Abstract
The identity of the gene encoding acyl coenzyme A dehydrogenase is a major remaining mystery of the Escherichia coli fatty acid degradation (fad) regulon. Our prior genome array analyses showed that transcription of the yafH gene is controlled by the FadR regulatory protein. We now report direct experimental proof that yafH and fadE are the same gene.
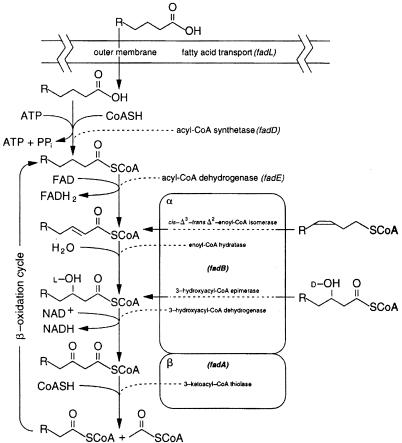
The first step of the β-oxidation cycle (Fig. 1) of fatty acid degradation in Escherichia coli is the oxidation of acyl coenzyme A (acyl-CoA) to 2-enoyl-CoA (reviewed in references 2 and 9). This reaction involves the transfer of two electrons from the substrate to a flavin adenine dinucleotide (FAD) cofactor that must be reoxidized in order for the dehydrogenase to have catalytic function. Although this is a key step in β-oxidation, it has received little attention. Much of the available genetic and biochemical information specific to the E. coli acyl-CoA dehydrogenase reaction comes from the doctoral dissertation of K. Klein (14), only part of which has been published in the primary literature (15). Klein (14) reported the isolation of several mutants lacking functional acyl-CoA dehydrogenase from cultures of the Ymel strain treated with N-methyl-N′-nitro-N-nitrosoguanidine and reported that these mutations mapped to at least two separate loci closely linked to proAB. Much of the confusion surrounding the genetics of acyl-CoA dehydrogenase can be traced back to the original phage P1 transductional mapping data (14, 15). The reported three-factor crosses and deletion data placed the fadE allele between proA and proB at min 5.62. However, the E. coli K-12 genome sequencing data show that only 12 bases lie between the two pro genes, and thus these mapping data are clearly in error. The current working map of the Coli Genetic Stock Center places fadE at min 4.78.
FIG. 1.
The fatty acid degradation pathway of E coli. The boxes labeled α and β represent the individual subunits of the β-oxidation complex.
Further confusion stemmed from the report of Klein (14) of two other acyl-CoA dehydrogenase mutant genes called fadF and fadG (although he suspected that these mutants might be allelic). These mutations were mapped between proA and the lac operon. However, these data also seem in error, since Δ(pro-lac) strains, which carry deletions of this chromosomal segment, grow well on fatty acids as the sole carbon source, which indicates the lack of essential fad genes within this region. Klein speculated that the fadF and fadG genes might encode proteins having different chain length specificities, similar to those found in the β-oxidation pathway of mammalian mitochondria (14) and concluded that at least two genes are necessary for acyl-CoA dehydrogenase activity (14). Only one mutant of the Klein collection has been widely used. This mutant, fadE62, is very stable and does not revert with any detectable frequency. The only other E. coli fadE mutant allele available is an insertion of Mud1 originally isolated by Clark (6) that is very unstable, since Mud1 insertion strains are prone to secondary transpositions and deletions. Clark (6) used strains carrying this allele to show that fadE transcription is negatively regulated by FadR. The Mud1 insertion was mapped to the 5-min region of the chromosome, but proved too unstable to map more precisely.
Our interest in fadE was rekindled by our transcriptional array analysis of the FadR regulon (4, 5), which showed that expression of the yafH gene at min 5.19 was significantly increased upon fatty acid addition or by disruption of the fadR gene. We report that yafH is the gene previously defined by the fadE62 mutation, and thus fadE maps at min 5.19 rather than min 4.78.
Transcriptional array analyses.
