Abstract
Dinoflagellate plastid genes are believed to be encoded on small generally unigenic plasmid-like minicircles. The minicircle gene complement has reached saturation with an incomplete set of plastid genes (18) compared with typical functional plastids (60–200). While some of the missing plastid genes have recently been found in the nucleus, it is still unknown if additional genes, not located on minicircles, might also contribute to the plastid genome. Sequencing of tailed RNA showed that transcripts derived from the known minicircle genes psbA and atpB contained a homogenous 3′ polyuridine tract of 25–40 residues. This unusual modification suggested that random sequencing of a poly(dA) primed cDNA library could be used to characterize the plastid transcriptome. We have recovered only 12 different polyuridylylated transcripts from our library, all of which are encoded on minicircles in several dinoflagellate species. The correspondence of all polyuridylylated transcripts with previously described minicircle genes thus supports the dinoflagellate plastid as harbouring the smallest genome of any functional chloroplast. Interestingly, northern blots indicate that the majority of transcripts are modified, suggesting that polyuridylylation is unlikely to act as a degradation signal as do the heterogeneous poly(A)-rich extensions of transcripts in cyanobacteria and other plastids.
INTRODUCTION
Although dinoflagellates are best known as the notorious cause of toxic red tides, they are also important contributors to the ocean's primary production. Photosynthesis in these organisms is typically carried out in plastids surrounded by three membranes (1), an evolutionary footprint reflecting their origin through secondary endosymbiosis (2). The evolutionary ancestor of the peridinin-containing plastids is suggested from molecular phylogenetic reconstructions using plastid-encoded genes to be a red alga (3,4), a conclusion supported by phylogeny of nuclear-encoded plastid-directed genes (5). In general, these findings are also consistent with phylogenetic reconstructions of the host cells as determined from non-plastid-directed genes (6,7).
Despite widespread acceptance of a red-algal origin for the peridinin-containing plastid, these organelles display a number of peculiar characteristics that share no homology with any known extant plastids. For example, the carotenoid peridinin itself (8) and the unusual light harvesting peridinin-chlorophyll a-protein to which it binds (9) are found in no other organisms. Furthermore, the Rubisco in peridinin-containing plastids, an unusual form II enzyme, is dissimilar from the form I protein employed by all other plastids (10). The evolutionary provenance of these proteins is thus unknown, and as they are derived from nuclear-encoded genes, it seems possible that their history may be distinct from that the plastid itself.
An additional issue having bearing on plastid evolution, and one more likely to reflect the plastids themselves, is the number and arrangement of the genes within the genome. Genome architecture may be different from the phylogeny of the plastid genes themselves, and in the case of the dinoflagellates, quite remarkably so. Indeed, the only known plastid genes so far identified have been located on generally unigenic plasmid-like minicircles. These minicircles have been found in at least five genera of peridinin-containing dinoflagellates including Amphidinium (11), Ceratium (12), Heterocapsa (13), Protoceratium (14) and Symbiodinium (15). Each minicircle has regions conserved within a species, and extensive PCR amplification of the genes located between these conserved regions has been performed in several species. Taken together, these studies have led to the conclusion that the known minicircle gene complement has reached saturation (16) with a total of sixteen protein encoding genes (atpA-B; petB and petD; psaA-B; psbA-E and psbI; ycf16 and ycf24; rpl28 and rpl33) in addition to the large (23S) and small (16S) ribosomal RNA. The identification of this highly reduced set of plastid genes as comprising the plastid genome is also supported by recent results demonstrating that at least some of the missing genes (i.e. ones normally found in plastids) are instead nuclear-encoded in several species (17–19). These experiments suggest that dinoflagellates do not obey for the rules normally governing plastid gene transfer to the nucleus (20).
