Abstract
The cap1 genes are required for the synthesis of type 1 capsular polysaccharide (CP1) in Staphylococcus aureus. We previously showed that the cap1 locus was associated with a discrete genetic element in S. aureus M. In this report, we defined the boundaries of the cap1 element by comparing its restriction pattern to that of a corresponding region from the CP1-negative strain Becker. The element was located in the SmaI-G chromosomal fragment of the standard mapping strain NCTC8325. The sequences of the entire cap1 element and the flanking regions were determined. We found that there were two additional cap1 genes not previously identified. The cap1 operon was located in a staphylococcal cassette chromosome (SCC) element similar to the resistance island SCCmec recently described for methicillin resistance in S. aureus. Notably, the SCCcap1 element was located at the same insertion site as all the SCCmec elements in the staphylococcal chromosome. The excision of SCCcap1 could be demonstrated only in the presence of the recombinase genes from an SCCmec element, verifying that SCCcap1 is a genuine SCC element but defective in mobilization. A novel enterotoxin gene, whose transcript was detected by Northern blotting, was found next to the SCCcap1 locus. We propose that the enterotoxin gene and SCCcap1 were inserted into this locus at the juxtaposition by independent events. Sequence comparison revealed numerous DNA rearrangements and mutations in SCCcap1 and the left flanking region, suggesting that the SCCcap1 had been inserted at the SCC attC site a long time ago. In addition, most genes in this region were incomplete, with the exception of the 15 cap1 genes, implying that the cap1 genes confer a survival advantage on strain M.
The ability of Staphylococcus aureus to cause a variety of diseases in humans and animals may be attributed to its ability to produce a plethora of virulence factors. Many of the virulence-factor-encoding genes are located in discrete genetic elements, which are strain specific. Genetic elements such as plasmids, transposable elements, converting bacteriophages, and, most recently, pathogenicity islands and resistance islands have all been described previously for S. aureus (20). These genetic elements account for most of the DNA sequence divergence among different strains.
One of the virulence factors of S. aureus is the capsular polysaccharide (CP). Most strains produce capsules of either serotype 5 (CP5) or serotype 8 (CP8). Three-quarters of the genes required for the biosynthesis of CP5 and CP8 are almost identical, and the cap5 and cap8 genetic loci are allelic (23). The cap5 or cap8 genes could be considered the basic genetic constituents of the staphylococcal chromosome. However, some strains of S. aureus possess an additional capsule locus that contains the cap1 genes responsible for producing type 1 CP (CP1). The CP1-producing strains are more resistant to phagocytosis and are more lethal to mice than are CP1-negative mutants (17-19, 21). Interestingly, the cap1 genes have been shown previously to reside in a discrete genetic element of 33 to 35 kb in size (15). However, the nature of this genetic element is not known.
In this study, we sequenced the entire cap1 element and its flanking regions. We found that the cap1 element was a staphylococcal cassette chromosome (SCC) element similar to the recently described SCCmec elements carrying the mecA methicillin resistance gene (8). The SCCcap1 element was located at the same site as all SCCmec elements (7) and was able to be excised in the presence of the functional recombinases from an SCCmec element. A novel enterotoxin gene whose transcript could be detected was found adjacent to the SCCcap1 site.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
S. aureus M is a CP1-producing strain which contains the cap1 locus and one other cap locus related to cap8 (18, 24). S. aureus Becker is a CP8-producing strain which contains the cap8 locus but not the cap1 locus (24). Escherichia coli strain XL1-Blue was used as a host strain for plasmid constructions. S. aureus RN4220 (12) was used as the recipient for plasmid electroporations. S. aureus strains were cultivated in Trypticase soy medium (Difco Laboratories, Detroit, Mich.). E. coli strains were routinely cultivated in Luria-Bertani medium (Difco Laboratories). Electroporation was carried out according to the procedure described by Kraemer and Iandolo (11). Plasmid vector pLI50 has been previously described (16).
DNA manipulations.
Standard DNA manipulations were carried out as described by Sambrook et al. (22). Rapid small-scale plasmid DNA isolation was performed according to the procedure of Holmes and Quigley (5). The Qiagen DNA isolation kits (Qiagen, Inc., Chatsworth, Calif.) were used for general plasmid purification. Chromosomal DNA from S. aureus was isolated by the method of Dyer and Iandolo (2). Enzymes used in DNA manipulation were obtained from GIBCO-BRL (Gaithersburg, Md.), New England Biolabs, Inc. (Beverly, Mass.), or Promega Corp. (Madison, Wis.). PCR amplification was carried out with the Advantage cDNA PCR kit (Clontech, Palo Alto, Calif.). The transfer of DNA to nitrocellulose membranes was done by the method of Southern (26). Field inversion gel electrophoresis was carried out as described by Goering and Winters (3).
PCR analyses of the SCCcap1 excision.
