Abstract
Actinobacillus actinomycetemcomitans is a member of the family Pasteurellaceae and a major causative agent of periodontitis. While several genera from this family are known to be competent for transformation, A. actinomycetemcomitans has yet to be fully characterized. Here we show that the competence of A. actinomycetemcomitans is remarkably similar to that of Haemophilus influenzae. In addition to having a similar frequency of transformation as H. influenzae, A. actinomycetemcomitans competence could also be induced at least 100-fold by cyclic AMP, suggesting that, as in H. influenzae, at least some competence genes are regulated by catabolite repression. Even more intriguing was the discovery of a putative A. actinomycetemcomitans DNA uptake signal sequence (USS) virtually identical to the USS of H. influenzae. Moreover, we provide evidence that this sequence functions in the same capacity as that from H. influenzae; the sequence appears to be required and sufficient for DNA uptake in a variety of assays. Finally, we have taken advantage of this system to develop a simple, highly efficient competence-based method for generating site-directed mutations in the wild-type fimbriated A. actinomycetemcomitans.
Actinobacillus actinomycetemcomitans, a gram-negative, facultative, nonmotile coccobacillus, is a major causative agent of localized juvenile periodontitis and other forms of chronic periodontitis (36). Moreover, A. actinomycetemcomitans is transmissible among family members and may persist for long periods (4). A. actinomycetemcomitans can also spread from the oral cavity, causing a variety of nonoral infections (31, 36). Indeed, the genomic DNA of A. actinomycetemcomitans was identified in atherosclerotic lesions (14), suggesting a possible role of this bacterium in cardiovascular disease.
Some A. actinomycetemcomitans strains are known to be naturally competent for transformation (23, 30). In many bacteria, transformation occurs when cells enter the state of competence. This enables them to take up DNA and incorporate it into their genomes (8, 18). In the best-studied naturally competent gram-negative bacteria, Haemophilus influenzae, Haemophilus parainfluenzae, Neisseria gonorrhoeae, and Neisseria meningitidis, double-stranded donor DNA fragments are preferentially bound into a DNase-resistant state if they contain a short recognition sequence abundant in their own genomes, the uptake signal sequence (USS) (10, 11). Although uptake has been demonstrated elsewhere for A. actinomycetemcomitans in two preliminary studies (23, 30), a quantitative analysis has yet to be performed. In particular, neither of these studies examined any of the requirements for the induction of competence. Nor was a specific DNA uptake sequence requirement for transformation investigated.
Because the function of the USS has been assumed to be exclusion of DNAs from other species, different genera were expected to use different USSs. However, only USSs from the very distantly related genera Haemophilus and Neisseria have been characterized; these use completely unrelated USSs. H. influenzae and A. actinomycetemcomitans are both members of the family Pasteurellaceae. In this report, we identified a highly competent, fimbriated wild-type A. actinomycetemcomitans clinical isolate and found that its transformation properties resemble those of H. influenzae. We also identified a USS that was remarkably similar to the H. influenzae USS. With this knowledge we have subsequently been able to develop a highly efficient method for gene replacement in wild-type A. actinomycetemcomitans via natural transformation.
MATERIALS AND METHODS
Bacteria and culture conditions.
A. actinomycetemcomitans strains HK1651 (ATCC 700685) (15), ATCC 33384, and Y4 (ATCC 43718) were purchased from the American Type Culture Collection, Manassas, Va. A. actinomycetemcomitans strain JP-2 was a gift from S. Asikainen of Umea University, Umea, Sweden. Clinical isolates of A. actinomycetemcomitans strains 29R, 29SS, H1S, H5P, D3S, D7S, D9S, D11S, D15S, D17S, D18P, D28S, and D37S were part of the collection of oral clinical isolates of A. actinomycetemcomitans in our laboratory. The A. actinomycetemcomitans clinical isolates were identified from subgingival plaque samples by the culture method as described previously (24) and further confirmed by a 16S rRNA-based PCR method (3). Spontaneous nonfimbriated smooth-colony mutants of A. actinomycetemcomitans were isolated by repeatedly transferring the bacteria into fresh medium until increased turbidity was noted and streaking the bacteria on agar medium for isolation of smooth-colony isolates. Escherichia coli plasmids were propagated in E. coli DH5α as previously described (32).
A. actinomycetemcomitans was routinely cultured on Trypticase soy broth supplemented with 0.1% yeast extract, 5% heat-inactivated horse serum, and 1.5% agar (sTSB agar) at 37°C in 5% CO2. When indicated, the medium was supplemented with spectinomycin (50 μg/ml), nalidixic acid (16 to 24 μg/ml), or rifampin (25 to 50 μg/ml). Additional media used for transformation included sTSB broth (same as above but without agar), TSB agar, Columbia agar (Difco), and Luria-Bertani (LB) agar. Bacteria were grown in broth without shaking at 37°C in 5% CO2.
Scanning electron microscopy.
