Abstract
Nitric oxide (NO.) is involved in neurotransmission, inflammation, and many other biological processes. Exposure of cells to NO. leads to DNA damage, including formation of deaminated and oxidized bases. Apurinic/apyrimidinic (AP) endonuclease-deficient cells are sensitive to NO. toxicity, which indicates that base excision repair (BER) intermediates are being generated. Here, we show that AP endonuclease-deficient cells can be protected from NO. toxicity by inactivation of the uracil (Ung) or formamidopyrimidine (Fpg) DNA glycosylases but not by inactivation of a 3-methyladenine (AlkA) DNA glycosylase. These results suggest that Ung and Fpg remove nontoxic NO.-induced base damage to create BER intermediates that are toxic if they are not processed by AP endonucleases. Our next goal was to learn how Ung and Fpg affect susceptibility to homologous recombination. The RecBCD complex is critical for repair of double-strand breaks via homologous recombination. When both Ung and Fpg were inactivated in recBCD cells, survival was significantly enhanced. We infer that both Ung and Fpg create substrates for recombinational repair, which is consistent with the observation that disrupting ung and fpg suppressed NO.-induced recombination. Taken together, a picture emerges in which the action of DNA glycosylases on NO.-induced base damage results in the accumulation of BER intermediates, which in turn can induce homologous recombination. These studies shed light on the underlying mechanism of NO.-induced homologous recombination.
Nitric oxide (NO.) is a versatile molecule that is critical for numerous biological processes. NO. is created intracellularly by nitric oxide synthetases through oxidation of the guanido group of arginine. At low concentrations, NO. mediates intracellular signaling and neurotransmission, while at higher concentrations, NO. is used by activated macrophages to fight invading pathogens and tumor cells (25). Although activated macrophages excrete a variety of agents, including reactive oxygen species, NO. is considered to be a critical agent, because inhibition of nitric oxide synthetases strongly diminishes the cytotoxic effect of macrophages (19). NO. and other reactive oxygen species are potent DNA-damaging agents, and exposure to NO. has been shown previously to be cytotoxic and mutagenic in Escherichia coli and a variety of other cell types (6, 12, 14, 37). Furthermore, NO. can induce homologous recombination in E. coli (34). While the mutagenic properties of NO. have been studied extensively (14, 29, 37), little is known about the underlying mechanism by which NO.-induced DNA damage leads to homologous recombination and puts cells at risk of genetic rearrangements.
NO. itself is not very reactive with DNA. However, NO. can react with oxygen and superoxide to create N2O3 and peroxynitrite, potent DNA-damaging agents. N2O3 can deaminate DNA bases to create mutagenic lesions, such as uracil, hypoxanthine, and xanthine. Peroxynitrite is an oxidizing agent, which reacts preferably with guanine. This reaction primarily results in 8-oxoguanine, which is potentially mutagenic, and 8-nitroguanine, which is susceptible to spontaneous depurination (22, 45). Interestingly, 8-oxoguanine is more susceptible to oxidation by peroxynitrite than is guanine, resulting in secondary oxidation products, which are potentially mutagenic and cytotoxic (5). Peroxynitrite has also been shown previously to induce single-strand breaks in plasmid DNA in vitro, most likely through the oxidative breakdown of deoxyribose (15).
The base excision repair (BER) pathway plays a major role in the removal of bases with NO.-induced damage in E. coli (34). DNA glycosylases initiate BER by cleaving the N-glycosylic bond between the base and the deoxyribose, resulting in an abasic (AP) site. Subsequently, AP endonucleases incise the DNA backbone immediately 5′ to the AP site, to create a 3′-OH terminus and a 5′-deoxyribose phosphate residue. The 5′-deoxyribose phosphate residue is removed by a deoxyribose phosphodiesterase, while the 3′-OH is extended by DNA polymerase I. Repair is then completed by a DNA ligase (31, 43). DNA glycosylases that can remove oxidative base damage have an associated lyase activity, so that removal of the base is thought to be coupled with nicking of the DNA backbone on the 3′ side of the lesion, resulting in a cleaved AP site. While this process eliminates the need for a deoxyribose phosphodiesterase, an AP endonuclease activity is still required to generate the 3′-OH terminus, necessary for DNA synthesis.
Several DNA glycosylases are potentially involved in the removal of NO.-induced lesions in E. coli. Uracil, xanthine, hypoxanthine, and 8-oxoguanine are substrates of the uracil DNA glycosylase (Ung), endonuclease V DNA glycosylase, AlkA 3-methyladenine DNA glycosylase (AlkA), and formamidopyrimidine DNA glycosylase (Fpg or MutM), respectively (18, 22, 30, 32). While Ung has been shown previously to protect against NO.-induced mutations, no DNA glycosylase-deficient strains, including the ung mutant, have been found to have enhanced sensitivity to NO. toxicity (34, 37). In contrast, E. coli strains deficient in AP endonuclease activity (double mutant in exonuclease III and endonuclease IV: xth nfo) are very sensitive to NO. toxicity (34). These results suggest that, upon exposure of cells to NO., BER is required to process AP sites that are formed either by DNA glycosylases or by spontaneous base loss.