The first experimental evidence that yafH might correspond to fadE came from total genomic, differential transcriptional array studies of a set of isogenic E. coli strain grown in the presence of exogenous long-chain fatty acids (4). These data are available online (http://www.life.uiuc.edu/∼jwcampbe). A simple method of reporting the significance of expression ratio data involves sorting genes based on expression ratio and dividing the position of each gene within the sorted list by the total number of genes (4, 24). This produces a relative ranking value between zero and 1, which can be expressed as a percentage, with the extreme high and low values representing genes that have the greatest degrees of differential expression. The relative rank of the differential transcriptional response of yafH in the wild-type strain grown in the presence or absence of oleate was 1.6%. When wild-type and isogenic fadR strains grown in the absence of fatty acid supplementation were compared, the relative rank of yafH was 0.023%. In contrast, when a fadR strain was grown in the presence or absence of oleate, no differences in yafH expression were observed (relative rank of 56%). Similar results were obtained when the wild-type strain was grown with acetate in place of oleate, where yafH had a relative rank of 55%. This transcription pattern shows that FadR negatively regulates yafH and that induction of yafH expression upon supplementation of cultures with fatty acids is mediated by FadR. The genes displaying the transcriptional profiles most similar to yafH in our genomic array experiments are fadBA, which together with fadE and fadD provide the core enzymes of the β-oxidation pathway. Analysis of the open reading frames (ORFs) immediately flanking yafH indicates that these genes have no significant transcriptional response to the presence of fatty acids or FadR, a conclusion consistent with the sequence of this genomic segment.
Disruption of yafH.
If yafH is fadE, then mutations that disrupt this ORF should result in a fad phenotype. We constructed yafH disruptions by phage λ Red-mediated recombinational replacement of the yafH coding sequence with a kanamycin resistance gene (7). Two of the colonies obtained were chosen for further study. Phage P1 vir was grown on the original isolates, and these lysates were used to transduce the wild-type strain MG1655 to kanamycin resistance, giving strains JWC265 and JWC266. These strains were unable to grow on oleate as the sole carbon source, but grew well on acetate (Table 1). This behavior demonstrates that the yafH gene is required for E. coli to utilize oleate. To investigate the chain length specificity of the yafH mutant, the fadR613::Tn 10 allele of strain CAG18497 (19) was transduced into JWC266 to give strain JWC267, which was then tested for the ability to grow on short-chain-length fatty acids. This strain was unable to grow on plates containing octanoate, decanoate, dodecanoate, or oleate as the sole carbon source (Table 1). The inability to metabolize fatty acids of any chain length is identical to the reported phenotype of fadE strains (14, 15)
TABLE 1.
Phenotypes of yafH disruption strainsa
| Strain | Genotype | Growth on carbon source:
|
||||
|---|---|---|---|---|---|---|
| Acetate | Octanoate | Decanoate | Dodecanoate | Oleate | ||
| MG1655 | Wild type | + | − | − | + | + |
| CAG18497 | fadR | + | + | + | + | + |
| JWC266 | yafH | + | − | − | − | − |
| JWC267 | fadR yafH | + | − | − | − | − |
Cultures were grown overnight on minimal medium M9 (16) glucose plates supplemented with the appropriate antibiotics. Single-well-isolated colonies were then streaked onto M9 plates containing the appropriate antibiotics and the carbon sources indicated. The plates were incubated at 37°C prior to scoring. Growth is denoted as +, and no growth is indicated by −. Growth on acetate and oleate was relatively rapid, and the plates could be scored within 2 days. Growth on octanoate and decanoate required longer incubation periods. In the course of prolonged incubations of strain MG1655 on plates containing octanoate or decanoate, spontaneous fadR mutants were selected. These growth responses were not scored as positive, since the vast majority of the cells remained unable to use these carbon sources. No spontaneous mutants capable of using any of the fatty acid substrates were observed from strain JWC266 or JWC267. Strain JWC266 was made by the method of Datsenko and Wanner (7). A primer with 42 bases of homology to the start codon region of yafH at its 5′ end and 22 bases of homology to the P1 site of plasmid pKD4 (7) at the 3′ end was synthesized. The sequence of this primer was 5′-GTGGTCAGACCTCCTAAAGTAAGGGGCTTTTCGTTATGATGTGTGTAGGCTGGAGCTGCTTCG-3′. A second primer with 42 bases of homology to the reverse complement of the stop codon region of yafH on its 5′ end, and 23 bases of homology to the P2 site of pKD4 on the 3′ end was also produced. The sequence of this primer was 5′-TTACGCGGCTTCAACTTTCCGCACTTTCTCCGGCAACTTTACCATATGAATATCCTCCTTAGTTC-3′. These primers were used to amplify the kanamycin resistance gene of plasmid pKD4 in a standard PCR. The 1.6-kbp PCR product was desalted on a Qiagen PCR purification column and resuspended in a final volume of 20 μl of water. This linear DNA fragment contains the kanamycin resistance gene flanked by 42 bases of sequence homologous to the yafH gene. The purified PCR product was electroporated into strain BW25113, which was then plated onto selective medium (7). Kanamycin-resistant colonies were isolated, and phage P1 lysates were made on these strains. The lysates were used to transduce strain MG1655 to kanamycin resistance. Phage transduction and other basic genetic techniques were generally carried out as described by Miller (16). All strains were derivatives of strain MG1655 made by transduction.