Scanning electron microscopy was used to examine the cell morphology of A. actinomycetemcomitans growing on coverslips. A starter culture of A. actinomycetemcomitans was prepared by growing the bacteria in sTSB broth overnight. The overnight culture (20 μl) was transferred to a tube containing 3 ml of sTSB broth and a piece of coverslip, and the bacteria were cultured for an additional 15 h. The coverslip was then gently rinsed once with cacodylate buffer (0.1 M cacodylate buffer with 0.02% CaCl2) to remove the nonadherent bacteria. The coverslip samples were then fixed for 20 min in fixative (2.5% glutaraldehyde, 2% paraformaldehyde in cacodylate buffer), rinsed three times with cacodylate buffer, and dehydrated in a graded ethanol series. The coverslip was then immersed in hexamethyldisilazane and air dried. After gold coating, the preparations were observed with a Cambridge 360 scanning electron microscope.
Isolation of antibiotic-resistant mutants.
A. actinomycetemcomitans cells were plated on sTSB agar containing rifampin (50 μg/ml) at ∼2 × 108 CFU per plate. Rifr variants exhibiting normal colony size were isolated for further use. Nalr Rifr double mutants were identified by plating Rifr mutants on agar containing nalidixic acid (24 μg/ml) and subsequently culturing them for 3 to 4 days.
DNA method and plasmids.
Chromosomal DNA was prepared as previously described (33). Plasmids and PCR products were purified with spin columns (Qiagen, Inc.). DNA sequencing and oligonucleotide synthesis were performed by the USC School of Medicine Microchemical Core Facility.
Plasmid pPK1 was a generous gift from P. Fives-Taylor. It is a 3.6-kb Sper E. coli-A. actinomycetemcomitans shuttle plasmid constructed from pUC19 and pVT736-1 (28). pPK1 used in experiments was always propagated in E. coli. The spectinomycin resistance cassette plasmid, pK-Spe, was constructed by replacing the 0.3-kb EcoRI fragment on pBSL237 (2) with the 1.2-kb SphI Sper gene from pVT1461 (a gift from P. Fives-Taylor) by standard recombinant DNA techniques (22).
Transformation assays.
Transformation assays were routinely performed on agar. Recipient bacteria were grown on sTSB agar overnight (approximately 15 to 20 h) and resuspended in TSB at 1 × 109 CFU/ml for the fimbriated wild-type strains or 5 × 109 CFU/ml for the nonfimbriated mutants. A 20-μl aliquot of the suspension was spotted onto a prewarmed sTSB agar plate and spread in a small area (diameter of ∼10 mm). After incubation for 2 h, 10 μl of donor DNA at a concentration of ∼100 μg/ml was added and mixed with the bacterial cells by using a loop. The mixture was incubated for an additional 6 h (for the fimbriated strain) or 5 h (for nonfimbriated mutants). The bacteria were scraped up, resuspended in LB broth, and plated onto selective medium for 2 to 7 days. Ten microliters of DNase I solution (200 μg/ml) was added simultaneously with DNA as a negative control or at designated times to quench DNA uptake. Unmarked chromosomal DNA from the wild-type strain D7S or Tris-EDTA buffer was also used as a negative control. Transformation frequency was calculated as the number of transformants per CFU, and transformation efficiency was calculated as the number of transformants per microgram of DNA.
Competition assays.
Transformation in the presence of competing DNA was performed on agar surfaces as described above. Transforming DNA (0.2 μg for each sample) and competing DNA (1.8 μg) were mixed in 10 μl of Tris-EDTA buffer and added to the competent bacteria. DNase I was added after 20 min, and the mixture was incubated for an additional 6 h for the wild-type strain or 5 h for the nonfimbriated strain. The relative transformation efficiency was the ratio of the number of Nalr transformants obtained from 0.2 μg of transforming DNA without competitor (the control) to the number of transformants obtained with the mixed DNA. Competing DNA included genomic DNA from A. actinomycetemcomitans strains D7S and HK1651, from E. coli strains DH5α and E44, and from H. influenzae Rd and two DNA fragments produced by PCR, designated pcr-USS-pilA (874 bp) and pcr-pilA (868 bp). Both pcr-USS-pilA and pcr-pilA contained a copy of the pilA gene, but they differed by the presence of a putative A. actinomycetemcomitans USS (which shares the 9-base USS of H. influenzae; pcr-USS-pilA) or a 5-base partial USS (pcr-pilA) at one end. Amplification used the cloned pilA gene of A. actinomycetemcomitans (unpublished data) and, for pcr-USS-pilA, primers pilB-B1, 5′-TTTCGTCGACTTTTTCGACGCGC, and USS-pilA, 5′-AAAAGTGCGGTGGAATTTTAC (the 9-base core USS is underlined). Amplification of pcr-pilA used primers pilB-B1 and pilA, 5′-GCGGTGGAATTTTACGGTGT.
Transformation in the presence of cAMP.
A. actinomycetemcomitans strains D7S and D7S-smooth were inoculated from overnight plate cultures into sTSB broth at an optical density value at 600 nm of 0.1 and incubated without shaking at 37°C in 5% CO2. After 30 min of incubation, cyclic AMP (cAMP) was added to the cultures to a 2 mM concentration, and incubation continued for an additional 50 min, followed by the addition of donor DNA to a final concentration of 2 μg/ml. After 20 min, DNA uptake was quenched by the addition of DNase I (50 μg/ml). Transformants were selected as described above.