However, it is still a formal possibility that additional genes may be encoded in the plastid in a form different from the minicircle format found so far. One method potentially applicable to the characterization of the plastid genome is to determine the spectrum of genes expressed, as gene expression should be independent from the form in which the genes are found. However, it is generally difficult to discriminate between organelle- and nuclear-encoded transcripts, as both can be modified by addition of a poly(A) tail at their 3′ termini (21). We have found that, unlike other transcripts in the dinoflagellates, those encoded by known minicircle genes carry a homogenous polyuridine tract at their 3′ termini. We have taken advantage of this unusual feature to characterize the dinoflagellate plastid transcriptome, and find that our analysis fully supports a highly reduced plastid genome for the peridinin-containing dinoflagellates. Furthermore, as it seems likely that polyuridylylation may be common in dinoflagellate plastids, it may be possible to rapidly characterize the transcriptome of many different species using the method described here.
MATERIALS AND METHODS
Amphidinium carterae (CCMP 1314) and Lingulodinium polyedrum (CCMP 1936, formerly Gonyaulax polyedra) were obtained from the Provasoli–Guillard Culture Center for Marine Phytoplancton (Boothbay Harbor, Maine) and cultured as described (22). Poly(A) RNA was purified from Lingulodinium and Amphidinium using oligo(dT) chromatography (23) and hybridized to a psbA and 23S probes as described (22). The 16S probe was prepared against an EST sequence from our library (see below), while the atpB probe was prepared using previously described amino acid microsequence data (22) to design two degenerate oligonucleotides 5′-TTYTICARGCIGGIWSIGARGT-3′ and 5′-ACYTCIGCIACRAARAAIGGYTG-3′; the 500 bp PCR product was confirmed as atpB by sequence analysis.
The 3′ end sequences of atpB and psbA transcripts were obtained from cDNA synthesized from poly(A) RNA tailed in vitro with rGTP by poly(A) polymerase. Specific sequences were amplified using a d(C)10 oligonucleotide and 5′-TATCCAATTGGACAAGGAAG-3′ (Lingulodinium psbA) 5′-GTGTAGCACAAGATGTAAGC-3′ (Lingulodinium atpB), 5′-GTATTCGGTCAAGAGGATG-3′ (Amphidinium psbA) and 5′-CTATCTCAGCCGTTCTTTG-3′ (Amphidinium atpB). The genomic psbA sequences from Lingulodinium were obtained using TAIL-PCR (24) using three nested internal psbA oligonucleotides (5′-GCTGCTTGGCCAGTTATTGGTATCTG-3′; 5′-TCTGGTTTACAGCACTTGGTGTTAG-3′; 5′-GTCCATAATTGATTCATCAGGTCATC-3′) to amplify a fragment from AT-rich DNA purified by bisbenzimide-CsCl gradients. To obtain the 3′ end of genomic sequences from Amphidinium, minicircle DNA was amplified by inverse PCR using outwardly directed oligonucleotides 5′-GTATTCGGTCAAGAGGATG-3′ and 5′-CAGGAGCAAGGAAGAAAG-3′ (psbA) or 5′-GTGGTCGTAAGATTGAGAGG-3′ and 5′-GATGAGAGCGTTGGCATAC-3′ (atpB).
For cDNA preparation, 10 µg poly(A)-enriched RNA was used as a template for first strand synthesis using a commercial kit (Stratagene) but replacing the usual oligo(dT) primer with 5′-(GA)10ACTAGTCTCGAG(A)18-3′. Sequences were identified using the BLAST algorithm (www.ncbi.nlm.nih.gov) and have been deposited in GenBank under the accession nos. DQ264844 through DQ264867. All sequence alignments and analyses were performed using MacVector (Accelrys) except for RNA secondary predictions that were made using a web server (www.bioinfo.rpi.edu/applications/mfold). Statistical analysis using rarefaction (25) to determine the likelihood that our sample size was sufficient to detect all different cDNAs in the library was made on a web server (www2.biology.ualberta.ca/jbrzusto/rarefact.php).
