomegaV) J Mol Biol. 2000;299:573–584. doi: 10.1006/jmbi.2000.3723. [DOI] [PubMed] [Google Scholar]
- 51.Taylor DJ, Krishna NK, Canady MA, Schneemann A, Johnson JE. Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J Virol. 2002;76:9972–9980. doi: 10.1128/JVI.76.19.9972-9980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. EMBO J. 2002;21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiller JT, Davies P. Delivering on the promise: HPV vaccines and cervical cancer. Nat Rev Microbiol. 2004;2:343–347. doi: 10.1038/nrmicro867. [DOI] [PubMed] [Google Scholar]
- 54.••.Buck CA, Thompson CD, Pang Y-YS, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol. 2005;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. This paper reports the existence of a maturation process for papilloma-viruses that involves large changes in size — shrinkage — in the opposite direction to bacteriophage expansion. Experiments were performed with ‘pseudo-viruses’ produced in a gene transfer vector expression system. The data indicate that papillomavirus maturation is a slow process based on the formation of networks of disulfide bonds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.•.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. Quantitative cryo-EM analysis of the polymorphic variability of CA cores inside mature HIV virions distinguished conical and tubular cores, and particles with more than one core. Nevertheless, the virions also contain substantial deposits of unassembled CA protein. The fine structure of the core matched that previously shown for CA particles assembled in vitro [56]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 57.Forster MJ, Mulloy B, Nermut MV. Molecular modelling study of HIV p17gag (MA) protein shell utilising data from electron microscopy and X-ray crystallography. J Mol Biol. 2000;298:841–857. doi: 10.1006/jmbi.2000.3715. [DOI] [PubMed] [Google Scholar]
- 58.•.Hewat EA, Blaas D. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J Virol. 2004. pp. 2935–2942. Based on cryo-EM comparisons of full and empty particles of rhinovirus, a picornavirus, the authors propose that a channel opens on the fivefold axis, allowing passage of the RNA. [DOI] [PMC free article] [PubMed]
- 59.•.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM: Structure of the poliovirus 135S cell-entry intermediate at 10Å resolution reveals the location of an externalized polypeptide that binds to membranes J Virol 2005, in press.Cryo-EM comparisons of the picornavirus poliovirus, with its cell entry intermediate, document rigid-body movements (‘tectonic shifts’) of the capsid protein subunits. A peptide is localized that is externalized in this transition, whereupon it interacts with the host cell membrane to facilitate entry. [DOI] [PMC free article] [PubMed]
- 60.Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoog SS, Smith WW, Qiu X, Janson CA, Hellmig B, McQueney MS, O’Donnell K, O’Shannessy D, DiLella AG, Debouck C. Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry. 1997;36:14023–14029. doi: 10.1021/bi9712697. [DOI] [PubMed] [Google Scholar]
- 62.Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the herpes simplex virus capsid and procapsid. J Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