Competence-based allelic replacement.
The competence-based allelic replacement strategy depends on homologous recombination of a chromosomal target gene with a recombinant DNA fragment consisting of a selectable marker inserted between two flanking regions of the target gene. The fragment was constructed either in E. coli by standard methods or by a PCR method developed in our laboratory. More details of this method will be published elsewhere. Briefly, the flp-rcp-tad gene cluster, which contains 14 genes coding for fimbria biogenesis and tight adherence (13, 16, 17), was deleted and replaced with the Sper cassette. A 1.1-kb DNA upstream of the flp gene was amplified with primers flp-UE (5′-CTGAATTCTCGCTCAGATACGGA) and flp-B1 (5′-AAGGATCCGACCGGTACTATAGTCCT; a BamHI site is underlined) with HK1651 DNA as template. Similarly, primers tadG-B2 (5′-CAGGATCCATACTATACAAACCCGT) and tadG-R (5′-GATTATGAGTCGACTGCTCT) were used to amplify a 1-kb DNA downstream of the tadG gene. Both of these PCR DNAs contain a copy of the USS. These two amplicons were purified through a spin column (Qiagen), digested with BamHI, and partially filled in with dGTP and dATP with the E. coli DNA polymerase Klenow fragment. pK-Spe was cleaved with SalI to produce a 2.7-kb vector and a 1.2-kb Sper cassette. The SalI-digested DNA was partially filled in with dTTP and dCTP without purification. Both PCR DNA and the marker DNA were combined, ethanol precipitated, and ligated at 15°C overnight. The ligation mixture was directly applied to competent bacteria on an agar plate. Transformants were isolated by selection for the inserted Spe marker.
All mutants were confirmed by PCRs to verify (i) the deletion of the target gene by using primers within the target gene, (ii) the insertion of the antibiotic cassette by using primers flanking the target site, and (iii) the direction of the inserted antibiotic cassette by using a primer upstream of the target site and a forward or reverse primer within the cassette.
RESULTS
Identification of naturally transformable A. actinomycetemcomitans.
A. actinomycetemcomitans strains ATCC 33384, JP-2, HK1651, and Y4 and 13 clinical isolates were examined by the agar-based transformation assay. The donor DNA was the genomic DNA of a Nalr Rifr mutant of A. actinomycetemcomitans strain D17S. Among 17 strains examined, only A. actinomycetemcomitans strain D7S was found to be transformable under the test conditions. The strain D7S was recovered from a deep periodontal pocket of an upper central incisor of a female African-American patient with aggressive periodontitis. D7S was found to be of serotype a.
Transformation frequencies for both chromosomal markers (Rifr and Nalr) were similar at (1.1 ± 0.8) × 10−3 transformants/CFU. However, since cells of strain D7S formed aggregates, the calculated transformation frequency is likely to be overestimated. Preliminary studies showed that the transformation frequency was higher with sTSB agar than with sTSB broth, TSB agar, Columbia agar, or LB agar (data not shown). Subsequent characterization of A. actinomycetemcomitans strain D7S and its derivatives was therefore routinely performed on sTSB agar. Transformation was also performed with DNA from a Rifr transformant of D7S. The transformation frequencies were similar whether the genomic DNA from strain D7S or that from D17S was used. The subsequent experiments were routinely performed with marked genomic DNA from strain D17S.
Characterization of natural transformation.
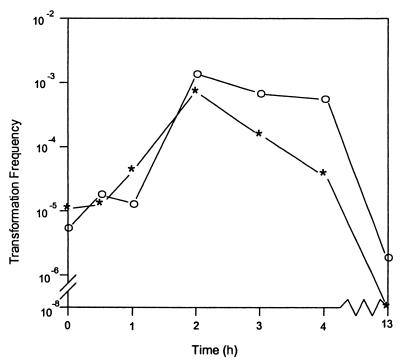
Figure 1 shows the development of competence by strain D7S (asterisks). Bacteria grown overnight on sTSB agar were transferred onto fresh agar. Transformation was initiated by adding DNA to the cells on the plate at the times indicated and stopped by addition of DNase I after 20 min of incubation with DNA. Although cells were immediately competent, the transformation frequency increased 100-fold after 2 h of incubation and then slowly decreased. Transformants were not detected (transformation frequency of <10−8/CFU) when DNA was added to overnight cultures.
FIG. 1.
Effect of incubation time on transformation frequency of A. actinomycetemcomitans. DNA (1 μg) was added at the indicated time after bacteria were placed on agar, and uptake was terminated by addition of DNase I after 20 min. Transformation frequency was defined as the number of Nalr transformants per CFU of recipient cells. Data are the averages of three experiments. ∗, D7S; ○, D7S-smooth.