RESULTS
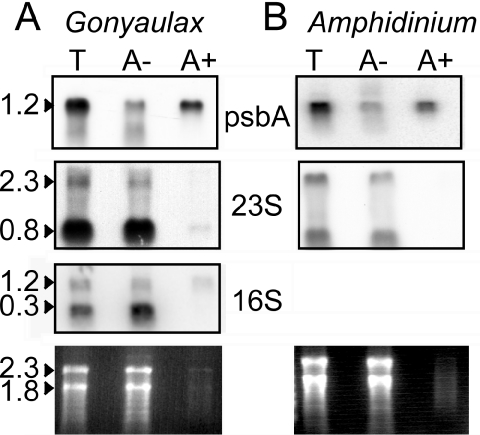
We began our analysis of the dinoflagellate plastid transcriptome by fractionation of RNA into poly(A)-enriched and depleted fractions by oligo(dT) chromatography. The plastid-encoded psbA (22) in RNA extracted from the dinoflagellates Lingulodinium (Figure 1A) and Amphidinium (Figure 1B), was found enriched by ∼10-fold in poly(A+) fractions, while 23S rRNA remained in the poly(A−) fraction. The small size of the 23S rRNA signal is suggestive of processing and has been previously observed in dinoflagellates (13). The 16S RNA was similarly processed, but the full-length form was found to a greater extent in the polyadenylated fraction than was the 23S RNA. The abundance of these transcripts in a poly(A) rich fraction was unexpected, as usually only a small fraction of plastid messages are polyadenylated (21). Furthermore, it seemed at odds with the lack of minicircle genes found in dinoflagellate EST libraries (17–19).
Figure 1.
Dinoflagellate plastid messages are located in poly(A)-enriched RNA. Total RNA samples (T) from the dinoflagellates Lingulodinium (A) and Amphidinium (B) were resolved into fractions enriched (A+) and depleted (A−) for poly(A) RNA by chromatography on oligo(dT) cellulose. RNA blots were challenged with gene probes for either psbA, 23S RNA or 16S. Lower panels show the ethidium bromide stained gels.
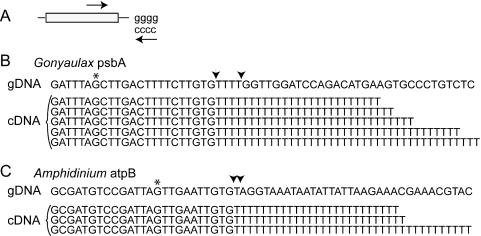
We originally thought that the tails might be heterogeneous, similar to the 3′ termini in chloroplasts and cyanobacteria RNA formed by polynucleotide phosphorylase (PNP) (21), since the presence of nucleotides other than adenine in the 3′ tails might inhibit cDNA synthesis more than oligo(dT) chromatography. To test this, we guanylated our poly(A)-enriched RNA using poly(A) polymerase, and performed RT–PCR with a primer pair allowing specific amplification of the psbA 3′ end (Figure 2A). Fourteen different psbA clones were sequenced, and all contained a homogeneous 3′ terminal stretch of thymidine residues. At least five of the sequences obtained were independent clones based on differences in tail length, which varied between 25 and 40 thymidine residues (Figure 2B). A comparison to the genomic DNA sequence, obtained by thermal asymmetric interlaced (TAIL) PCR (24), shows that these residues are added post-transcriptionally, and all appeared to have been added at the same site (between arrows, Figure 2B). Poly(T) tracts of similar length were found on the 3′ termini of atpB transcripts from Lingulodinium (data not shown) and on atpB transcripts from the dinoflagellate Amphidinium (Figure 2C). We propose that binding of these polyuridylylated mRNAs to the longer and more prevalent poly(A) tails of nuclear transcripts is responsible for their presence in poly(A)-enriched RNA fractions obtained by oligo(dT) cellulose chromatography.
Figure 2.