In addition to AP endonucleases, recombinational repair plays a pivotal role in preventing the genotoxic effects of NO. (34). Homologous recombinational repair processes can be initiated by DNA double-strand breaks (DSBs) or single-stranded DNA regions (17). In E. coli, the RecBCD complex processes DSBs (1), while the RecFOR proteins bind single-stranded DNA regions and facilitate resumption of replication (9, 41). To survive NO. toxicity, E. coli is dependent on RecBCD, but not on RecF, which suggests that the requirement for homologous recombination following NO. exposure is not due to lesions that induce single-stranded regions or lesions that inhibit the replication fork but rather to the formation of DSBs (34). As neither NO., N2O3, nor peroxynitrite efficiently creates DSBs by direct reaction with DNA in vitro (4, 37), it is likely that NO. induces other types of damage that are subsequently converted into DSBs by enzymatic processing and/or DNA replication.
DNA damage created by the exposure of cells to NO. consists mostly of base damage (34, 37). Although some single-strand breaks appear immediately after exposure, the majority of single-strand breaks do not appear until hours after NO. exposure (at least in Chinese hamster ovary cells) (37). This delayed appearance suggests that the majority of the single-strand breaks are created enzymatically. We therefore hypothesized that NO.-induced recombination may be stimulated by the action of DNA glycosylases. NO. induces DNA base damage that is removed by DNA glycosylases, thereby protecting the cell against mutations. However, the processing of DNA base damage by DNA glycosylases leads to the creation of AP sites and single-strand breaks. In their turn, AP sites and single-strand breaks may be converted into DSBs, possibly during DNA replication. Such DSBs can induce genetic rearrangements and make the cell dependent on recombinational repair.
In this work, we show that disruption of the DNA glycosylase Ung or Fpg rescues AP endonuclease-deficient E. coli from NO. toxicity. We infer that these two DNA glycosylases are active in the production of AP sites, which are toxic to an AP endonuclease-deficient cell. Furthermore, we show that upon exposure of E. coli to NO. these DNA glycosylases lead to a dependence on RecBCD, indicating DSB repair. While overexpression of DNA glycosylases has been shown previously to sensitize recombinational repair-deficient E. coli (8, 27, 28), here we show that cells with normal expression levels of DNA glycosylases are sensitive to NO.-induced DNA DSBs. The results presented in this work shed light on the effects of DNA glycosylases on the maintenance of genomic stability. While DNA glycosylases may protect against NO.-induced mutations, they simultaneously put cells at risk of genetic rearrangements.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in this study are listed in Table 1. All strains are derived from AB1157 or GM7330, except for KL16 fpg (derived from BW35) and MV3880 (derived from MV1161). Strains constructed for this study were generated by standard P1vir transduction techniques (24). GM7624 was created by transducing the SM1004 alkA::Tet locus (33) into GM7330, while the BD2314 ung-152::Tn10 (10) and the CC104 mutM::mini-Tn10 (23) loci were transduced into GM7330 to make GM7617 and GM7619, respectively. The KM21 ΔrecBCD::Kan locus (26) was moved into GM7330 to create GM7346, while transduction of this locus into GM7617 and GM7619 resulted in the creation of GM7628 and GM7630, respectively. GM7635, GM7637, and GM7643 were constructed by the transfer of the DNA glycosylase mutant loci BD2314 ung-152::Tn10, CC104 mutM::mini-Tn10, and SM1004 alkA::Tet, respectively, into BW528. The Δfpg::Amp locus of KL16 fpg (2) was transferred into GM7617 to create EJ101. Transduction of the KM21 ΔrecBCD::Kan locus into EJ101 resulted in EJ103. EJ120 was created by moving the RW82 Δ(umuDC)595::Chl locus (44) into AB1157. Subsequent transduction with the KM21 ΔrecBCD::Kan locus resulted in EJ121. The antibiotic markers associated with the gene disruptions were confirmed by growth on the appropriate selection medium (20). Disruptions in ΔrecBCD were confirmed by UV sensitivity, while alkA and xth nfo disruptions were confirmed by sensitivity to methyl methanesulfonate. Transduction of the ung-152::Tn10 locus is 100% linked between Tetr and ung phenotypes when selecting for Tetr recombinants (10). Transduction of ung-152::Tn10 into GM7330 and GM7346 was also confirmed by observing ∼5-fold-increased mutation frequency on rifampin plates. Transductions of the mutM::mini-Tn10, Δ fpg::Amp, Δ(umuDC)595::Chl, and ΔrecBCD::Kan loci were confirmed by PCR (data not shown). Plasmids listed in Table 1 were verified by restriction analysis. The pTr vector was created by deleting ∼70% of the Ung coding sequence (between two BamHI sites) that had been cloned into a pTrc99A plasmid (42).
TABLE 1.