The chromosomal DNAs were subjected to PCR analysis with the protocol from the Advantage cDNA PCR kit with the annealing-extension temperature set at 63 to 65°C. To amplify the attC site, two primers were used: cp1e22 (AACGCATGACCCAAGGGCAA) and cp1e24 (GGTTTCATTCCTAGGTGTTAG). The amplified fragments were either purified with the Qiagen PCR kit or cloned into the pGEMT-Easy vector (Promega) and sequenced directly with nested primers cp1e26 (CTGCACAAGGACGTCTTACA) and cp1e28 (CATGAAGTACCAAAGATTGTCC). To amplify attSCCcap1, two primers were used: cp1e29 (GCATTTGAGTTTTGGAGGAG) and cp1e30 (GACATTACTTCGATGAATAACC). The amplified fragments were cloned into pGEMT-Easy, and two independent recombinant plasmids were sequenced with the standard T7 primer.
Recombinant PCR.
To alter the nonsense mutation of CM34/35, the two-step overlapping PCR technique as described by Higuchi (4) was used. In the first step, two PCR fragments were obtained by two pairs of primers (pair 1, cr1 [GGTACCGGATCCAGACGATGAGGCATTAGAT] and cr2 [CTGAAAATGGCTTTTGATTAATAGTACGATA]; pair 2, cr3 [TATCGTACTATTAATCAAAAGCCATTTTCAG] and cr4 [TCTAGAGGATCCTCCTAATGTATTTGGTTG]). Both cr2 and cr3 contain the nucleotide base that alters the nonsense mutation. Pair 1 was used to amplify the N-terminal end and the upstream sequence of the CM35/34 open reading frame (ORF), whereas pair 2 was used to amplify the C-terminal end. In the second step, the two PCR fragments were annealed (cr2 and cr3 are complementary to each other) and used as a template for a second PCR amplification by cr1 and cr4 (both with BamHI adapter, underlined). The resultant 2,028-bp BamHI fragment, which contains the reconstituted CM35/34 ORF, was cloned into vector pLI50 and verified by sequencing.
RNA extraction and Northern hybridization.
The Blue FastRNA Kit (Bio 101, Inc., Vista, Calif.) was used in RNA extraction (1). Northern hybridization was carried out as described by Sambrook et al. (22). Total RNAs were resolved in a 1% agarose gel containing formaldehyde and transferred to a nitrocellulose membrane by using a TurboBlotter (Schleicher & Schuell, Keene, N.H.). The conditions for hybridization and washing had been previously described (27)
Cloning of the ent gene.
A 1.5-kb BamHI-HindIII fragment containing the ent gene identified in this study was obtained by restriction digestion of a recombinant cosmid clone from a previously constructed cosmid library (14). The fragment was ligated to the similarly digested shuttle vector pLI50. The resultant plasmid, pCL8418, was electroporated into strain RN4220.
Nucleotide sequence accession number.
The nucleotide sequence has been deposited in the EMBL/GenBank databases under accession no. U10927.
RESULTS
Physical mapping of cap1 element.
To map the cap1 locus on the staphylococcal chromosome, DNA from S. aureus NCTC8325-4, a prophage-free strain derived from the standard mapping strain NCTC8325 (6), was isolated and digested with SmaI. The fragments were separated by field inversion gel electrophoresis and transferred to nitrocellulose paper. Because strain NTCC8325-4 does not harbor the cap1 gene cluster, a 5.4-kb EcoRI fragment (Fig. 1), which is located downstream of the cap1 element from the strain M chromosome and conserved in all S. aureus strains tested (15), was used as the probe. The 5.4-kb EcoRI fragment hybridized to a 175-kb DNA fragment that corresponds to the SmaI-G fragment of NCTC8325 (results not shown), implying that the cap1 locus is located in the same SmaI-G fragment. A single band was also detected from strain M. Interestingly, the cap8/cap5 locus was also mapped in the SmaI-G fragment, indicating that the cap1 and cap8/cap5 loci are not far apart (23).
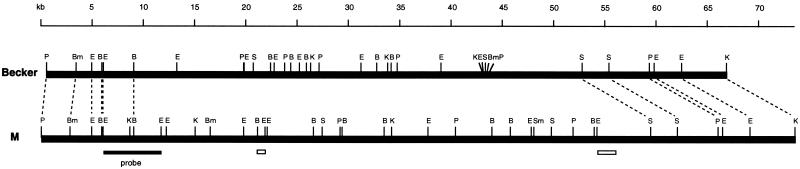
FIG. 1.
Comparison of the restriction map of the chromosome region containing the cap1 locus and the flanking regions in strain M and that of the allelic region in strain Becker. Dashed lines represent matched restriction sites. The probe used for physical mapping of the cap1 element is indicated. The boundaries defined previously by Southern hybridization are indicated by open bars. Restriction sites: B, BglII; Bm, BamHI; E, EcoRI; K, KpnI; P, PstI; S, SalI; Sm, SmaI.
Defining the boundaries of the cap1-associated element.