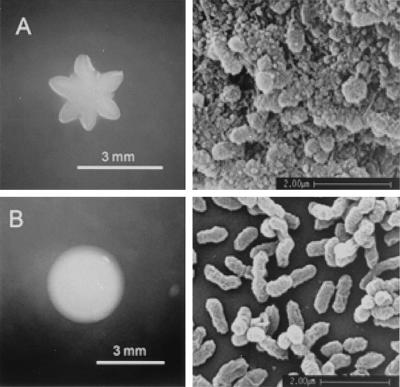
A. actinomycetemcomitans strain D7S exhibited rough colony morphology and was fimbriated (Fig. 2A). To determine whether fimbriae participate in DNA uptake, as they do in many other gram-negative bacteria, a spontaneous nonfimbriated smooth-colony mutant of strain D7S (designated D7S-smooth [Fig. 2B]) was isolated and its competence was examined (Fig. 1, open circles). The time course of competence development was similar to that of strain D7S, but cells remained competent longer. Transformants of strain D7S-smooth could be detected even when DNA was added after overnight growth. This extension of competence for the smooth phenotype may be indicative of the physical exclusion of the transforming DNA by the accumulation of extracellular matrix produced by the wild-type D7S strain as the cells age. Alternatively, there may be other pleiotropic effects linked to the smooth phenotype that are as yet unknown.
FIG. 2.
Morphology of A. actinomycetemcomitans strain D7S and its smooth-colony mutant. Left side, colonies on sTSB agar; right side, scanning electron micrographs of bacteria on the coverslip surface. (A) D7S; (B) D7S-smooth.
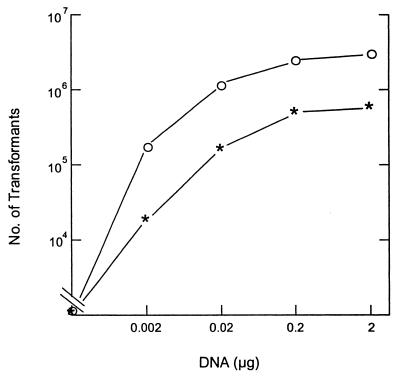
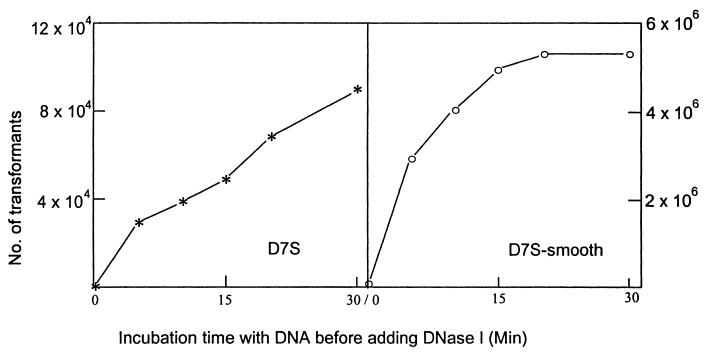
Uptake of DNA is saturable in most other naturally competent bacteria, and where it has been investigated, it has been found to be an absolute saturation (cells cannot take up any more DNA) rather than a kinetic one (cells cannot take up DNA any faster). Figure 3 shows that amounts of marked chromosomal DNA less than 0.2 μg are limiting for transformation and confirms that the 1.0 μg of DNA used in the standard transformation assay is saturating. Figure 4 shows that uptake of DNA by the nonfimbriated D7S-smooth became absolutely saturated within 20 min after the addition of the donor DNA. Cells were given 1.0 μg of DNA, and uptake was assayed by conversion of DNA to a state of DNase I resistance. In contrast, the transformation frequency for the wild-type stain D7S continued to increase for at least 30 min. The dense cell aggregations typical of D7S may physically hinder interaction of DNA with cells, so that prolonged incubation is needed for donor DNA to reach competent cells. It should be noted that the degree of aggregation of strain D7S varied between experiments. It was difficult to compare experiments conducted on different occasions. This may explain the unexpected differences in the number of transformants of strain D7S in Fig. 3 and Fig. 4.
FIG. 3.
Transformation efficiencies of A. actinomycetemcomitans strains D7S and D7S-smooth with different concentrations of DNA. Donor DNA (0.002 to 2 μg) was added to the competent bacteria on 2-h plate cultures and incubated for 30 min before addition of DNase I. Nalr transformants were selected. ∗, D7S; ○, D7S-smooth. Note that total CFU for D7S was ∼5 × 108 and that for D7S-smooth was ∼5 × 109.
FIG. 4.
Transformation efficiency versus time of incubation with donor DNA. DNA uptake was terminated by addition of DNase I at the indicated time. Nalr transformants were selected.
Identification of the putative USS and competition assays.