E. coli strains and plasmids used
| Strain or plasmid | Relevant genotype | Source |
|---|---|---|
| Strains | ||
| AB1157 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(As) galK2(Oc) hisG4(Oc) rfbD1 mgl-51 rpoS396(Am) rpsL31(Strr) kdgK51 xylA5 mtl-1 argE3(Oc) thi-1 | Lab stock |
| BD2314 | ung-152::Tn10 lacY1 gal-6 trpC45 his-68 purC50 tyrA2 rpsL125(Strr) malT1 xylA7 mtl-2 thi-1 [fluA2 tsx-70 supE44(As)] | B. Duncan |
| BW35 | Hfr KL16 PO-45::[lysA(61)-serA(62)]thi relA spoT1 | S. Wallace |
| BW528 | As AB1157 but Δxth-pncA nfo-1::Kan | B. Weiss |
| CC104 | [F′ proAB+lacI378 lacZ461] ara Δ(gpt-lac)5rpsL mutM::mini-Tn10 | J. Miller |
| GC4803 | As AB1157 but tagA1::Tn5 his+alkA1 | S. Boiteux |
| KL16 fpg | As KL16 but Δfpg::Amp | S. Wallace |
| KM21 | As AB1157 but ΔrecBCD::Kan | K. Murphy |
| MV1161 | As AB1157 but rfa-550 | M. Volkert |
| MV3880 | As MV1161 but ung-152::Tn10 | M. Volkert |
| RW82 | Δ(umuDC)595::Chl uvrA6 | G. Walker |
| SM1004 | As CC106 but uvrA alkA::Tet | J. Laval |
| GM7330 | ΔlacMS286 φ80dIIlacBK1 ara thi(?) | Lab stock |
| GM7617 | As GM7330 but ung-152::Tn10 | This work |
| GM7619 | As GM7330 but mutM::mini-Tn10 | This work |
| GM7624 | As GM7330 but alkA::Tet | This work |
| EJ101 | As GM7330 but ung-152::Tn10 Δfpg::Amp | This work |
| GM7346 | As GM7330 but ΔrecBCD::Kan | This work |
| GM7628 | As GM7330 but ΔrecBCD::Kan ung-152::Tn10 | This work |
| GM7630 | As GM7330 but ΔrecBCD::Kan mutM::mini-Tn10 | This work |
| EJ103 | As GM7330 but ΔrecBCD::Kan ung-152::Tn10 Δfpg::Amp | This work |
| GM7635 | As BW528 but ung-152::Tn10 | This work |
| GM7637 | As BW528 but mutM::mini-Tn10 | This work |
| GM7643 | As BW528 but alkA::Tet | This work |
| EJ120 | As AB1157 but Δ(umuDC)595::Chl | This work |
| EJ121 | As AB1157 but Δ(umuDC)595::Chl ΔrecBCD::Kan | This work |
| Plasmids | ||
| pSL-Fpg | Fpg sequence in pSL | L. Samson |
| pSL | pSL301; IPTGa inducible, Ampr | Lab stock |
| pTr-Ung | Ung sequence in pTrc99A | J. van de Sande |
| pTr | Ung deleted in pTr-Ung | This work |
| pTrc99A | IPTG inducible, Ampr | J. van de Sande |
IPTG, isopropyl-β-d-thiogalactopyranoside.
Cytotoxicity assay.
The cytotoxicity assay has been described previously (34). Briefly, overnight cultures were diluted 100-fold into Luria-Bertani medium (LB) and grown to early log phase. Cultures were exposed to NO. gas via silastic tubing at rates of ∼35 nmol/ml/min. Delivery was confirmed by measuring the concentration of nitrate plus nitrite formed after 2 h or by including a strain of known sensitivity as an internal control (38). After exposure, cells appropriately diluted in M9 salts were plated onto LB plates. Surviving colonies were scored after overnight incubation at 37°C. Strains bearing plasmids were inoculated from a single colony and grown overnight in LB-ampicillin (50 μg/ml) medium. The next day, the presence of the plasmid was confirmed by plasmid isolation (Qiagen miniprep kit) and agarose gel analysis (20). Cultures carrying appropriate vectors were diluted 100-fold into LB-ampicillin medium supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside to induce expression. Cultures were grown until early log phase, exposed to NO., and analyzed as described above.
Recombination assay.
Strains derived from GM7330 (GM7617, GM7619, GM7624, GM7346, GM7628, GM7630, EJ101, and EJ103) carry two different nonfunctional alleles of the lac operon, each with a different deletion. A homologous recombination event between the mutant alleles can restore the Lac+ phenotype (16). A quantitative recombination assay has been described previously (34). Briefly, early-log-phase cells were exposed to NO. for 2 h, as described above. After exposure, cells were incubated in M9 salts for 30 min, pelleted, resuspended in M9 salts, appropriately diluted, and plated. LB agar plates were used to determine the number of survivors. Recombinant colonies on lactose plates (M9 salts, 1% lactose, 1 μg of thiamine/ml, and 15 g of agar/liter) were scored after incubation for 3 days at 37°C.
RESULTS
DNA glycosylases remove NO.-induced base damage in E. coli.
When cells are exposed to NO., damaged bases are the most prevalent form of DNA damage (37). Our first objective was to identify DNA glycosylases that remove NO.-induced base damage. We were unable to determine which DNA glycosylases were active by assaying DNA glycosylase mutant strains for sensitivity to NO. toxicity, since none of the strains examined showed any increased NO. sensitivity (34). However, AP endonuclease-deficient E. coli strains are sensitive to NO.-induced toxicity (34), and DNA glycosylases generate AP sites, which are toxic if they are not processed by AP endonucleases. Thus, if a particular DNA glycosylase is active, disruption of that DNA glycosylase may rescue AP endonuclease-deficient cells from NO. toxicity.