Plastid transcripts contain a homogenous poly(U) tail at a specific site. (A) Lingulodinium RNA samples enriched for poly(A) RNA were tailed with guanine residues, and transcript 3′ end sequences amplified using a specific internal oligonucleotide and an oligo(dC) oligonucleotide. (B) Sequences of the 3′ end of fourteen psbA cDNAs yielded five clones with different numbers of thymidine residues. The cDNA sequences are aligned with genomic psbA sequences obtained by TAIL-PCR. The polyuridylylation site is defined to the stretch of thymidine residues encoded by the genomic sequence (arrows). The asterisk indicates the termination codon. (C) Amphidinium RNA samples were treated similarly except that atpB specific primers were used in the amplification. The cDNA sequences are aligned with minicircle atpB sequences obtained by inverse PCR.
The unusual 3′ terminal polyuridylylation, if a common feature of dinoflagellate plastid transcripts, suggested that analysis of cDNA synthesized using an oligo(dA) instead of the usual oligo(dT) primer would provide a straightforward method to catalog the plastid gene complement. Roughly 60 ng of cDNA was synthesized from 10 µg poly(A)-enriched RNA, a yield 30-fold less than that obtained with an oligo(dT) primer in similar experiments. Polyuridylylated transcripts are thus a small but significant proportion of cellular RNA. To characterize the library, several hundred clones were selected at random and sequenced. Some GC-rich and unidentified sequences were found, but none were polyuridylylated and were presumably derived from hairpin priming of the predominant GC-rich nuclear-encoded transcripts. In contrast, all polyuridylylated sequences were AT-rich and were identified as transcripts from the minicircle genes found in other dinoflagellates (16) (Table 1). The majority of the transcripts correspond to photosystem II components (psbA-D) and 16S RNA. This latter may appear more frequently in our oligo(dA)-primed library than the 23S RNA because the full-length16S RNA appears more abundant in a poly(A) enriched sample (Figure 1A).
Table 1.
Only transcripts from known minicircle genes are polyuridylylated in Lingulodinium
| Gene | Size (kb) | % Clones (n) |
|---|---|---|
| AtpA | 1.5 | 5.7% (17) |
| AtpB | 1.4 | 0.7% (2) |
| PsaA | 2.5 | 3.7% (11) |
| PsaB | 2.3 | 2.0% (6) |
| PsbA | 1.2 | 23.0% (69) |
| PsbB | 1.6 | 9.3% (28) |
| PsbC | 1.4 | 12.7% (38) |
| PsbD | 1.3 | 17.0% (51) |
| PetB | 0.9 | 0.7% (2) |
| PetD | 0.5 | 2.3% (7) |
| 16S rRNA | 1.2 | 21.0% (63) |
| 23S rRNA | 2.3 | 2.0% (6) |
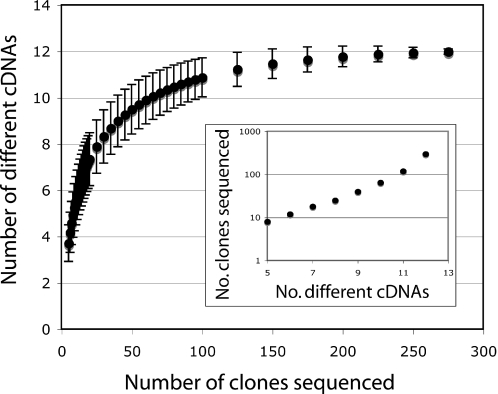
To assess the likelihood that other low abundance polyuridylylated transcripts might be present in the library, we performed rarefaction analysis (25). Originally developed to compare species richness in biodiversity collections of different sizes, this technique estimates the number of different cDNAs that would be obtained if smaller sample sizes were taken. The analysis of the data in Table 1 indicates that the identification of twelve different sequences could have been possible with only 120 random clones sequenced; the three hundred clones reported here represent a significant excess of this minimum value. The calculations can also be used to illustrate the progression in the number of random sequences required to reveal each additional sequence: while 10 different cDNAs are expected in 65 random sequences, almost twice as many sequences are required to uncover the eleventh sequence, and roughly six times more sequences to yield the twelfth cDNA (Figure 3 inset). This analysis suggests that to recover a potential new plastid transcript, well over a thousand random clones would have to be sequenced.