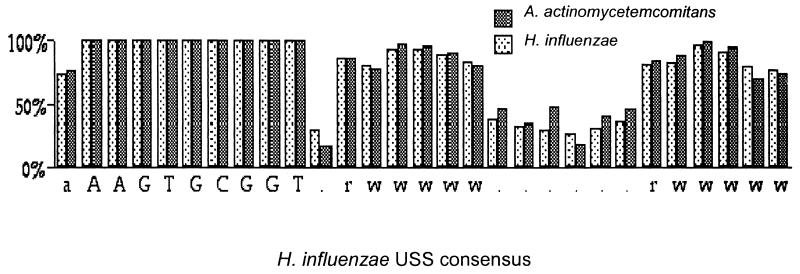
Competent cells of H. influenzae (like A. actinomycetemcomitans, a member of the family Pasteurellaceae) preferentially take up DNA fragments containing a 9-bp sequence which is abundant in H. influenzae DNA (6, 25). Interestingly, DNA of A. actinomycetemcomitans has been reported elsewhere to compete efficiently with DNA of H. influenzae in H. influenzae transformation experiments (1). This suggests that the two genomes may share the same uptake sequence. We examined 133 kb of randomly chosen A. actinomycetemcomitans genomic sequence (downloaded from www.genome.ou.edu/act.html) and found 52 occurrences of the H. influenzae USS or its complement, a frequency of 0.4/kb, much higher than the frequency of 6 × 10−3 copies per kb expected for a random sequence of this base composition. Alignment of the putative USS of A. actinomycetemcomitans, shown in Fig. 5, revealed a flanking consensus very similar to that seen in H. influenzae.
FIG. 5.
Comparison of 52 occurrences of the putative USS of A. actinomycetemcomitans to the USS consensus of H. influenzae, aligned at the 9-bp core. Capital letters are invariant bases; for others, r, A or G; w, A or T.
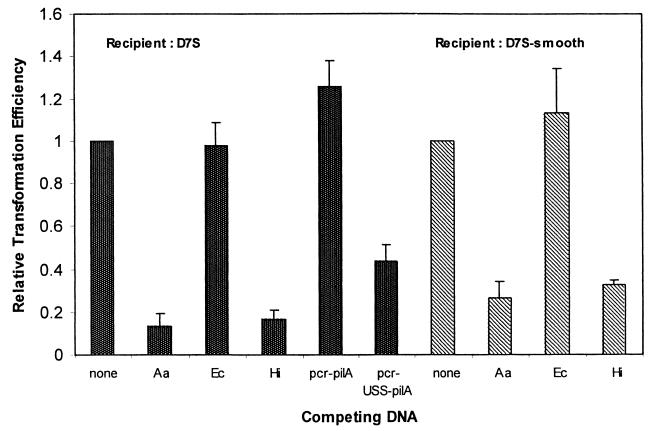
We assessed the specificity requirements of transforming DNA by performing competition experiments. We tested whether the uptake of A. actinomycetemcomitans chromosomal markers could out-compete homologous and heterologous sources of DNA. Figure 6 shows that both A. actinomycetemcomitans and H. influenzae DNA competed for uptake with genetically marked A. actinomycetemcomitans DNA but that E. coli DNA did not. Furthermore, an 874-bp PCR fragment containing the H. influenzae USS (pcr-USS-pilA) competed for uptake, while a nearly identical fragment lacking the USS (pcr-pilA) did not. This is the first clear evidence not only that an A. actinomycetemcomitans USS exists but also that it is likely highly similar to, if not indistinguishable from, the USS of H. influenzae.
FIG. 6.
Relative transformation efficiencies of strains D7S and D7S-smooth in the presence of competing DNA. Competing DNAs used: Aa, A. actinomycetemcomitans genomic DNA; Hi, H. influenzae Rd genomic DNA; Ec, E. coli chromosomal DNA; pcr-pilA, an 868-bp PCR DNA containing pilA; pcr-USS-pilA, an 874-bp PCR DNA containing the same sequence as that in pcr-pilA and a USS site at one end. The relative transformation efficiency was expressed as the ratio between the number of transformants of control (no competing DNA) to the number of transformants in the presence of mixed DNA; data are the averages of three experiments ± standard deviations.
Competence stimulated by cAMP.
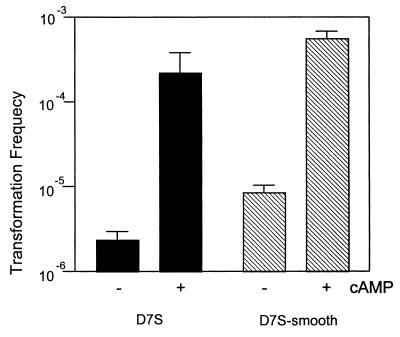
Transcription of some H. influenzae competence genes depends on the cAMP-cAMP receptor protein complex (5, 19), and addition of cAMP to cells in exponential growth phase increases transformation frequencies from the threshold of detectability (∼10−8) to about 10−4. Therefore, we tested the effect of cAMP on A. actinomycetemcomitans competence development in exponential-phase cells grown in broth. The doubling time of the nonfimbriated strain D7S-smooth was approximately 2 h under our culture conditions. Although the culture was less homogenous, the growth phase of the fimbriated wild-type strain D7S was estimated to be exponential phase as well. Figure 7 illustrates that, although the broth cultures of A. actinomycetemcomitans strains D7S and D7S-smooth were competent without cAMP, the transformation frequencies increased by approximately 100-fold in the presence of cAMP. Furthermore, addition of cAMP to the plate cultures also increased competence (data not shown). The results indicate that A. actinomycetemcomitans competence, like that of H. influenzae, is likely regulated by catabolite repression.