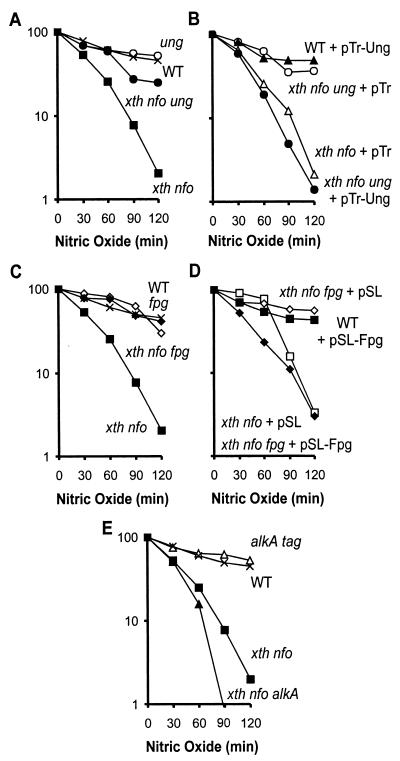
Here we show that disruption of the ung gene in the AP endonuclease-deficient E. coli strain renders the cells significantly more resistant to the toxic effects of NO. (compare xth nfo and xth nfo ung strain data in Fig. 1A). The rescue of the xth nfo double mutant afforded by disruption of ung suggests that Ung creates a significant portion of the AP sites that make AP endonuclease-deficient cells sensitive to NO.. To confirm that the phenotype of the xth nfo ung triple mutant strain was due specifically to a defect in Ung, we complemented this strain by expressing Ung from an isopropyl-β-d-thiogalactopyranoside-inducible promoter (see Materials and Methods). Reintroduction of Ung restored the sensitivity of the xth nfo ung triple mutant to NO. toxicity, confirming that Ung is responsible for sensitization of AP endonuclease-deficient cells. As expected, additional expression of Ung did not sensitize wild-type (WT) cells to NO. toxicity (Fig. 1B) (27).
FIG. 1.
Relative NO. sensitivities of E. coli strains with varied levels of expression of BER genes. Percent survival is shown on the y axis in all panels. (A) Survival of WT (×), xth nfo (▪), ung (○), and xth nfo ung (•) E. coli strains. (B) Survival of WT cells expressing ung from pTr-Ung (▴), xth nfo E. coli carrying control pTr vector (▵), xth nfo ung E. coli expressing ung from pTr-Ung (•), and xth nfo ung E. coli carrying control pTr vector (○). (C) Survival of WT (×), xth nfo (▪), fpg (⋄), and xth nfo fpg (♦) E. coli strains. (D) Survival of WT cells expressing fpg from pSL-Fpg (▪), xth nfo cells carrying control pSL (□), xth nfo fpg cells expressing fpg from pSL-Fpg (♦), and xth nfo fpg cells carrying control pSL (⋄). (E) Relative survival of WT (×), alkA tag (▵), xth nfo (▪), and xth nfo alkA (▴) E. coli strains. Data shown in panels A and C are the averages of three or more experiments. Experiments represented in panels B and D were repeated at least two times. Data shown in panel E are the averages of two or more experiments.
As in the case of Ung, we reasoned that, if Fpg is active following NO. exposure, disruption of fpg expression could protect xth nfo double mutants from NO. toxicity. Indeed, disruption of fpg in an xth nfo double mutant made that strain as resistant to NO. as were WT cells, implying that Fpg converts a significant proportion of NO.-induced base damage into potentially toxic BER intermediates (Fig. 1C). Reintroduction of Fpg restored the sensitivity of the xth nfo fpg triple mutant to NO. toxicity (Fig. 1D), confirming that the phenotype of the xth nfo fpg triple mutant was due to a defect in Fpg. It is noteworthy that additional expression of Fpg in WT cells did not increase cellular sensitivity to NO. toxicity (Fig. 1D). While inactivation of Ung or Fpg rescued AP endonuclease-deficient E. coli from the toxic effects of NO., inactivation of AlkA did not rescue the xth nfo double mutant (Fig. 1E). In contrast, inactivation of AlkA seemed to slightly sensitize the AP endonuclease-deficient E. coli strain to NO. toxicity.
Remarkably, inactivation of either Ung or Fpg can rescue AP endonuclease-deficient E. coli from the toxic effects of NO.. Apparently, abolishing the activity of just one of these DNA glycosylases is sufficient to prevent BER intermediates from reaching toxic levels in an AP endonuclease-deficient strain. These results imply that the number of AP sites must be above a certain threshold in order to be toxic.
Ung and Fpg convert NO.-induced base damage into substrates for recombinational repair.