We have previously shown by Southern hybridization that the cap1 element is about 33 to 35 kb in length (15). Because the element is specific to CP1-producing strains, we postulated that the element could be originally acquired by S. aureus M through insertion of the element at a specific site in the chromosome. To test this hypothesis, we cloned the potential chromosomal cap1 element insertion site from strain Becker, which contains no cap1 genes, by screening an E. coli cosmid library of strain Becker (24) with the chromosomal DNA regions flanking the cap1 element as probes. The screening identified several clones with inserts ranging from 23 to 27 kb. However, those clones identified by the probe from the region flanking one side of the cap1 element did not hybridize to the probe prepared from the region flanking the other side of the element. These results indicate that strain Becker also contains a sizable DNA sequence in its chromosome corresponding to the cap1 element insertion site of the strain M chromosome. To clone this DNA region from strain Becker corresponding to the cap1 element of strain M, chromosomal walking experiments were performed. Several clones were identified, and the overlapping inserts of these clones were mapped with a set of seven restriction enzymes. The restriction maps of all the inserts from strain Becker were combined, and the composite map was constructed (Fig. 1). By comparing the restriction maps of the chromosome region containing the cap1 element of strain M and the corresponding region of Becker, we estimated that the cap1 element was between 42 and 50 kb (kb 9 to 12 at the left and kb 54 to 59 at the right) and that the corresponding region in Becker was between 37 and 45 kb in size. The size of the cap1 element based on the restriction map comparison was larger than the size of 33 to 35 kb estimated previously by Southern hybridization (15), which mapped the left and the right junctions at about kb 21 and 54, respectively (Fig. 1). The right boundary was in good agreement between the two methods. However, the left junction mapped previously by Southern blotting was about 10 to 15 kb away from that mapped by the restriction pattern comparison.
Sequencing of the cap1-associated element.
To precisely define the cap1 element, we determined the nucleotide sequences of the DNA region containing the cap1 element between kilobase coordinates ∼8 (KpnI) and ∼55. Sequences beyond these points were not determined because they appeared to be conserved between strains based on restriction pattern comparisons. In addition, in order to determine the boundaries of the cap1 element at the nucleotide level, the Becker chromosomal DNA fragments corresponding to the boundaries of the cap1 element were subcloned and subjected to sequencing. However, during the course of this work, genome sequences of two S. aureus strains, NTCC8325 and COL, had been made available before publication, whereas the complete genomes of strains N315 and Mu50 had just been published (13). All of these strains do not contain the cap1 operon and thus can serve as cap1-negative strains for localizing the cap1 element. The sequences corresponding to the cap1 element from these strains were then compared. For clarity, only the comparison between strains M and NCTC8325 is shown in Fig. 2. The sequence comparisons allowed us to define the junctions precisely. Remarkably, we found that the right junction, attR, of the cap1 element almost exactly matched those of the SCCmec elements (7, 8), the staphylococcal chromosomal cassette mec elements responsible for methicillin resistance in S. aureus (Fig. 3). These results suggest that the cap1 element is likely a chromosome cassette type of element similar to SCCmec. However, the left junction was much more difficult to define because, unlike in strains COL, N315, and Mu50, a sizable fragment immediately adjacent to the left side of the SCC insertion site in strain 8325 was deleted in strain M (Fig. 2). Nevertheless, by careful alignment, we found a stretch of sequence of ∼40 bp in length, located about 1 kb downstream of the cap1 operon (at approximately kb 18.5 in Fig. 2), that matched well with the left junction of the type III SCCmec (7). The left junction sequence of the cap1 element contained a direct repeat and an inverted repeat that almost perfectly matched the 15-bp direct repeat and 7-bp inverted repeat, respectively, found in the consensus sequences of the SCCmec att sites (Fig. 3). Thus, on the basis that the cap1 element and the SCCmec elements shared similar features with respect to the insertion site, the conserved repeats in the att sites, and the coding ORFs (see below), we hypothesize that the cap1 element is a chromosome-cassette-type genetic element. Accordingly, we renamed the cap1 element SCCcap1.
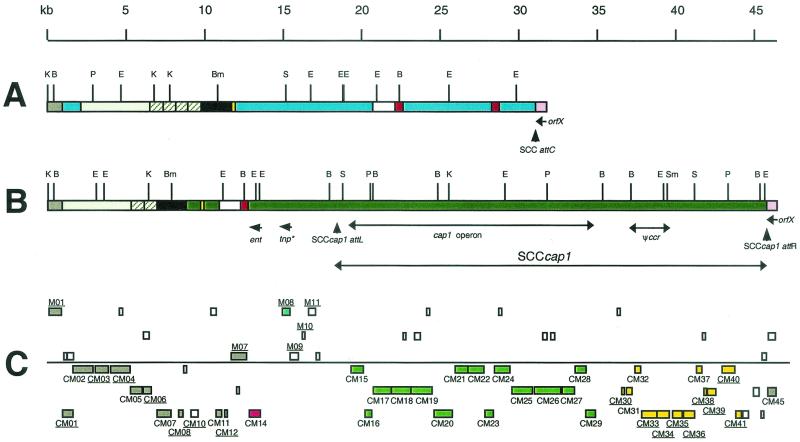
FIG. 2.