FIG. 7.
Transformation frequencies in the presence or absence of cAMP. Data are the averages of two experiments ± standard deviations.
Plasmid transformation.
DNA uptake pathways that have been characterized in naturally competent bacteria cannot transport closed circular molecules such as intact plasmids into the cytoplasm. Because only single strands of broken or cleaved plasmid molecules enter the cytoplasm, transformation by plasmid DNA depends on either reassociation of strands derived from molecules taken up independently or recombinational repair with a homologous DNA segment already resident in the cell. Consistent with this, in our standard transformation assay the transformation frequency of 1 μg of the supercoiled shuttle plasmid pPK1 was only ∼10−8/CFU in both strain D7S and strain D7S-smooth. Plasmid isolated from two of the transformants appeared indistinguishable in size from the donor pPK1 isolated from E. coli.
Competence-based gene replacement.
We exploited the natural competence of A. actinomycetemcomitans strain D7S to develop a gene replacement protocol. Transformation by a linear DNA molecule containing a disrupted gene was sufficiently efficient such that any cloning steps could be eliminated, and cells were directly transformed with ligation mixtures. The method was simple and typically produced the desired specific deletion mutants in less than 1 week. One example was the deletion of the 11-kb flp-rcp-tad gene cluster of 14 genes in D7S. Twenty-three Sper transformants were obtained from ∼0.6 μg of ligated DNA. All these transformants exhibited the expected smooth-colony phenotype and lacked fimbria. PCR examination of four selected colonies indicated that both flp and tadG sequences were absent while expected PCR products were amplified from D7S (data not shown), and the Spe gene was found to be inserted at either direction in these mutants.
DISCUSSION
Two previous studies have observed natural transformation in A. actinomycetemcomitans. Tøonjum et al. (30) used a broth-based method and identified two naturally competent strains among five study strains of A. actinomycetemcomitans with transformation frequencies of 1.3 × 10−3 and 6.2 × 10−5. In our present study, we identified a single highly competent A. actinomycetemcomitans strain among 17 strains examined. Although this is an apparently lower prevalence of naturally competent A. actinomycetemcomitans strains than was reported by Tøonjum et al., the transformation frequencies reported in this study were similar.
Sato et al. (23) generated transposon insertional mutants from the A. actinomycetemcomitans strain Y4 by natural transformation. The transformation was performed with broth-cultured exponential-phase A. actinomycetemcomitans cells. However, strain Y4 was not transformable in our study. While the loss of competence is sometimes a consequence of laboratory propagation, it is also possible that A. actinomycetemcomitans strains may spontaneously acquire and lose transformability in their natural habitat, as has been observed previously for Pseudomonas fluorescens (7).
Neither of the studies by Tøonjum et al. and Sato et al. examined the requirement for the induction of competence or the sequence specificity of DNA uptake. This study showed that competence development in A. actinomycetemcomitans resembles that in H. influenzae. Moderate levels of competence develop in H. influenzae cultures at late exponential phase and early stationary phase, and exponentially growing cultures become fully competent after transfer to a starvation medium (26). Cells growing in colonies on agar plates are also moderately competent (34). A. actinomycetemcomitans competence developed within 2 h after ∼108 cells were plated on a small area of fresh agar. We are not certain whether the growth phase of the competent A. actinomycetemcomitans was comparable to that of the competent H. influenzae.
We also showed that addition of cAMP induced competence in A. actinomycetemcomitans, similar to the effect of cAMP on H. influenzae competence (35). This strongly suggests that catabolite repression regulates the development of competence. Consistent with this, A. actinomycetemcomitans possesses homologs of the H. influenzae genes mediating catabolite repression (20). Recently, Finkel and Kolter (9) have demonstrated that E. coli possesses orthologs to H. influenzae competence genes and that they appear to function as a means to derive sustenance from DNA. According to this model, one of the roles of competence is to enable activation of pathways for the utilization of other energy sources when simple sugars are not available. Hence, competence would serve a dual function as either a means to derive energy from DNA or a means of horizontal transfer of genetic information. Our data are consistent with such a model.
Fresh clinical isolates of A. actinomycetemcomitans invariably express fimbriae and display rough colony morphology (21). The fimbriated strains may yield smooth-colony, nonfimbriated mutants after extended in vitro subculturing (21). In preliminary studies, we noted that the transformation efficiency of the fimbriated wild-type A. actinomycetemcomitans was higher if the transformation assay was performed on agar. Furthermore, fimbriated wild-type A. actinomycetemcomitans isolates grew poorly in broth, formed microcolonies on the surface of the test tubes, and were generally difficult to manipulate. For the above reasons, we chose to perform transformation assays on agar instead of in broth. However, transformation of the nonfimbriated strain D7S-smooth can be easily performed in broth.