Figure 3.
The library is likely to contain only 12 different cDNA sequences. Estimates of the number of different sequences expected for different numbers of random clones sequenced were made by rarefaction analysis of the data in Table 1. The estimated number of different cDNA sequences expected with smaller sample sizes shows a statistical possibility that the twelve different clones could have been identified with only 120 different sequences. The progression of the number of random sequences as a function of the number of different clones (inset) suggests over a thousand sequences would be required to identify any potentially new clone in the library.
To further test the contention the twelve genes recovered are likely to represent saturation coverage of the oligo(dA) primed library, we employed a virtual subtraction protocol (26). Here, roughly a thousand different cDNA clones were randomly selected and streaked on a new Petri plate in a grid configuration. Colony lifts were then hybridized with a probe prepared against a mixture of psbA-D or 16S sequences. From this, 50 cDNAs that hybridized weakly or not at were selected and sequenced. Only 14 sequences among the 50 sequenced were AT-rich and polyuridylylated, and these included two atpA, four 23S RNA, a petB, three psbB, three psbC and a psbD. These later three photosystem II components may have been recovered following the virtual subtraction because of poor bacterial growth or poor transfer to the membrane. More importantly, no new polyuridylylated genes were identified, again suggesting that our coverage of the library had reached saturation. As it seems unlikely that alternative 3′ modifications might be found within the same organelle, we conclude that the genome of this photosynthetically active chloroplast is likely to encode only the transcripts identified here.
To address the mechanism underlying site selection for poly(U) addition, we evaluated a potential role for both secondary structure and primary structure elements. Secondary structure is the principal determinant of polyadenylation site selection in prokaryotes (27). However, computer generated secondary structure predictions from RNA complementary to the genomic sequence surrounding the polyuridylylation site of either Lingulodinium psbA or Amphidinium atpB did not show any reasonably stable stem–loop structures (data not shown). However, two loosely conserved primary structure motifs are located within 50 bp of the modification site in all twelve transcripts (Figure 4). These motifs (AGAAA and AAUUA) might thus constitute primary structure elements signalling the polyuridylylation site in a manner similar to the use of AAUAAA in determining polyadenylation sites in nuclear encoded transcripts (28).
Figure 4.
Plastid transcripts may contain primary structure motifs for polyuridylylation. All different polyuridylylated 3′ terminal sequences corresponding to sequences identified in the library were aligned at the site of the poly(U) modification and at each of two potential conserved sequences (underlined).
We were also curious about the identity of the enzyme that might be used to catalyze the polyuridylylation reaction. As polyuridylylation of protein coding transcripts has been observed in organelles undergoing extensive uridine insertion editing (29,30), we thus checked for uridine insertion in comparisons of genomic and cDNA sequences using Lingulodinium psbA, petB and 16S RNA (∼2.5 kb total sequence). Our data reveals no evidence for uridine insertion although numerous examples of substitutional editing in dinoflagellate plastid transcripts (principally A to G) were observed (Table 2, Supplementary Figures S1 and S2). These results agree with a previous report of substitutional editing in the dinoflagellate Ceratium (31).
Table 2.
Various patterns of substitutional editing are found in dinoflagellate plastid transcripts
| Number of modifications | ||||
|---|---|---|---|---|
| Modification | PetB | PsbA | 16S | Total |
| A→G | 10 | 6 | 16 | 32 |
| G→A | 2 | 0 | 1 | 3 |
| C→T | 2 | 1 | 1 | 4 |
| T→C | 1 | 0 | 0 | 1 |
| T→G | 0 | 0 | 1 | 1 |
| G→C | 0 | 3 | 0 | 3 |
DISCUSSION
We have found that the plastid transcripts in two species of peridinin-containing dinoflagellates are characterized by an unusual 3′ polyuridylylation. This modification differs dramatically from the poly(A) tails of nuclear-encoded transcripts, and so provides a facile method for cataloging the plastid transcriptome. We report here that an oligo(dA)-primed cDNA library has a remarkably low complexity, with only 12 different clones identified in 300 randomly selected polyuridylylated sequences (Table 1). Interestingly, all the sequences identified from the library were previously identified as minicircle genes in other dinoflagellate species. This concordance between two independent methods (characterization of the minicircular genome and the transcriptome analysis reported here) suggests that the minicircular gene format is likely to be the only genome architecture in the plastid. Our results thus strongly support the contention that the dinoflagellate genome is the smallest of any functional chloroplast. Furthermore, since the dinoflagellate Amphidinium also contains polyuridylylated transcripts, it is possible that the technique described here could be used to catalog the plastid transcriptome from a range of other species.