Structural features of the SCCcap1 element and the flanking regions. (A) The SCCcap1 allelic region from NCTC8325 based on the unpublished S. aureus genome sequence obtained from the University of Oklahoma Health Sciences Center. (B) SCCcap1 and the flanking regions from strain M based on nucleotide sequences deposited in GenBank. Regions shown in the same color for the two strains are at least 93% identical, except for the ∼770-bp repeated sequences shown in hatched boxes, which are between 81 and 87% identical. Blue indicates regions found in NCTC8325 but not in M. Green indicates a region found in M but not in NCTC8325. Vertical arrowheads represent attachment sites of the SCCcap1 element. orfX, the enterotoxin gene (ent), and the truncated transposase (tnp∗) are indicated by short arrows. The gene complexes (cap1 genes and ψccr genes; see text) are shown by double-headed arrows. (C) The ORFs derived from the sequence shown in panel B that can encode a peptide larger than 65 amino acids in six possible reading frames are shown by boxes. Those above the line are transcribed from the left, and those below the line are transcribed from the right. Labeled ORFs indicate those with significant homology to sequences in the databases. The ORFs that contain mutations are underlined. Those that are identical to ORFs in NCTC8325 are labeled in gray. The ent and tnp∗ genes are denoted by red and blue, respectively. The cap1 genes are shown in green, and those ORFs with significant homology to genes found in type III SCCmec of strain 85/2082 are indicated by yellow.
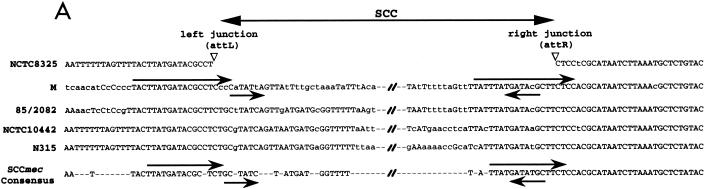
FIG. 3.
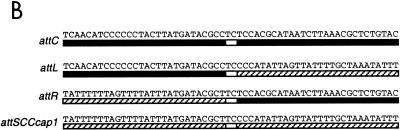
(A) Alignment of DNA sequences of SCC-chromosome junctions between SCC elements. The attL and attR sites are indicated by inverted open triangles. Direct arrows above the sequence indicate direct repeats. Inverted arrows below the sequence indicate inverted repeats. Uppercase letters denote those nucleotides that match the consensus sequence. Sequences of NCTC8325 are from the University of Oklahoma Health Sciences Center. Sequences of 85/2082, NCTC10442, and N315 and the SCCmec consensus sequence are from the work of Ito et al. (7). (B) Sequences of the att sites. Solid bars represent bacterial chromosomal DNA; hatched bars represent SCCcap1 element DNA; open bars denote the core sequence.
Genomic organization of SCCcap1.
The genomic organization of the SCCcap1 genetic element is shown in Fig. 2. The ORFs longer than 65 amino acids are shown. There were 27 ORFs encoded within the SCCcap1 element that could be inferred from the data bank. Thirteen of these ORFs were cap1 genes previously identified and shown to be required for CP1 biosynthesis in strain M (18). However, the previously reported sequence contained two errors just downstream of the cap1M gene originally thought to be the last gene of the cap1 operon. From the new sequence data, we found two ORFs, CM15 and CM16, with significant homology to the CP8 synthesis genes, cap8N and cap8O, of S. aureus, respectively (Table 1). These two ORFs were located just downstream of cap1M and were closely linked to the cap1 operon. We therefore named these two ORFs cap1N (CM16) and cap1O (CM15) and grouped them as part of the cap1 gene cluster, which brought the total number of cap1 genes from 13 to 15. These cap1 genes comprise the largest group of genes in the SCCcap1 element and represent the only virulence genes carried in the element. The total length of the coding region of the cap1 operon, therefore, is 15,535 bp, which is more than half of the size of the SCCcap1 element. As indicated above, SCCcap1 attL was located about 1 kb downstream of the cap1 operon. Interestingly, we found no ORF of significant length within this 1-kb region. Similarly, a region of ∼1.5 kb immediately upstream of the cap1 operon contained no ORF longer than 65 amino acids (Fig. 2).
TABLE 1.