The fadE62 mutation is a frameshift within yafH.
Since loss of yafH resulted in a fad phenotype, fadE62 strains should have a mutation within yafH. The fadE62 mutation behaves as though it is a small deletion; it has no detectable reversion frequency, but is readily transferred by transduction. The yafH ORFs of both strain MG1655 and strain K19 were isolated by PCR amplification with a mixture of the Pfu and Taq polymerases. Two separate amplification reactions were performed with the strain K19 yafH allele to generate independent samples. Several restriction endonucleases were used to digest the PCR products, and no differences were detected between the wild-type and K19 yafH digestion products. The intact PCR products were cloned into pCR2.1 (Invitrogen), and the orientations of the inserts were determined by restriction mapping (13). Clones having the insert in the orientation that allowed expression of yafH from the vector lac promoter were retained for sequencing and further study. Both strands of the DNA molecules encoding the two yafH alleles were sequenced in their entirety by primer walking. The sequence of the wild-type yafH allele was an exact match to the GenBank sequence. The two independently produced PCR fragments from strain K19 genomic DNA had identical sequences and showed a deletion of one of the C nucleotides of the CC sequence at positions 2204 to 2205 of the wild-type coding sequence. No other differences from the wild-type sequence were detected in the two PCR products from the mutant strain. The single base deletion converted one alanine codon (GCC) to another (GCG) and shifted the reading frame. This results in the YafH protein of strain K19 being 51 residues shorter than the wild-type protein. Since the last 28 residues of the mutant protein are abnormal residues resulting from the frameshift, the normal yafH reading frame was truncated by 79 residues, and the loss of this small segment of an 814-residue protein resulted in the complete loss of biological activity. The fact that the sequences of two independently produced PCR products were identical precludes an amplification artifact as the source of this mutation.
yafH and fadE are the same gene .
The presence of a mutation in yafH of the fadE62 K19 strain indicated that yafH is fadE. To test if yafH could complement a fadE62 mutant strain, plasmids carrying a kanamycin resistance cassette and either the wild-type yafH or K19 yafH allele were transformed into a panel of fad mutants. The resulting transformants were selected on minimal glucose medium containing kanamycin and then tested for the ability to grow on oleate or acetate as the sole carbon and energy source. Table 2 shows that the only combination of plasmid and fad mutation that resulted in a strain capable of growth on oleate was strain K19 transformed with the plasmid encoding the wild-type yafH allele. Equally significant was the finding that the plasmid carrying the yafH allele of strain K19 failed to rescue the K19 fadE62 defect, thereby eliminating gene dosage or bypass models of complementation. We therefore, conclude that yafH and fadE are the same gene.
TABLE 2.