Is it likely that the transcriptome contains other low abundance transcripts that were undetected in our relatively small sample size? The chance of finding a specific transcript in a single random sample is a function of its relative proportion, or frequency of occurrence within the bank. However, if many other sequences were present, then the chance of finding any other sequence would also depend on the number of additional sequences present and on their relative proportions within the library. To estimate our coverage of the library, we used rarefaction analysis to determine if smaller sample sizes would also have recovered the same twelve genes. Our analysis suggests that it is likely the same twelve genes would have been recovered using a smaller sample size, indicating that the number of clones sequence was in excess of that required. Furthermore, the progression in the number of clones sequenced as a function of the number of different clones identified (Figure 3) suggests that well over a thousand random clones would have to be sequenced to find an additional clone if it were indeed present in the library. Taken together with the virtual subtraction of psbA-D and 16S RNA sequences, these results strongly suggest that coverage of the library has reached saturation.
The selective forces that serve to maintain genes in the chloroplast are hotly debated, and thought to reflect either difficulties in targeting or transport of proteins that are extremely hydrophobic or a relationship between control of gene expression and the redox state of the organelle (32). With respect to the former, the twelve protein encoding genes found in our library are not the most hydrophobic of known thylakoid proteins. Indeed, a better explanation for the retention of this particular gene set in the plastid may lie in the length of the protein rather than hydrophobicity. As recently shown by analysis of Arabidopsis thylakoid proteins (33), the proteins encoded by the dinoflagellate plastid genes are generally the longest of the thylakoid proteins. However, even a combination of length and hydrophobicity is insufficient to explain transfer of some genes to the nucleus, such as the shorter yet relatively hydrophobic protein encoded by the atpI gene recently reported in an oligo(dT) primed EST bank from the dinoflagellate Alexandrium. If instead of hydrophobicity, the redox control of gene expression were the determining factor, this would suggest the set of proteins encoded by the dinoflagellate plastids are most sensitive to changes in redox potential. This hypothesis could potentially be tested in other plastids. The proposal that it is genes requiring editing may that are conserved in plastids (34) is supported by analysis of the petB gene of Lingulodinium, as editing removes a stop codon in the middle of the derived protein sequence (Supplementary Figure S2).
With respect to the unusual nature of the modification itself, there is precedent for polyuridylylation in organelles experiencing extensive editing, such as the mitochondria of trypanosomes or myxomycetes. In some cases, 3′ polyuridine tails have been observed not only for the guide RNAs used to determine the site of uridine insertion but for rRNA and mRNA as well (29,30). However, a comparison of ∼2.5 kb genomic and cDNA sequences showed only substitutional editing (Table 2). Our experiments thus provide the first example of extensive polyuridylylation occurring in the absence of RNA editing. It is tempting to speculate that the poly(U) tracts may result from a terminal uridylyltransferase (TUT) (35) acting in the absence of guide RNA to define a site for uridine insertion. However, an alternative possibility is that the plastids may contain a type poly(A) polymerase with specificity for uridine residues instead of adenine.