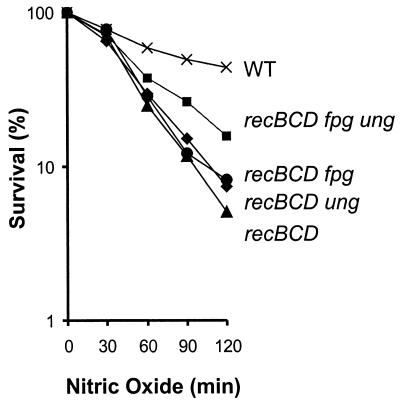
Upon NO. exposure, Ung and Fpg create BER intermediates (see above). While Fpg can create cleaved AP sites, Ung can create only uncleaved AP sites (although these AP sites are likely to be rapidly converted into single-strand breaks by AP endonucleases). Both AP sites and single-strand breaks can induce recombination, because they block DNA replication and/or can be converted into DSBs when encountered by a replication fork (17, 27). Furthermore, DNA glycosylases can create DSBs by removing damaged bases that are in close proximity on opposite strands of the DNA (3, 35). To survive DSBs, E. coli employs RecBCD, which initiates DSB recombinational repair. Previous studies show that recBCD mutant cells are sensitive to NO. toxicity (Fig. 2) (34). If Ung and Fpg are responsible for creating recombinogenic lesions, inactivating these DNA glycosylases should rescue recBCD mutant cells from NO. toxicity. Inactivation of either Ung or Fpg has little effect on the recBCD mutant cells and, if anything, provides slight protection. However, inactivation of both Ung and Fpg provided protection of recBCD E. coli from NO. toxicity (Fig. 2). These results suggest that, when cells are challenged with NO., Ung and Fpg create BER intermediates that are potentially toxic if they are not repaired via homologous recombination.
FIG. 2.
Relative NO. sensitivities of WT E. coli and strains deficient in recombinational repair and DNA glycosylases. All strains were derived from GM7330. The figure shows survival of WT (×), recBCD (▴), recBCD ung (♦), recBCD fpg (•), and recBCD fpg ung (▪) E. coli strains. The data are the averages of at least three independent experiments.
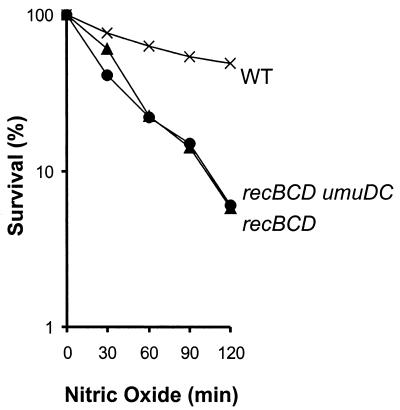
Inactivation of umuDC does not render recBCD mutants more sensitive to NO..
The gene product of umuDC is involved in translesion synthesis of a variety of DNA lesions, including AP sites (39), and cells deficient in umuDC are sensitive to the toxic effects of high levels of uncleaved AP sites (27). If uncleaved AP sites lead to replication fork collapse, which would induce recombinational repair, then inactivation of umuDC in a recBCD mutant is expected to lead to additional sensitization of the recBCD mutant. However, inactivation of umuDC did not render recBCD mutants more sensitive to NO. (Fig. 3). (Inactivation of umuDC in WT E. coli did not sensitize the cells to NO. [data not shown].) Uncleaved AP sites are therefore unlikely to play a major role in initiating recombination. We therefore conclude that cleaved abasic sites, rather than uncleaved abasic sites, lead to DSBs that are toxic to recBCD mutant cells.
FIG. 3.
Relative NO. sensitivities of WT E. coli and strains deficient in recombinational repair and translesion DNA synthesis. The figure shows survival of WT (×), recBCD (▴), and recBCD umuDC (•) E. coli strains. The data are the averages of at least three independent experiments.
Ung and Fpg induce recombination in E. coli exposed to NO..
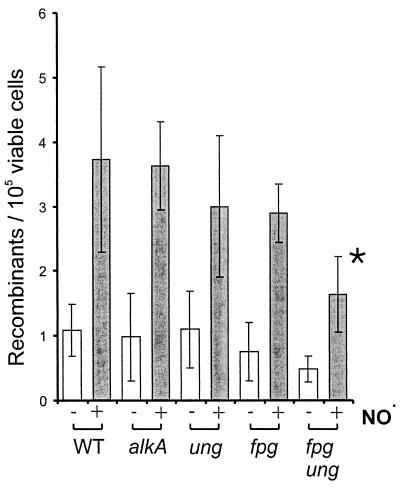
The bacterial strain GM7330 carries two inactive copies of the lac operon, which can be reconstituted via homologous recombination (16). An earlier study showed that exposure of GM7330 to NO. results in an increase in Lac+ cells, indicating that NO. induces homologous recombination (34). Since Ung and Fpg create substrates for recombinational repair, it seemed likely that they are also involved in promoting NO.-induced recombination. We tested this hypothesis by inactivating the Fpg, Ung, and AlkA DNA glycosylases in GM7330. While an alkA mutant showed the same level of NO.-induced recombination as that of WT E. coli (Fig. 4), the recombination frequency appeared to be slightly lower in strains deficient in either ung or fpg, though the difference was not statistically significant. However, a statistically significant decrease in NO.-induced recombination was observed when both ung and fpg were inactivated. This implies that DNA glycosylase activity is the underlying cause of a significant fraction of NO.-induced recombination.