Complementation of fad mutants with cloned yafH allelesa
| Strain | Genotype | Growth on:
|
|||
|---|---|---|---|---|---|
| pCR2.1-yafHMG1655
|
pCR2.1-yafHK19
|
||||
| Oleate | Oleate + acetate | Oleate | Oleate + acetate | ||
| MG1655 | Wild type | + | + | + | + |
| CAG18483 | fadL | − | + | − | + |
| K27 | fadD | − | + | − | + |
| K19 | fadE | + | + | − | + |
| CAG18496 | fadBA | − | + | − | + |
Each strain was transformed with the indicated plasmid and selected for kanamycin resistance on minimal glucose media. The plasmids differed only in the source of the insert. Plasmid pCR2.1-yafHMG1655 carried the minimal yafH gene from MG1655, while pCR2.1-yafHK19 carried the minimal yafH gene from strain K19. In both constructs, the insert was in the orientation in which the lac promoter of plasmid vector pCR2.1 would transcribe the yafH gene. Individual colonies were plated onto minimal medium M9 (16) supplemented with oleate or plates containing oleate plus acetate (4) and incubated at 37°C for 3 days prior to scoring of growth. The yafH ORFs of strains MG1655 and K19 were cloned by ligating PCR amplification products directly into the pCR2.1 TA cloning vector (Invitrogen). These plasmids were used for both sequencing and complementation analysis. The PCR primers were designed to amplify only the minimal coding sequence of the gene. The 5′-specific primer had the sequence 5′-GTGGTCAGAGCTCCTACAAGTAAGG-3′, whereas the 3′-specific primer had the sequence 5′-AGACTGGTGTACGCGGCTTCAACTTTC-3′. About 100 ng of purified chromosomal DNAs from strain MG1655 or the fadE62 mutant strain K19 was used as amplification templates. Amplification reactions used the LA-Taq system of Sigma, which is a mixture of Taq and Pfu DNA polymerases. The 3′-to-5′ exonuclease activity of the Pfu polymerase serves to suppress PCR-generated mutations (1). The fadD88 mutant K27 and the fadE62 mutant strain K19 were originally isolated in strain Ymel (14, 15). The strain MG1655-derived fadBA, fadL, and fadR strains CAG18496, CAG18483, and CAG18497, respectively, are from the ordered Tn10 collection of Singer and Gross (19). These strains are available from the Coli Genetic Stock Center (Yale University). The media used were described previously (4).
FadR binds upstream of fadE.
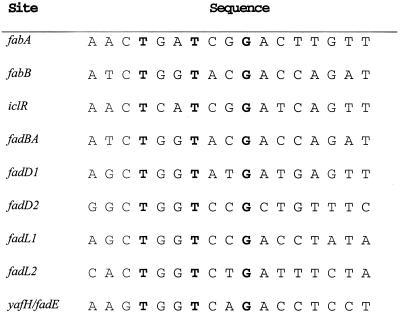
FadR is known to negatively regulate expression of fadE (6, 15), and thus a FadR binding site should be located upstream of the yafH/fadE coding sequence. Indeed, a candidate sequence centered 31 bp upstream of the yafH/fadE initiation codon closely matched the FadR consensus binding site (Fig. 2). Surprisingly, computer-assisted sequence inspection (17) failed to detect this sequence, and it is not listed as a potential FadR binding site on the DNA Motifs web page (http://arep.med.harvard.edu/ecoli_matrices/fadR.html), although a very recent search using different criteria detected the site (18).
FIG. 2.
FadR binding sites. All binding sites except that of yafH/fadE have all been confirmed by published FadR footprinting or gel shift experiments. FadR positively regulates transcription of fabA, fabB, and iclR. The other sites are from genes that are negatively regulated by FadR. The strictly conserved nucleotides are in boldface type. The last line shows the putative FadR binding site located upstream of yafH/fadE. This sequence shows greater homology to the sites of the other negatively regulated genes than to the sites of the positively regulated genes.