Recently, it has also been shown that microRNA-directed cleavage can result in addition of non-encoded oligonucleotides (mostly uridine) to 3′ termini (36). However, in this case the polyuridylylated transcripts are intermediates in an RNA degradation pathway. It seems unlikely that the extensive polyuridylylation of the plastid transcripts is used as a signal for RNA turnover, since the majority of psbA transcripts appear modified as judged by their co-purification on oligo(dT) chromatography. In contrast to the full-length protein coding transcripts, the small fragments hybridizing to 16S and 23S RNA on northern analyses appear unmodified by these criteria (Figure 1). Interestingly, the apparent stability of the polyuridylylated plastid transcripts thus suggests that this particular 3′ end modification has a different function from that in either cyanobacteria or the plastids of higher plants, where transcripts are marked for degradation by 3′ end polyadenylation (21).
Despite extensive molecular phylogenetic studies pointing to a common evolutionary origin for both host cells (7) and plastids (37) of the chromalveolates, no other plastids or cyanobacteria are known to polyuridylylate transcripts. Thus the mechanism and function of the dinoflagellate plastid transcript 3′ end modification are as unique as their form II Rubisco (10) and peridinin–chlorophyll a-protein (9). The major challenge for plastid phylogeny underscored by our results is to reconcile the many unique features of the dinoflagellate plastids with their phylogenetic relationships to red algae. In addition, given the concurrence of several lines of evidence supporting the highly reduced nature of the dinoflagellate plastid genome, it will be of interest to reinvestigate the nature of the selective forces maintaining the current plastid genome size in higher plants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank M. Lapointe, A. Lukombo and A. Tran for technical assistance, Drs B. F. Lang for helpful discussion, P. Legendre for suggesting the rarefaction analysis, and M. Hijri and M. Cappadocia for critical reading of the manuscript. The present work was funded by the National Science and engineering Research Council of Canada (171382-03 to D.M.). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dodge J.D. A survey of chloroplast ultrastructure in Dinophyceae. Phycologia. 1975;14:253–263. [Google Scholar]
- 2.Keeling P.J. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 2004;91:1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z., Green B.R., Cavalier-Smith T. Phylogeny of ultra-rapidly evolving dinoflagellate chloroplast genes: a possible common origin for sporozoan and dinoflagellate plastids. J. Mol. Evol. 2000;51:26–40. doi: 10.1007/s002390010064. [DOI] [PubMed] [Google Scholar]
- 4.Yoon H.S., Hackett J.D., Pinto G., Bhattacharya D. The single, ancient origin of chromist plastids. Proc. Natl Acad. Sci. USA. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fast N.M., Kissinger J.C., Roos D.S., Keeling P.J. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 6.Baldauf S.L., Roger A.J., Wenk-Siefert I., Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 7.Harper J.T., Waanders E., Keeling P.J. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. Syst. Evol. Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- 8.Jeffrey S., Seilicki M., Haxo F. Chloroplast pigment patterns in dinoflagellates. J. Phycol. 1975;11:374–384. [Google Scholar]
- 9.Hofmann E., Wrench P.M., Sharples F.P., Hiller R.G., Welte W., Diederichs K. Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae. Science. 1996;272:1788–1791. doi: 10.1126/science.272.5269.1788. [DOI] [PubMed] [Google Scholar]
- 10.Morse D., Salois P., Markovic P., Hastings J.W. A nuclear encoded form II rubisco in dinoflagellates. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 11.Barbrook A.C., Howe C.J. Minicircular plastid DNA in the dinoflagellate Amphidinium operculatum. Mol. Gen. Genet. 2000;263:152–158. doi: 10.1007/s004380050042. [DOI] [PubMed] [Google Scholar]