FIG. 4.
NO.-induced recombination in WT E. coli and strains deficient in DNA glycosylases. All strains are derived from GM7330. Recombination frequencies (Lac+ E. coli per 105 viable cells) were determined for control (open bars) and cells exposed to NO. for 2 h (gray bars). Recombination frequencies at equitoxic doses are shown (survival is between 20 and 40%). The error bars denote 95% confidence intervals. The asterisk indicates that the difference between GM7330 and the fpg ung double mutant is statistically significant (Student's t test).
DISCUSSION
During DNA replication, lesions that inhibit DNA polymerases and single-strand breaks in the template DNA can lead to unreplicated single-stranded regions and/or DSBs at collapsed replication forks. Both single-strand gaps and DSBs are a threat to genomic integrity and cellular survival. Homologous recombinational repair offers critical protection against these inevitable problems (17). An example of an endogenously produced lesion that blocks DNA replication is 3-methyladenine, which is normally removed by 3-methyladenine DNA glycosylases (43). Saccharomyces cerevisiae and mammalian cells deficient in 3-methyladenine DNA glycosylase activity are sensitive to the lethal and recombinogenic effects of unrepaired 3-methyladenine, which is consistent with the protective role generally attributed to DNA glycosylases (7, 11). Ironically, DNA glycosylases generate repair intermediates that may themselves require recombinational repair. During BER, AP sites and single-strand breaks are created that can be converted into DSBs during DNA replication (17). The effects of DNA glycosylases on homologous recombination are therefore complex, as the ability of DNA glycosylase substrates to induce recombination must be weighed against that of the repair intermediates generated by the action of DNA glycosylases.
In this work we show that Ung and Fpg remove NO.-induced base damage by demonstrating that AP endonuclease-deficient cells can be rescued from NO. toxicity by disruption of ung and fpg. Using a similar approach, Galhardo et al. have shown that, under specific conditions, inactivation of Fpg in an AP endonuclease-deficient strain can partially protect that strain from hydrogen peroxide toxicity (13). Furthermore, in a classic study Taylor and Weiss showed that the nonviable dut xth double mutant strain can be rescued by inactivating Ung (40). The dut xth double mutant strain incorporates large amounts of uracil into the genome, which subsequently is repaired by Ung, leading to an accumulation of repair intermediates and inviability of the strain. Interestingly, our studies show that inactivation of either Ung or Fpg suffices to rescue the AP endonuclease mutant cells from NO. toxicity, which is remarkable since none of the major NO.-induced lesions are thought to be substrates for both Ung and Fpg. However, both Ung and Fpg generate AP sites, and cells have several alternative pathways for tolerating AP sites, including translesion synthesis and recombinational repair (27, 36). Although these alternative defense mechanisms apparently suffice if either Fpg or Ung is inactivated in xth nfo double mutants, the combined activity of both Fpg and Ung in an AP endonuclease-deficient strain can overwhelm these defense mechanisms.
In contrast to disruption of Ung or Fpg, inactivation of the AlkA 3-methyladenine DNA glycosylase activity in AP endonuclease-deficient E. coli does not rescue the xth nfo double mutant from NO. toxicity. If anything, the xth nfo alkA strain seems to be slightly more sensitive to NO. than the xth nfo strain. One explanation for this observation lies in a difference in the availability of damage tolerance pathways in WT cells and xth nfo mutant cells. In AP endonuclease-deficient cells, recombinational repair and translesion synthesis are likely to be recruited to process the large numbers of BER intermediates that accumulate. Consequently, in xth nfo mutants exposed to NO., these tolerance pathways may not be available for lesions normally removed by AlkA, and inactivating AlkA can increase the sensitivity of the xth nfo mutant.
Accumulation of AP sites and other BER intermediates makes cells dependent on homologous recombinational repair for survival (27, 36). Several studies have shown that overexpression of a DNA glycosylase leads to the induction of homologous recombination and/or sensitizes DSB-repair-deficient strains to the effects of BER intermediates (8, 27, 28). Furthermore, the alkylation sensitivity of an Schizosaccharomyces pombe strain deficient in recombinational repair could be partially relieved by inactivation of its 3-methyladenine DNA glycosylase, which suggests that BER intermediates are fed into the recombinational repair pathway in this species (21). Here, we show that recBCD mutant E. coli, deficient in the repair of DSBs, can be partially rescued from NO. toxicity by inactivating Ung and Fpg, which demonstrates that a significant number of NO.-induced DSBs are created by DNA glycosylases. A similar consequence of DNA glycosylase activity has been reported by Blaisdell and Wallace, who have shown that inactivation of Fpg can protect E. coli from radiation damage by decreasing the amount of radiation-induced DSBs (3). Consistent with the result that recBCD mutant cells can be significantly protected from NO. toxicity by inactivation of Ung and Fpg, we find that inactivation of these same DNA glycosylases suppresses NO.-induced homologous recombination. Our results imply that, in WT cells, Ung and Fpg are responsible for the promotion of a significant portion of NO.-induced recombination events (Fig. 4).