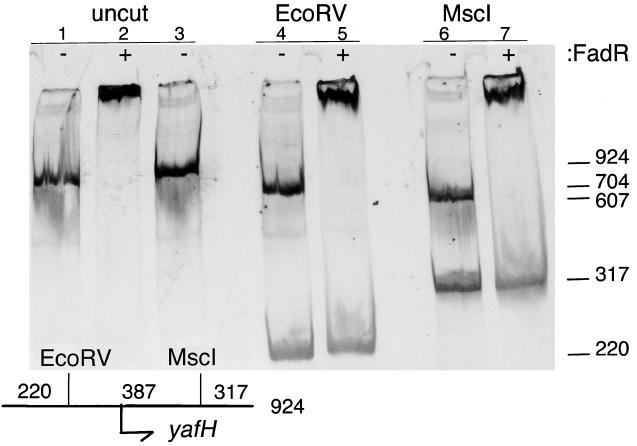
To test if FadR could bind this putative site, we performed in vitro gel shift analyses (Fig. 3). A 924-bp 33P-labeled DNA fragment containing the putative yafH FadR binding site was used in these experiments. FadR protein was purified by nickel chelate chromatography of a polyhistidine-tagged fusion protein known to be fully functional in vivo (5, 21). When incubated with FadR prior to electrophoresis, the electrophoretic mobility of the DNA fragment was significantly slowed, indicating DNA binding by FadR. The specificity of the FadR DNA binding was demonstrated by the finding that, upon digestion of the yafH DNA with various restriction enzymes, only the DNA fragment that contained the putative binding site was shifted by FadR (Fig. 3). These results effectively precluded nonspecific FadR-DNA binding under these conditions. The shifted fragments of the MscI and EcoRV digests (Fig. 3) shared only 387 bp of common sequence, and additional digests with FokI and DraIII localized the binding site to a 128-bp fragment (data not shown). Since 60 bp of this fragment is coding sequence, the binding site must lie within 68 bp of the yafH/fadE initiation codon, a location consistent with the site deduced by sequence inspection.
FIG. 3.
Gel shift analyses of FadR binding to the region upstream of yafH/fadE. The experiments test the ability of FadR to specifically bind and retard the mobility of a 924-bp 33P-labeled DNA fragment containing the yafH/fadE-associated, putative FadR binding site. The presence or absence of FadR in the binding assay is denoted by + or −, respectively, at the top of each lane. Lane 1 is the yafH/fadE DNA fragment without added FadR and shows the unrestricted mobility of the labeled DNA fragment. Lane 2 contains the products resulting from mixing the yafH/fadE DNA fragment with FadR. Lane 3 is the same as lane 2, except that the FadR protein was incubated at 95°C for 2 min prior to addition to the binding reaction. Lanes 4 and 5 are the products of an EcoRV restriction digest of the labeled fragment. In the presence of FadR, only the 704-bp fragment was shifted (lane 5), indicating that FadR specifically bound to sequences present only on that DNA fragment. Likewise, when the 924-bp fragment was digested with MscI (lanes 6 and 7), only one of two restriction fragments was bound by FadR. The relative positions of the EcoRV and MscI sites plus the yafH/fadE start codon are shown on the map in the lower left of the figure. The sizes of the fragments are shown on the scale at the right of the figure. A 924-bp DNA fragment centered on the yafH/fadE start codon was produced by PCR amplification. The primers used in this reaction were 5′-CACGGTAGCGACCGGTCAACTCTT and 5′-CGCCCACAACTCCGGCGGCAGATC. A unique 924-bp product was obtained and was purified on a Qiagen PCR column. The gel retardation conditions used and the method for purification of FadR are described in references 5, 11, and 21.
Conclusions.
The data reported here show that yafH and fadE are the same gene. However, the enzymatic activities of FadE remain to be demonstrated by biochemical analysis, which should be facilitated by the identification of the fadE gene. It should be noted that the location of yafH (now fadE) is consistent with the mapping data of Klein and coworkers (14, 15), albeit not with their interpretations of the data. These authors reported that fadE cotransduced with the proA and proB loci. These findings are consistent with the location of yafH, although a considerably higher cotransduction frequency might have been expected. However, this section of the E. coli K-12 linkage map has long been known to give variable transductional mapping results (22), presumably due to sequence differences among strains resulting from the many insertion elements present in this region of the genome (8). Klein and coworkers (15) also reported that the proAB deletion strain χ711 was unable to use oleate as sole carbon source. The upstream endpoint of this deletion has been recently mapped (3). The endpoint is within yafT or yafF, and a novel IS 5 insertion is located upstream of the endpoint, consistent with an IS 5-mediated deletion event. The deletion of strain χ711 is about 34 kbp in length and extends through yafH to a point downstream of proAB that is very likely to be the IS 5 element inserted between yhfC and yhfD in many E. coli K-12 strains (8, 23). The deletion has been shown to include lpcA, a gene located only 140 bp from yafH (3).