Similarities of ORFs in the SCCcap1 element and the adjacent region of the M chromosome
| ORFa | Locationb | Sizec | Identityd | Genee | Description | Speciesf |
|---|---|---|---|---|---|---|
| M01 | 175-993 | 272 | 100 (264) | SA0100 | Conserved hypothetical protein | S. aureus |
| M07 | 11816-12685 | 289 | 100 (253) | SA0085 | Conserved hypothetical protein | |
| M08 | 14851-15375 | 174 | 37 (169) | tnp | Transposase (IS1181) | |
| M09 | 15603-16079 | 158 | 43 (144) | Trans-Golgi membrane protein p230 | Helicobacter pylori | |
| M10 | 16103-16327 | 74 | 40 (59) | Trans-Golgi membrane protein p230 | Helicobacter pylori | |
| M11 | 16645-17151 | 168 | 35 (118) | SV0404 | Hypothetical protein | S. aureus |
| CM01 | 965-1714 | 249 | 97 (249) | SA0099 | Hypothetical protein | S. aureus |
| CM02 | 1716-2903 | 395 | 96 (392) | SA0098 | Hypothetical protein | S. aureus |
| CM03 | 3045-3965 | 306 | 97 (306) | SA0097 | Hypothetical protein | S. aureus |
| CM04 | 4092-5282 | 396 | 97 (396) | SA0097 | Hypothetical protein | S. aureus |
| CM05 | 5380-6150 | 256 | 83 (256) | SA0094 | Hypothetical protein | S. aureus |
| CM06 | 6202-6744 | 180 | 85 (180) | SA0096 | Hypothetical protein | S. aureus |
| CM07 | 7193-8179 | 328 | 95 (328) | SA0091 | h-Phosphatidylinositol phosphodiesterase precursor | S. aureus |
| CM08 | 8606-8866 | 86 | 96 (58) | SA0090 | Hypothetical protein | S. aureus |
| CM10 | 9416-9751 | 111 | 51 (93) | Hypothetical protein | Streptococcus agalactiae | |
| CM11 | 10959-11255 | 98 | 82 (97) | SA0087 | Hypothetical protein | S. aureus |
| CM12 | 11304-11507 | 67 | 81 (33) | SA0086 | Hypothetical protein | S. aureus |
| CM14 | 12987-13766 | 259 | 46 (263) | sec3 | Enterotoxin type C3 | S. aureus |
| CM15 | 19515-20378 | 287 | 42 (267) | cap8N | Type 8 capsule synthesis gene | S. aureus |
| CM16 | 20378-20935 | 185 | 67 (185) | cap8M | Type 8 capsule synthesis gene | S. aureus |
| CM17 | 20935-22077 | 380 | 100 (380) | cap1M | Type 1 capsule synthesis gene | S. aureus |
| CM18 | 22105-23379 | 424 | 100 (424) | cap1L | Type 1 capsule synthesis gene | S. aureus |
| CM19 | 23392-24741 | 449 | 100 (449) | cap1K | Type 1 capsule synthesis gene | S. aureus |
| CM20 | 24734-25909 | 391 | 100 (391) | cap1J | Type 1 capsule synthesis gene | S. aureus |
| CM21 | 25929-26933 | 334 | 100 (334) | cap1I | Type 1 capsule synthesis gene | S. aureus |
| CM22 | 26952-28019 | 355 | 100 (355) | cap1H | Type 1 capsule synthesis gene | S. aureus |
| CM23 | 28019-28537 | 172 | 100 (172) | cap1G | Type 1 capsule synthesis gene | S. aureus |
| CM24 | 29548-29738 | 396 | 100 (396) | cap1F | Type 1 capsule synthesis gene | S. aureus |
| CM25 | 29740-31062 | 440 | 100 (440) | cap1E | Type 1 capsule synthesis gene | S. aureus |
| CM26 | 31090-32889 | 599 | 100 (599) | cap1D | Type 1 capsule synthesis gene | S. aureus |
| CM27 | 32914-33681 | 255 | 100 (255) | cap1C | Type 1 capsule synthesis gene | S. aureus |
| CM28 | 33684-33670 | 228 | 100 (228) | cap1B | Type 1 capsule synthesis gene | S. aureus |
| CM29 | 34285-35050 | 332 | 100 (332) | cap1A | Type 1 capsule synthesis gene | S. aureus |
| CM30 | 36571-36816 | 81 | 64 (96) | CZ068 | Hypothetical protein | S. aureus |
| CM31 | 36955-37314 | 119 | 84 (91) | CZ069 | Hypothetical protein | S. aureus |
| CM32 | 37410-37748 | 112 | 91 (112) | CZ070 | Hypothetical protein | S. aureus |
| CM33 | 37847-38593 | 248 | 93 (204) | CZ072 | Site-specific recombinase | S. aureus |
| CM34 | 38879-39529 | 216 | 92 (216) | CZ072 | Site-specific recombinase | S. aureus |
| CM35 | 39971-40489 | 172 | 82 (169) | CZ073 | Hypothetical protein | S. aureus |
| CM36 | 40529-41359 | 276 | 73 (277) | CZ073 | Hypothetical protein | S. aureus |
| CM37 | 41391-41729 | 112 | 63 (101) | CZ074 | Hypothetical protein | S. aureus |
| CM38 | 41722-41973 | 83 | 79 (83) | CZ075 | Hypothetical protein | S. aureus |
| CM39 | 41986-42549 | 187 | 90 (187) | CZ075 | Hypothetical protein | S. aureus |
| CM40 | 43035-43883 | 282 | 26 (148) | CZ076 | Hypothetical protein | S. aureus |
| CM41 | 43913-44224 | 103 | 31 (102) | CZ076 | Hypothetical protein | S. aureus |
| CM45 | 45802-46281 | 159 | 98 (159) | SA023 | Conserved hypothetical protein orfX | S. aureus |
ORFs with potential mutations are underlined.
Positions are based on the nucleotide sequence deposited in EMBL/GenBank under accession no. U10927.
Sizes of ORF-encoded proteins in amino acids.
Percent identities of amino acid sequences; numbers in parentheses indicate lengths of the homologous regions.
Genes with best match from the databases.
Species containing the best-matched gene.