Ung and Fpg do not create DSB substrates but create uncleaved and cleaved AP sites, respectively. However, both kinds of AP sites can be converted into DSBs (uncleaved AP sites may stall the replication fork, and cleaved AP sites may directly cause fork collapse [17]). In the case of exposure of E. coli to NO., uncleaved AP sites do not seem to play a role in the induction of recombination, since inactivation of umuDC, which can suppress the toxicity of uncleaved AP sites, did not further sensitize a recBCD mutant strain to NO. toxicity. Although Ung is actively creating uncleaved AP sites upon NO. exposure, these may be rapidly converted into single-strand breaks by the action of AP endonucleases. Alternatively, uncleaved AP sites may be less easily converted into substrates for recombination than cleaved AP sites or may be far less abundant when the cell processes NO. damage.
The studies presented here show that NO.-induced recombination is promoted by DNA glycosylases. In the case of Ung and Fpg, the lesions normally repaired by these enzymes (which are likely to include uracil and 8-oxoguanine, respectively) are potentially mutagenic, but they are not thought to inhibit DNA replication. We observed that cells deficient in Ung or Fpg have increased susceptibility to NO.-induced mutations (data not shown; for Ung, see also the work of Tamir et al. [37]). However, inactivation of Ung and Fpg also resulted in a decrease in recombination frequency (Fig. 4). Thus, leaving NO.-induced base damage in the genome puts cells at risk of mutations, while removing these lesions can put cells at risk of genetic rearrangements (Fig. 5). It is noteworthy that, in these studies, cells were exposed to levels of NO. that approximate conditions of inflammation (38). Furthermore, in contrast to many other studies, DNA glycosylases are expressed at normal levels. Thus, these studies demonstrate that, under conditions where cells are exposed to physiologically relevant levels of NO., DNA glycosylases, expressed under their endogenous promoters, can create high-enough levels of repair intermediates to put cells at risk of genetic rearrangements. These findings shed light on the effects of exposure of bacteria to macrophages during inflammation and also provide a framework for the study of the relationship between inflammation and genetic stability in mammalian cells.
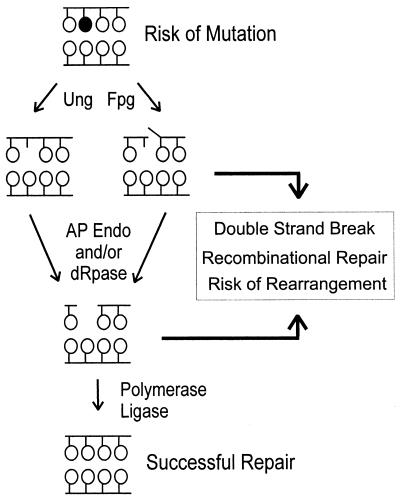
FIG. 5.
Model for the promotion of genetic changes upon exposure of cells to NO.. Genetic changes can be avoided by successful repair of base damage by BER enzymes. However, NO.-induced base damage can lead to mutations if encountered by the replication fork prior to BER. Recombinational repair is required when repair intermediates are converted into DSBs, possibly during DNA replication.
Acknowledgments
This work was primarily supported by NIH grant R01CA79827-0 with partial support from NIH grant CA26371-21. M.G.M. is supported by a Howard Hughes Medical Institute Research Resources Program for Medical Schools Award to the University of Massachusetts Medical School.
We thank Leona Samson, Steven Tannenbaum, and Gerald Wogan for valuable discussions and comments on the manuscript. The help of Teresa Wright and Joe Glogowski is gratefully acknowledged. We thank S. Boiteux, B. Duncan, J. Laval, J. Miller, K. Murphy, L. Samson, J. van de Sande, M. Volkert, G. Walker, S. Wallace, and B. Weiss for strains and plasmids listed in Table 1.