During the course of our work, yafH mutants of Salmonella enterica serovar Typhimurium were isolated by transposon mutagenesis with a lacZ fusion element (the S. enterica serovar Typhimurium yafH gene encoding a protein 96% identical to E. coli YafH) (20). However, these mutants were designated as fadF, and the possibility of the identity of yafH and fadE was not mentioned. The S. enterica serovar Typhimurium yafH gene was not tested for complementation of the known E. coli mutants (to our knowledge, the fadF strains having been lost, although E. coli fadE mutants are readily available), and thus the choice of fadF as the gene designation seems arbitrary and (given our data) is incorrect. Moreover, as discussed above, the original data supporting the existence of fadF are equivocal and have not been published in the primary literature. Indeed, the fadF designation has been mentioned only in reviews (2, 9, 10), whereas the fadE data are readily available and the fadE62 allele has been widely used. For these reasons, it seems clear that the new name for yafH should be fadE in both E. coli and S. enterica serovar Typhimurium. Spector and coworkers (20) reported that S. enterica serovar Typhimurium yafH encoded a medium- or long-chain-length acyl-CoA dehydrogenase, which implied that the encoded enzyme was inactive with short-chain acyl-CoA substrates (although no data on short chains were reported). E. coli fadE62 mutant strains are defective in both the β-oxidation and the utilization of fatty acids of all chain lengths from C18 to C4 (15). Given the very high amino acid identity between the E. coli and S. enterica serovar Typhimurium FadE proteins, it seems most unlikely that the specificity of the S enterica serovar Typhimurium enzyme is limited to medium- and long-chain-length acyl-CoAs.
The only data supporting the existence of fadF and fadG in E. coli are the problematical mapping data of Klein (14, 15), which led to the hypothesis that that at least two genes are necessary for acyl-CoA dehydrogenase activity. This hypothesis was based on the β-oxidation pathway of mammalian mitochondria in which several acyl-CoA dehydrogenases of different chain length specificities use a common intermediate protein called electron transfer factor (ETF) to transfer electrons to the respiratory chain (12). Since fadE mutants lacked the ability to oxidize fatty acids of any chain length, they had the phenotype expected of ETF mutants (15). However, our array analysis and prior genetic analysis give no candidates for an ETF homologue in the FadR regulon. Identification of the fadE gene together with the fact that the encoded protein contains the motifs expected of an acyl-CoA dehydrogenase (this is the only protein containing acyl-CoA dehydrogenase motifs encoded by the E. coli genome according to the EcoCyc annotation [http://ecocyc.org/] of the M54 version of the genome database) leads to our belief that fadE encodes the sole acyl-CoA dehydrogenase of E. coli. Since in vivo this protein is involved in the utilization of fatty acids of chain lengths from C4 to C18 (12) (Table 2), it seems clear that FadE catalyzes the dehydrogenation reaction required in each cycle of acyl chain shortening by the β-oxidation pathway. Note that FadE is 814 residues in length, more than twice the size of the mammalian acyl-CoA dehydrogenases of known structure. The first 150 and last 400 or so residues of FadE lie outside the sequences that align with the mammalian proteins and therefore are available to perform additional functions, such as transfer of electrons from the dehydrogenase domain to the E. coli electron transport chain. Although these “extra” protein sequences are conserved in several other putative bacterial acyl-CoA dehydrogenases, motif searches give no clues to their function. However, it is clear that the C-terminal residues play an essential role in FadE function, since loss of only 79 resides from the end of the protein (the fadE62 mutant) destroys biological activity. An insertion that removes the last 266 residues of S. enterica serovar Typhimurium FadE also inactivates the protein in vivo (20). It seems very likely (based on the genome sequence and our array analyses) that the fadF and fadG mutants were fadE mutants that were inaccurately mapped, perhaps due to insertion element heterogeneity among the strains used.