The region adjacent to the right side of the cap1 operon (kb 36.5 to 42.8) contained sequence with high homology to a region of the type III SCCmec DNA from strain 85/2082 (7). Furthermore, the relative positions of the region with respect to the SCC attR site were also comparable between the two elements (Table 1). However, most of these ORFs found in SCCcap1 were incomplete due to mutations within the coding region. Among these ORFs, CM35 and CM34 were highly homologous to the N-terminal and the C-terminal portions, respectively, of the type III SCCmec CZ072 gene of strain 85/2082. These two ORFs apparently resulted from a nonsense mutation of an otherwise intact ORF whose translated product was 93% identical to the CZ072 gene product. The type III SCCmec CZ072 gene shares significant homology to the ccrB gene, which is one of two site-specific recombinase genes found in all SCCmec elements. For type II SCCmec, both the ccrB and ccrA genes have been shown elsewhere to be required for site-specific integration and excision of the SCCmec element (10). It is not known whether CZ072 is required for the movement of type III SCCmec, in which both the ccrB and ccrA homologs with high identity to those of type II SCCmec were also found. Since CM35/34, the CZ072 homolog of SCCcap1, had a nonsense mutation and we found no other ccr genes within the element, we expect that SCCcap1 may not be a live genetic element. In strain 85/2082, the region containing CZ072 and the adjacent three ORFs (equivalent to CM30 to CM32 of SCCcap1) had been designated the ψccr complex by Ito et al. (7) due to their conservation in amino acid sequences among all SCCmec elements. We therefore also labeled these homologs in SCCcap1 as the ψccr complex in Fig. 2.
Precise excision of the SCCcap1.
To test whether SCCcap1 is a live element, nested PCR experiments described by Ito et al. (8) for demonstrating the SCCmec excision were employed. However, we failed to show excision of SCCcap1. Because our sequencing analyses above showed that SCCcap1 possessed a ccrB homolog (CM35/34) with a nonsense mutation, we hypothesized that this gene initially mediated the insertion of SCCcap1 but acquired the mutation later. To test this, we altered the nonsense mutation to reconstitute the gene according to the CZ072 sequence of the type III SCCmec. The restored gene was cloned into a plasmid and transferred to strain M. The chromosomal DNA of the resultant strain was subjected to PCR analyses with two primers (cp1e22 and cp1e24) flanking the SCCcap1 element with the 3′ end toward the SCCcap1 element. Again, we failed to detect excision of SCCcap1. We next tested whether ccrA and ccrB genes could promote excision of SCCcap1. The plasmid pSR (10), which contains the ccrA and ccrB genes of type II SCCmec from strain N315 (provided by K. Hiramatsu), was transferred to strain M. The chromosomal DNA was isolated from strain M(pSR) and subjected to PCR analysis with cp1e22 and cp1e24 primers. We detected a 1.2-kb fragment (data not shown), which matched the expected size if SCCcap1 was excised from the predicted att sites. Sequencing of the 1.2-kb fragment showed that it contained the expected attC site formed by joining the left half of attL and the right half of attR (Fig. 3B). To detect the excised circular SCCcap1 element, two primers located within the SCCcap1 element and near att sites were synthesized. PCR results showed that a fragment of ∼0.52 kb was detected from strain M(pSR) but not from strain M (data not shown). Sequencing of the fragment showed that it contained attSCCcap1, consisting of the left half of attR and the right half of attL (Fig. 3B). These results therefore confirmed the locations of the att sites predicted by sequence comparison and showed that SCCcap1 belonged to the SCC genetic element family. According to the att site, the size of SCCcap1 was calculated to be 27,377 bp.
Genomic organization of the SCCcap1 flanking regions.
The sequence flanking SCC attR was highly conserved among all strains examined. As in all SCCmec elements, the stop codon of the orfX gene was located within the direct repeat, and thus, insertion of SCCcap1 would not alter the reading frame (7, 8). In contrast, the sequence flanking the left junction, which is conserved in type I and type II but not in type III SCCmec, was lost in strain M. Instead, we found several remnants of transposases and incomplete ORFs within the 4-kb region to the left of attL (Table 1), indicating that this region of the DNA has undergone significant rearrangement and mutation.
As shown in Fig. 2, SCCcap1 was within the ∼33-kb contiguous region from kb 12.8 to 45.7 of strain M DNA that was not found in the corresponding region of the NCTC8325 chromosome. Near the left end of this region, we found an ORF (CM14) with significant homology to several staphylococcal enterotoxin genes, most notably to the S. aureus enterotoxin C serotype 3 genes (sec3), with 46% identity over 256 amino acids, and the canine type C enterotoxin of Staphylococcus intermedius, with 48% identity over 234 amino acids. Since staphylococcal enterotoxin genes typically share 26 to 84% identity between serotypes and over 93% between subtypes (28), this newly identified enterotoxin gene may be considered a gene for a new type of enterotoxin. The ent gene is preceded by a potential Shine-Dalgarno sequence and an apparent prokaryotic promoter, suggesting that the gene is likely to be expressed. In fact, a 1.4-kb RNA band was detected from strain M by Northern hybridization when a 200-bp internal DNA fragment from ORF CM14 was used as the probe (Fig. 4). However, the transcript was much larger than the 780-bp coding region of ORF CM14, suggesting that there might be a large untranslated region of the transcript. We also cloned a 1.5-kb fragment containing CM14 into plasmid vector pLI50 and transferred it to strain RN4220. Northern blotting of the RNA from RN4220(pCL8418) showed a strong band with the same size as the 1.4-kb RNA band and two smaller RNA fragments, which are likely the degradation products of the 1.4-kb transcript. These results indicate that CM14 is a novel enterotoxin gene, which is most likely expressed in strain M.