REFERENCES
- 1.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 90:77-86. [DOI] [PubMed] [Google Scholar]
- 2.Blaisdell, J. O., Z. Hatahet, and S. S. Wallace. 1999. A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J. Bacteriol. 181:6396-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaisdell, J. O., and S. S. Wallace. 2001. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:7426-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burney, S., J. L. Caulfield, J. C. Niles, J. S. Wishnok, and S. R. Tannenbaum. 1999. The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat. Res. 424:37-49. [DOI] [PubMed] [Google Scholar]
- 5.Burney, S., J. C. Niles, P. C. Dedon, and S. R. Tannenbaum. 1999. DNA damage in deoxynucleosides and oligonucleotides treated with peroxynitrite. Chem. Res. Toxicol. 12:513-520. [DOI] [PubMed] [Google Scholar]
- 6.Burney, S., S. Tamir, A. Gal, and S. R. Tannenbaum. 1997. A mechanistic analysis of nitric oxide-induced cellular toxicity. Nitric Oxide 1:130-144. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., B. Derfler, and L. Samson. 1990. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 9:4569-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coquerelle, T., J. Dosch, and B. Kaina. 1995. Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents—a case of imbalanced DNA repair. Mutat. Res. 336:9-17. [DOI] [PubMed] [Google Scholar]
- 9.Courcelle, J., D. J. Crowley, and P. C. Hanawalt. 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and RecF protein function. J. Bacteriol. 181:916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan, B. K. 1985. Isolation of insertion, deletion, and nonsense mutations of the uracil-DNA glycosylase (ung) gene of Escherichia coli K-12. J. Bacteriol. 164:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelward, B., A. Dreslin, J. Christensen, D. Huszar, C. Kurahara, and L. Samson. 1996. Repair deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 15:945-952. [PMC free article] [PubMed] [Google Scholar]
- 12.Gal, A., and G. N. Wogan. 1996. Mutagenesis associated with nitric oxide production in transgenic SJL mice. Proc. Natl. Acad. Sci. USA 93:15102-15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galhardo, R. S., C. E. Almeida, A. C. Leitao, and J. B. Cabral-Neto. 2000. Repair of DNA lesions induced by hydrogen peroxide in the presence of iron chelators in Escherichia coli: participation of endonuclease IV and Fpg. J. Bacteriol. 182:1964-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juedes, M. J., and G. N. Wogan. 1996. Peroxynitrite-induced mutation spectra of pSP189 following replication in bacteria and in human cells. Mutat. Res. 349:51-61. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, L. J., K. Moore, Jr., J. L. Caulfield, S. R. Tannenbaum, and P. C. Dedon. 1997. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem. Res. Toxicol. 10:386-392. [DOI] [PubMed] [Google Scholar]
- 16.Konrad, E. B. 1977. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J. Bacteriol. 130:167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl, T. 1974. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 71:3649-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacMicking, J., Q. W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323-350. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Memisoglu, A., and L. Samson. 2000. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 182:2104-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaels, M. L., L. Pham, C. Cruz, and J. H. Miller. 1991. MutM, a protein that prevents G · C—-T · A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 19:3629-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Moncada, S., R. M. Palmer, and E. A. Higgs. 1991. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 43:109-142. [PubMed] [Google Scholar]
- 26.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otterlei, M., B. Kavli, R. Standal, C. Skjelbred, S. Bharati, and H. E. Krokan. 2000. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 19:5542-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posnick, L. M., and L. D. Samson. 1999. Imbalanced base excision repair increases spontaneous mutation and alkylation sensitivity in Escherichia coli. J. Bacteriol. 181:6763-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Routledge, M. N. 2000. Mutations induced by reactive nitrogen oxide species in the supF forward mutation assay. Mutat. Res. 450:95-105. [DOI] [PubMed] [Google Scholar]
- 30.Saparbaev, M., and J. Laval. 1994. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl. Acad. Sci. USA 91:5873-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharer, O. D., and J. Jiricny. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23:270-281. [DOI] [PubMed] [Google Scholar]
- 32.Schouten, K. A., and B. Weiss. 1999. Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res. 435:245-254. [DOI] [PubMed] [Google Scholar]
- 33.Sidorkina, O., M. Saparbaev, and J. Laval. 1997. Effects of nitrous acid treatment on the survival and mutagenesis of Escherichia coli cells lacking base excision repair (hypoxanthine-DNA glycosylase-AlkA protein) and/or nucleotide excision repair. Mutagenesis 12:23-28. [DOI] [PubMed] [Google Scholar]
- 34.Spek, E. J., T. L. Wright, M. S. Stitt, N. R. Taghizadeh, S. R. Tannenbaum, M. G. Marinus, and B. P. Engelward. 2001. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol. 183:131-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland, B. M., P. V. Bennett, O. Sidorkina, and J. Laval. 2000. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry 39:8026-8031. [DOI] [PubMed] [Google Scholar]
- 36.Swanson, R. L., N. J. Morey, P. W. Doetsch, and S. Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamir, S., S. Burney, and S. R. Tannenbaum. 1996. DNA damage by nitric oxide. Chem. Res. Toxicol. 9:821-827. [DOI] [PubMed] [Google Scholar]
- 38.Tamir, S., R. S. Lewis, T. de Rojas Walker, W. M. Deen, J. S. Wishnok, and S. R. Tannenbaum. 1993. The influence of delivery rate on the chemistry and biological effects of nitric oxide. Chem. Res. Toxicol. 6:895-899. [DOI] [PubMed] [Google Scholar]
- 39.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, A. F., and B. Weiss. 1982. Role of exonuclease III in the base-excision repair of uracil-containing DNA. J. Bacteriol. 151:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umezu, K., N. W. Chi, and R. D. Kolodner. 1993. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl. Acad. Sci. USA 90:3875-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varshney, U., T. Hutcheon, and J. H. van de Sande. 1988. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J. Biol. Chem. 263:7776-7784. [PubMed] [Google Scholar]
- 43.Wilson, D. M., III, B. P. Engelward, and L. Samson. 1998. Prokaryotic base excision repair, p. 29-64. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair: biochemistry, genetics, and cell biology, vol. I. Humana Press Inc., Totowa, N.J.
- 44.Woodgate, R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]
- 45.Yermilov, V., J. Rubio, M. Becchi, M. D. Friesen, B. Pignatelli, and H. Ohshima. 1995. Formation of 8-nitroguanine by the reaction of guanine with peroxynitrite in vitro. Carcinogenesis 16:2045-2050. [DOI] [PubMed] [Google Scholar]