Finally, it should be noted that different initiation codons have been assigned to the E. coli fadE/yafH sequences in the various databases. Although, direct evidence must await analysis of the purified FadE protein, we have chosen the first ATG codon downstream of the FadR binding site, whereas others have chosen upstream GTG and TTG codons. We believe that our assignment is the most likely to be correct, because all of the available S. enterica serovar sequences have an additional base inserted upstream of the ATG, and thus the other putative upstream initiation codons are out of frame with the fadE coding sequence. Moreover, the other putative initiation codons lie either within or upstream of the putative FadR binding site.
Nucleotide sequence accession number.
The sequence of the fadE62 allele has been communicated to GenBank (accession no. AF486265).
Acknowledgments
This work was supported by National Institutes of Health grant AI15650.
REFERENCES
- 1.Barnes, W. M. 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. USA 91:2216-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, P. N., and C. C. DiRusso. 1994. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210:123-145. [DOI] [PubMed] [Google Scholar]
- 3.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, J. W. 2000. Total genomic transcriptional analysis of the fatty acid degradation regulon of Escherichia coli. Ph.D. thesis. University of Illinois at Urbana-Champaign. Urbana.
- 5.Campbell, J. W., and J. E. Cronan, Jr. 2001. Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 183:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, D. 1981. Regulation of fatty acid degradation in Escherichia coli analysis by operon fusion. J. Bacteriol. 148:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deonier, R. C. 1987. Locations of native insertion elements, p. 982-989. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 9.DiRusso, C. C., P. N. Black, and J. D. Weimar. 1999. Molecular inroads into the regulation and metabolism of fatty acids: lessons from bacteria. Prog. Lipid Res. 38:129-197. [DOI] [PubMed] [Google Scholar]
- 10.DiRusso, C. C., and T. Nystrom. 1998. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol. Microbiol. 27:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Henry, M. F., and J. E. Cronan, Jr. 1992. A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell 70:671-679. [DOI] [PubMed] [Google Scholar]
- 12.Hiltunen, J. K., and Y. Qin. 2000. β-Oxidation—strategies for the metabolism of a wide variety of acyl-CoA esters. Biochim. Biophys. Acta 1484:117-128. [DOI] [PubMed] [Google Scholar]
- 13.Holton, T. A., and M. W. Graham. 1991. A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res. 19:1156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein, K. 1973. Acyl-CoA-Dehydrogenasen und ETF in Escherichia coli: Studien zum Fettsäureabbau. Ph.D. thesis. Universität zu Köln, Köln, Germany.
- 15.Klein, K., R. Steinberg, B. Fiethen, and P. Overath. 1971. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur. J. Biochem. 19:442-450. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Robison, K., A. M. McGuire, and G. M. Church. 1998. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 284:241-254. [DOI] [PubMed] [Google Scholar]
- 18.Sadovskaya, N. S., O. N. Laikova, A. A. Mironov, and M. S. Gelfand. 2001. Study on regulation of long-chain fatty acid metabolism with the use of computer analysis of complete bacterial genomes. Mol. Biol. 35:862-866. [PubMed] [Google Scholar]
- 19.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spector, M. P., C. C. DiRusso, M. J. Pallen, F. Garcia del Portillo, G. Dougan, and B. B. Finlay. 1999. The medium-/long-chain fatty acyl-CoA dehydrogenase (fadF) gene of Salmonella typhimurium is a phase 1 starvation-stress response (SSR) locus. Microbiology 145:15-31. [DOI] [PubMed] [Google Scholar]
- 21.Subrahmanyam, S. 1998. Fatty acid and biotin metabolism in Escherichia coli. Ph.D. thesis. University of Illinois at Urbana-Champaign, Urbana.
- 22.Taylor, A. L., and C. D. Trotter. 1967. Revised linkage map of Escherichia coli. Bacteriol. Rev. 31:332-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timmons, M. S., A. M. Bogardus, and R. C. Deonier. 1983. Mapping of chromosomal IS 5 elements that mediate type II F′ plasmid excision in Escherichia coli K-12. J. Bacteriol. 153:395-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei, Y., J.-M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]