- 12.Laatsch T., Zauner S., Stoebe-Maier B., Kowallik K.V., Maier U.G. Plastid-derived single gene minicircles of the dinoflagellate Ceratium horridum are localized in the nucleus. Mol. Biol. Evol. 2004;21:1318–1322. doi: 10.1093/molbev/msh127. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z., Green B.R., Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z., Cavalier-Smith T., Green B.R. Evolution of dinoflagellate unigenic minicircles and the partially concerted divergence of their putative replicon origins. Mol. Biol. Evol. 2002;19:489–500. doi: 10.1093/oxfordjournals.molbev.a004104. [DOI] [PubMed] [Google Scholar]
- 15.Moore R.B., Ferguson K.M., Loh W.K.W., Hoegh-Guldberg O., Carter D.A. Highly organized structure in the non-coding region of the psbA minicircle from clade C Symbiodinium. Int. J. Syst. Evol. Microbiol. 2003;53:1725–1734. doi: 10.1099/ijs.0.02594-0. [DOI] [PubMed] [Google Scholar]
- 16.Koumandou V.L., Nisbet R.E., Barbrook A.C., Howe C.J. Dinoflagellate chloroplasts–where have all the genes gone? Trends Genet. 2004;20:261–267. doi: 10.1016/j.tig.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Bachvaroff T.R., Concepcion G.T., Rogers C.R., Herman E.M., Delwiche C.F. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist. 2004;155:65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
- 18.Hackett J.D., Yoon H.S., Soares M.B., Bonaldo M.F., Casavant T.L., Scheetz T.E., Nosenko T., Bhattacharya D. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr. Biol. 2004;14:213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Patron N.J., Waller R.F., Archibald J.M., Keeling P.J. Complex protein targeting to dinoflagellate plastids. J. Mol. Biol. 2005;348:1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Martin W., Herrmann R.G. Gene transfer from organelles to the nucleus: how much, what happens, and Why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rott R., Zipor G., Portnoy V., Liveanu V., Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Jensen L., Hojrup P., Morse D. Synthesis and degradation of dinoflagellate plastid-encoded psbA proteins are light-regulated, not circadian-regulated. Proc. Natl Acad. Sci. USA. 2005;102:2844–2849. doi: 10.1073/pnas.0406522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Vol. 1,2,3. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Liu Y.G., Chen Y., Zhang Q. Amplification of genomic sequences flanking T-DNA insertions by thermal asymmetric interlaced polymerase chain reaction. Methods Mol. Biol. 2005;286:341–348. doi: 10.1385/1-59259-827-7:341. [DOI] [PubMed] [Google Scholar]
- 25.Hurlbert S.H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 26.Germain H., Rudd S., Zotti C., Caron S., O'Brien M., Chantha S.C., Lagace M., Major F., Matton D.P. A 6374 unigene set corresponding to low abundance transcripts expressed following fertilization in Solanum chacoense Bitt, and characterization of 30 receptor-like kinases. Plant Mol. Biol. 2005;59:515–532. doi: 10.1007/s11103-005-0536-8. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 28.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 29.Adler B.K., Harris M.E., Bertrand K.I., Hajduk S.L. Modification of Trypanosoma brucei mitochondrial rRNA by post-transcriptional 3′ polyuridine tail formation. Mol. Cell Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton T.L., Landweber L.F. Mitochondrial RNAs of myxomycetes terminate with non-encoded 3′ poly(U) tails. Nucleic Acids Res. 2000;28:4750–4754. doi: 10.1093/nar/28.23.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zauner S., Greilinger D., Laatsch T., Kowallik K.V., Maier U.G. Substitutional editing of transcripts from genes of cyanobacterial origin in the dinoflagellate Ceratium horridum. FEBS Lett. 2004;577:535–538. doi: 10.1016/j.febslet.2004.10.060. [DOI] [PubMed] [Google Scholar]
- 32.Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Rev. Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 33.Friso G., Giacomelli L., Ytterberg A.J., Peltier J.B., Rudella A., Sun Q., Wijk K.J. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bungard R.A. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. Bioessays. 2004;26:235–247. doi: 10.1002/bies.10405. [DOI] [PubMed] [Google Scholar]
- 35.Aphasizhev R. RNA uridylyltransferases. Cell Mol. Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen B., Goodman H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 37.Yoon H.S., Hackett J.D., Van Dolah F.M., Nosenko T., Lidie K.L., Bhattacharya D. Tertiary endosymbiosis driven genome evolution in dinoflagellate algae. Mol. Biol. Evol. 2005;22:1299–1308. doi: 10.1093/molbev/msi118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.