The persistent aggregates formed by wild-type A. actinomycetemcomitans interfere with determination of transformation frequencies. However, the number of CFU is a lower limit to the number of viable cells, and so the calculated transformation frequency of D7S is an upper limit. This increases the significance of the finding that the transformation frequency and efficiency of the nonfimbriated A. actinomycetemcomitans strain D7S-smooth were higher than those of the fimbriated, wild-type parental strain D7S.
In many naturally competent bacteria, components of type IV pilins are essential for DNA uptake (8, 12). However this study showed that the spontaneous nonfimbriated mutant of A. actinomycetemcomitans strain D7S was more competent than was the fimbriated parental wild type. We also noted that the nonfimbriated, flp-deletion mutant of A. actinomycetemcomitans strain D7S was naturally competent (data not shown). As Inoue et al. have shown that the A. actinomycetemcomitans flp gene encodes a fimbrial protein with some similarity to type IV pilins (16), this suggested that flp-encoded fimbriae are not required for the natural competence of this bacterium.
H. influenzae Rd preferentially takes up DNA containing the USS. The USS was originally identified as an 11-bp sequence (6), but the 9-bp sequence 5′-AAGTGCGGT (plus orientation) or 5′-ACCGCACTT (minus orientation) was later found to be highly conserved and most important for uptake (10). A total of 1,465 copies of this 9-bp site were found by searching the 1.8 Mb of genome sequence of H. influenzae Rd. The alignment of these sequences revealed an extended conserved 29-bp sequence containing the core 9-bp site (25).
A. actinomycetemcomitans genomic DNA anneals poorly to H. influenzae DNA in DNA-DNA hybridization experiments (1), and the 16S rRNAs of the two species have only 94% identity (E. coli and Salmonella enterica serovar Typhimurium have 97%). Nevertheless, the finding that A. actinomycetemcomitans DNA competes efficiently with H. influenzae DNA for uptake by H. influenzae indicated that A. actinomycetemcomitans might contain an abundant sequence resembling the H. influenzae USS (1). Consistent with this, Thomson et al. found several copies of an 11-bp sequence similar to the H. influenzae USS in intergenic regions near the katA gene of A. actinomycetemcomitans (29). They further identified 848 copies of this 11-bp sequence in the A. actinomycetemcomitans Genome Sequencing Project database and postulated that this sequence may be used as a DNA uptake signal (29). We confirmed these observations by showing that the A. actinomycetemcomitans genome does contain a great excess of repeated sequences remarkably similar to the H. influenzae USS. Furthermore, we showed that DNA of various sources containing this USS competed for uptake for transformation in A. actinomycetemcomitans, suggesting that A. actinomycetemcomitans does preferentially take up DNA containing this USS.
These findings call into question the assumed function of the USS. It is unlikely to serve as a genetic barrier between related species, as DNAs of many members of the Pasteurellaceae compete for uptake by H. influenzae. Nor does it prevent uptake of potentially harmful DNAs: the genome of the H. influenzae phage HP1 has a high density of H. influenzae USSs. Other functions have been suggested for the USS, but none stand up under close scrutiny. The H. influenzae USS is not the Chi sequence (27), and most USSs cannot be transcription terminators, as they lie within coding sequences. Their abundance may simply be a consequence of the molecular drive imposed by the strong sequence bias of the DNA receptor. This bias in turn may result from constraints on DNA binding imposed by the uptake process, perhaps needed to overcome the difficulty of translocating highly charged DNA across the cell membranes.
Our studies of natural transformation in A. actinomycetemcomitans provided a new avenue for genetic engineering in this bacterium. Construction of strains with new combinations of existing genes is facilitated by recombinant DNA techniques and natural transformation. Over 30 gene replacement mutants were generated in the strain D7S or D7S-smooth. The PCR DNAs used were between 0.4 and 1.9 kb, and the 1-kb length appears to be optimal. Transformation efficiencies for single-gene deletions in D7S were found in the range of 3 × 102 to 2 × 104/μg of ligated DNA when the PCR DNA contained one or more USSs and about 100-fold less if the DNA did not have USSs (unpublished results). Incorporation of the USS in one of the PCR primers appeared to increase the transformation efficiency. A much higher efficiency was obtained when D7S-smooth was used as the recipient. A detailed study of these conditions is under way in our laboratory.
Acknowledgments
This research was supported by NIDCR grant R01 DE12212 and American Heart Association grant-in-aid 0150743Y.
We thank O. Kay for reading and providing useful comments on the manuscript, C. Ma for searching A. actinomycetemcomitans genome sequences for USSs, S. Asikainen for serotyping the D7S strain, and P. Fives-Taylor and M. F. Alexeyev for plasmids. We are grateful to B. Roe, F. Najar, S. Clifton, T. Ducey, L. Lewis, and D. Dyer for the use of unpublished nucleotide sequence data from the A. actinomycetemcomitans Genome Sequencing Project at the University of Oklahoma.