FIG. 4.
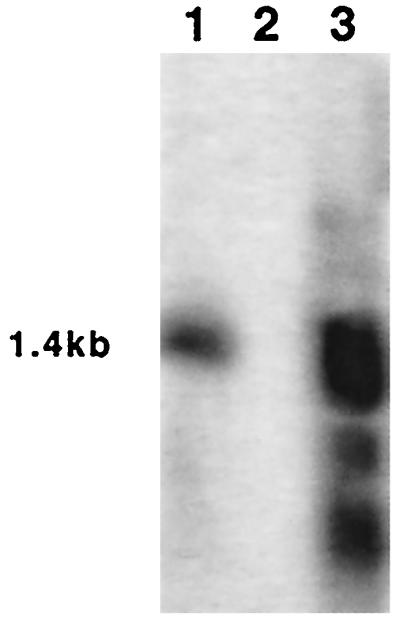
Northern hybridization of the ent gene. Total RNA was isolated from strains M (lane1), RN4220(pLI50) (lane 2), and RN4220(pCL8418) (lane 3); resolved by gel electrophoresis; and transferred to a nitrocellulose membrane. The membrane was probed with a 200-bp internal fragment of the ent gene.
The region of the M chromosome from kb 0 to ∼12.8 contained 20 potential ORFs. The majority of these ORFs, however, carried some types of mutations, and only five ORFs (CM02, CM05, CM07, CM11, and CM12) were likely intact, indicating that this region of the strain M chromosome has significantly accumulated mutations (Table 1). From Fig. 2, it is also apparent that this region of the M chromosome had undergone significant DNA insertions and deletions. Most notably, four blocks of NCTC8325 DNA totaling nearly 20 kb were not present in the M chromosome, whereas a region of about 2 kb of DNA (kb 9 to 11) found in M was not present in NCTC8325. In addition, two ∼770-bp tandem repeat sequences found in strain M (kb 5.5 to 7) were found repeated four times in tandem in NCTC8325. As a result, 22 ORFs found in this region of the NCTC8325 chromosome were not found in the corresponding M chromosome. In comparison, this region of the NCTC8325 chromosome (∼31 kb [Fig. 2]) was totally conserved in strain COL and was also well conserved in strains N315 and Mu50, except that the latter two strains had a 5-kb deletion corresponding to kb 15.7 to 20.7 of NCTC8325 (Fig. 2) and contained one extra copy of the ∼770-bp direct repeat of the tandem repeats.
DISCUSSION
Our previous results showed that the cap1 gene cluster and its flanking regions were specific to CP1 strains. In this study, sequencing revealed that this region of the strain M chromosome was composed of an SCC genetic element, SCCcap1, carrying the cap1 operon, a segment with remnants of transposases, and a novel enterotoxin gene. The SCCcap1 element was initially identified based on its similarity to SCCmec elements. However, it lacks a pair of ccr genes, ccrA and ccrB, required for recombination of SCCmec elements. By complementation with the ccrA and ccrB genes of SCCmec, excision could be detected by PCR. Thus, SCCcap1 is a bona fide SCC genetic element, which at one time must have been a live genetic element but lost its recombination genes and became defective over time. The element, nonetheless, still contains biologically functional attR and attL sites. By comparing all four att sites, we found a 2-bp core sequence, TC, suggesting that the crossover mediated by the ccrA and ccrB genes occurs within these two bases (Fig. 3B).
Sequence analysis of SCCcap1 and its left flanking region showed that there was a high occurrence of mutations in this region of the strain M chromosome. Indeed, among 46 ORFs in this region shown in Fig. 2 that could be inferred from a homology search, only 25 of them appeared intact. Within SCCcap1, the mutations concentrated in the region near the attR site, where only three of the eight genes (CM30 to CM42, which represent eight possible genes [Table 1]) appeared intact. The region to the left of SCCcap1 also contained several DNA deletions and insertions, which include the newly identified enterotoxin gene inserted immediately to the left of attL (Fig. 2). Thus, in this region, not only did mutational events occur, resulting in numerous incomplete ORFs, but also gross DNA rearrangements had taken place. The fact that there are several remnants of transposase abutting attL indicates that the rearrangements could have been initially promoted by transposable elements.
Compared to the three types of SCCmec elements, SCCcap1 most resembled the type III SCCmec. However, our results showed that SCCcap1 was a highly degenerate element in its present form. In contrast, strains containing SCCmec showed little rearrangement, especially the type I and type II strains. Furthermore, there is a lower proportion of the genes in SCCmec elements that are defective. Thus, the higher degree of degeneracy in DNA associated with SCCcap1 than SCCmec suggests that strain M acquired SCCcap1 much earlier than the methicillin-resistant strains acquired the SCCmec elements. The first methicillin-resistant strain was isolated in 1961, after the introduction of methicillin antibiotics in 1960 (9). Accordingly, the acquisition of the SCCcap1 element by strain M would have occurred earlier than 1960, although strain M was isolated in 1969 (25).
In light of the high incidence of mutations occurring within SCCcap1 and the left flanking region, it is intriguing to find that all the virulence genes in this region including the cap1 genes and the enterotoxin gene are still intact. In particular, the 15 cap1 genes, which occupy a large block of DNA 15.5 kb in length, were all functional. The sharp contrast in the occurrence of mutations in genes with close proximity suggests that the cap1 genes and maybe the enterotoxin gene confer a selective advantage for the survival of the organism. However, it is equally puzzling that CP1 strains have not been isolated frequently. Perhaps the CP1 strains had been the major pathogenic strains in the past in certain environments, which had since changed due to advances in health care management such as the advent of wide antibiotic usage.
The high number of incomplete ORFs and the deletion of 22 ORFs adjacent to the left side of the SCCcap1 insertion site in the M chromosome imply that these genes are not required for the survival of strain M in its environmental niche. In strain Becker, this region of the chromosome was also highly variable, as revealed by restriction maps shown in Fig. 1. Furthermore, Southern hybridization of 17 laboratory strains by using various segments of DNA from this region of the Becker chromosome as probes also showed that these strains were highly variable in hybridization pattern (data not shown). These results therefore suggest that most of this region of the staphylococcal chromosome is dispensable and thus may serve as sites for insertions such as SCC elements and other insertions like the novel enterotoxin gene found in strain M.
SCCs have been shown up to now to be associated with antibiotic resistance. Nonetheless, Ito et al. (7) speculated that SCC could be the conveyor of not only methicillin or other antibiotic resistance genes but also virulence genes in S. aureus. In this study, our finding that SCCcap1 had been inserted at the same location of the chromosome with a mechanism apparently identical to that of the SCCmec elements has provided strong support for their proposal. To our knowledge, this is the first report that an SCC element carries virulence genes. It remains to be determined how frequently SCC can serve as an element through which S. aureus strains can acquire other virulence genes.
Acknowledgments
We thank K. Hiramatsu for supplying plasmid pSR. The sequence of NCTC8325 was obtained from the University of Oklahoma S. aureus Genome Sequencing Project.
This work was supported by grant AI37027 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from Gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 2.Dyer, D. W., and J. J. Iandolo. 1983. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 46:283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goering, R. V., and M. A. Winters. 1992. Rapid method for epidemiological evaluation of gram-positive cocci by field inversion gel electrophoresis. J. Clin. Microbiol. 30:577-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higuchi, R. 1989. Using PCR to engineer DNA, p. 61-70. In H. A. Erlich (ed.), PCR technology. Stockton Press, New York, N.Y.
- 5.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 6.Iandolo, J. J. 2000. Genetic and physical map of the chromosome of Staphylococcus aureus 8325, p. 317-325. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 7.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jevons, M. P. 1961. “Celbenin”-resistant staphylococci. Br. Med. J. 1:124-125. [Google Scholar]
- 10.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 12.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature (London) 305:709-712. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Lee, C. Y. 1992. Cloning of genes affecting capsule expression in Staphylococcus aureus strain M. Mol. Microbiol. 6:1515-1522. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C. Y. 1995. Association of staphylococcal type-1 capsule encoding-genes with a discrete genetic element. Gene 167:115-119. [DOI] [PubMed] [Google Scholar]
- 16.Lee, C. Y., S. L. Buranen, and Z.-H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 17.Lee, J. C., M. J. Betley, C. A. Hopkins, N. E. Perez, and G. B. Pier. 1987. Virulence studies, in mice, of transposon-induced mutants of Staphylococcus aureus differing in capsule size. J. Infect. Dis. 156:741-750. [DOI] [PubMed] [Google Scholar]
- 18.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melly, M. A., L. J. Duke, D.-F. Liau, and J. H. Hash. 1974. Biological properties of the encapsulated Staphylococcus aureus M. Infect. Immun. 10:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novick, R. P., P. Schlievert, and A. Ruzin. 2001. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 3:585-594. [DOI] [PubMed] [Google Scholar]
- 21.Peterson, P. K., B. J. Wilkinson, Y. Kim, D. Schmeling, and P. G. Quie. 1978. Influence of encapsulation on staphylococcal opsonization and phagocytosis by human polymorphonuclear leukocytes. Infect. Immun. 19:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395-2405. [DOI] [PubMed] [Google Scholar]
- 24.Sau, S., and C. Y. Lee. 1996. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J. Bacteriol. 178:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott, A. C. 1969. A capsulated Staphylococcus aureus. J. Med. Microbiol. 2:253-260. [DOI] [PubMed] [Google Scholar]
- 26.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 27.Ye, Z.-H., and C. Y. Lee. 1993. Cloning, sequencing, and genetic characterization of regulatory genes rinA and rinB, required for the activation of staphylococcal phage φ11 int expression. J. Bacteriol. 175:1095-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, S., and G. C. Stewart. 2001. Staphylococcal enterotoxins, p. 117-136. In A. L. Honeyman, H. Friedman, and M. Bendinelli (ed.), Staphylococcus aureus infection and disease. Kluwer Academic/Plenum Publishers, New York, N.Y.