REFERENCES
- 1.Albritton, W., J. Setlow, M. Thomas, and F. Sottnek. 1986. Relatedness within the family Pasteurellaceae as determined by genetic transformation. Int. J. Syst. Bacteriol. 36:103-106. [Google Scholar]
- 2.Alexeyev, M. F., and I. N. Shokolenko. 1995. RP4 oriT and RP4 oriT-R6K oriV DNA cassettes for construction of specialized vectors. BioTechniques 19:22-26. [PubMed] [Google Scholar]
- 3.Ashimoto, A., C. Chen, I. Bakker, and J. Slots. 1996. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 4.Asikainen, A., and C. Chen. 1999. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontology 2000 20:65-81. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, M. S. 1992. The gene encoding cyclic AMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc. Natl. Acad. Sci. USA 89:1626-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danner, D. B., R. A. Deich, K. L. Sisco, and H. O. Smith. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311-318. [DOI] [PubMed] [Google Scholar]
- 7.Demanèche, S., E. Kay, F. Gourbière, and P. Simonet. 2001. Natural transformation of Pseudomonas fluorescens and Agrobacterium tumefaciens in soil. Appl. Environ. Microbiol. 67:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 9.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzmaurice, W. P., R. C. Benjamin, P. C. Huang, and J. J. Scocca. 1984. Characterization of recognition sites on bacteriophage HP1c1 DNA which interact with the DNA uptake system of Haemophilus influenzae Rd. Gene 31:187-196. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graupner, S., and W. Wackernagel. 2001. Pseudomonas stutzeri has two closely related pilA genes (type IV pilus structural protein) with opposite influences on natural genetic transformation. J. Bacteriol. 183:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haraszthy, V. I., J. J. Zambon, M. Trevisan, M. Zeid, and R. J. Genco. 2000. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71:1554-1560. [DOI] [PubMed] [Google Scholar]
- 15.Haubek, D., J. M. Dirienzo, E. M. B. Tinoco, J. Westergaard, N. J. López, C.-P. Chung, K. Poulsen, and M. Kilian. 1997. Racial tropism of a highly toxic clone of A. actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 35:3037-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, T., I. Tanimoto, H. Ohta, K. Kato, Y. Murayama, and K. Fukui. 1998. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42:253-258. [DOI] [PubMed] [Google Scholar]
- 17.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macfadyen, L. P. 2000. Regulation of competence development in Haemophilus influenzae. J. Theor. Biol. 207:349-359. [DOI] [PubMed] [Google Scholar]
- 20.Macfadyen, L. P., I. R. Dorocicz, J. Reizer, M. H. Saier, Jr., and R. J. Redfield. 1996. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol. Microbiol. 21:941-952. [DOI] [PubMed] [Google Scholar]
- 21.Rosan, B., J. Slots, R. J. Lamont, M. A. Listgarten, and G. M. Nelson. 1988. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol. Immunol. 3:58-63. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sato, S., N. Takamatsu, N. Okahashi, N. Matsunoshita, M. Inoue, T. Takehara, and T. Koga. 1992. Construction of mutants of Actinobacillus actinomycetemcomitans defective in serotype b-specific polysaccharide antigen by insertion of transposon Tn916. J. Gen. Microbiol. 138:1203-1209. (Erratum, 138:1992.) [DOI] [PubMed] [Google Scholar]
- 24.Slots, J. 1982. Selective medium for isolation of Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 15:606-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, H. O., J.-F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 26.Solomon, J. M., and A. D. Grossman. 1996. Who's competent when: regulation of natural genetic competence in bacteria. Trends Genet. 12:150-155. [DOI] [PubMed] [Google Scholar]
- 27.Sourice, S., V. Biaudet, M. El Karoui, S. D. Ehrlich, and A. Gruss. 1998. Identification of the Chi site of Haemophilus influenzae as several sequences related to the Escherichia coli Chi site. Mol. Microbiol. 27:1021-1029. [DOI] [PubMed] [Google Scholar]
- 28.Sreenivasan, P. K., and P. Fives-Taylor. 1994. Isolation and characterization of deletion derivatives of pDL282, an Actinobacillus actinomycetemcomitans/Escherichia coli shuttle plasmid. Plasmid 31:207-214. [DOI] [PubMed] [Google Scholar]
- 29.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tøonjum, T., G. Bukholm, and K. Bøovre. 1990. Identification of Haemophilus aphrophilus and Actinobacillus actinomycetemcomitans by DNA-DNA hybridization and genetic transformation. J. Clin. Microbiol. 28:1994-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Winkelhoff, A. J., and J. Slots. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in nonoral infections. Periodontology 2000 20:122-135. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y., S. H. Huang, C. A. Wass, M. F. Stins, and K. S. Kim. 1999. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 67:4751-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams, P. M., L. A. Bannister, and R. J. Redfield. 1994. The Haemophilus influenzae sxy-1 mutation is in a newly identified gene essential for competence. J. Bacteriol. 176:6789-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wise, E. M., S. Alexander, and M. Powers. 1973. Adenosine 3′:5′-cyclic monophosphate as a regulator of bacterial transformation. Proc. Natl. Acad. Sci. USA 70:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]