Abstract
The HIV-1 nucleocapsid (NC) protein is a small, basic protein containing two retroviral zinc fingers. It is a highly active nucleic acid chaperone; because of this activity, it plays a crucial role in virus replication as a cofactor during reverse transcription, and is probably important in other steps of the replication cycle as well. We previously reported that NC binds with high-affinity to the repeating sequence d(TG)n. We have now analyzed the interaction between NC and d(TG)4 in considerable detail, using surface plasmon resonance (SPR), tryptophan fluorescence quenching (TFQ), fluorescence anisotropy (FA), isothermal titration calorimetry (ITC) and electrospray ionization Fourier transform mass spectrometry (ESI-FTMS). Our results show that the interactions between these two molecules are surprisngly complex: while the Kd for binding of a single d(TG)4 molecule to NC is only ∼5 nM in 150 mM NaCl, a single NC molecule is capable of interacting with more than one d(TG)4 molecule, and conversely, more than one NC molecule can bind to a single d(TG)4 molecule. The strengths of these additional binding reactions are quantitated. The implications of this multivalency for the functions of NC in virus replication are discussed.
INTRODUCTION
The nucleocapsid (NC) protein of human immunodeficiency virus type 1 (HIV-1) is a very small, basic protein containing two retroviral zinc fingers. These zinc fingers exhibit the following conserved retroviral-type arrangement of zinc-coordinating residues: C-X2-C-X4-H-X4-C. NC is initially synthesized as part of the Gag polyprotein (1). As a domain of Gag, it participates in selection of the genomic RNA for encapsidation into the nascent virus particle (2) and its interactions with RNA are somehow critical in the structure of the particle (3,4). It also acts as a nucleic acid chaperone, unwinding a cellular tRNA molecule and catalyzing the hybridization of a portion of the tRNA to the viral genome, where it serves as a primer for reverse transcription (5,6). After the virus particle is released from the virus-producing cell, the Gag polyprotein is cleaved by the viral protease, resulting in the release of NC from the polyprotein (1). NC is complexed with genomic RNA in the mature, infectious virion; the nucleic acid chaperone activity of the protein causes a stabilization of the dimeric linkage between the genomic RNA molecules within the particle (7,8). When the particle infects a new host cell, this chaperone activity plays a crucial role in reverse transcription (9,10) and possibly in integration as well (11,12).
Thus, NC interacts with nucleic acids in remarkably diverse ways during the retroviral replication cycle. Because its functions are so critical for retrovirus replication, it is important to understand its interactions with nucleic acids in as much detail as possible. As one approach to this goal, we have studied the binding of this protein to very short, simple oligonucleotides. We and others previously reported that in 0.15 M NaCl, NC binds with significantly higher affinity to the repeating sequence, d(TG)n, than to other tested sequences (13,14). We have now extended this analysis in several ways. The binding of NC to d(TG)4 and related oligonucleotides was characterized by tryptophan fluorescence quenching (TFQ), fluorescence anisotropy (FA), isothermal titration calorimetry (ITC) and electrospray ionization Fourier transform mass spectrometry (ESI-FTMS), as well as in further experiments using surface plasmon resonance (SPR). All the results were subjected to a quantitative analysis. The results of these studies show that both NC and d(TG)4 are multivalent, despite their small sizes. We have quantitated the binding affinities for formation of higher-order, as well as 1:1, complexes between these two molecules. We propose that these properties of the system make important contributions to the nucleic acid chaperone activity of NC, and thus to HIV-1 replication.
MATERIALS AND METHODS
Mutant and wild-type NC proteins
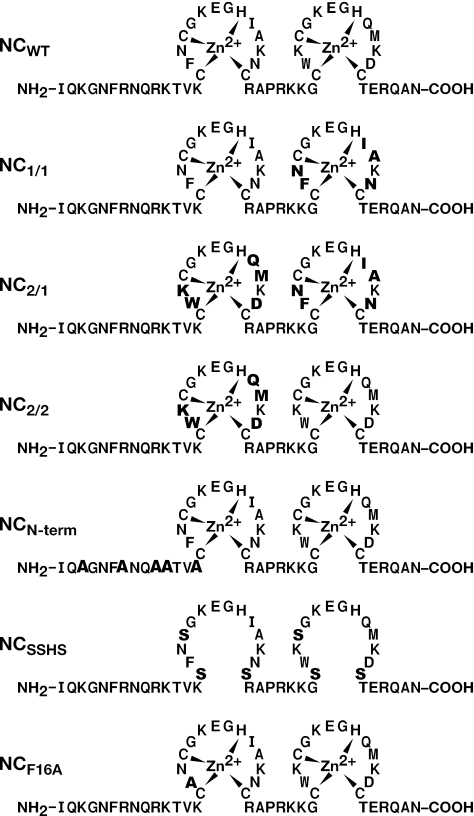
All NC proteins are the mature, 55 amino acid form of NC from pNL4-3 (GenBank accession no. AF324493). Procedures for the preparation of wt NC, NC1:1, NC2:1, NC2:2, NCSSHS and NCF16A mutant recombinant proteins have been published previously (11,15). The NC N-term mutant (‘N-term ala’), in which all positive charges are removed from the N-terminus before the first zinc finger, was made by introducing the following nucleotide changes in the NC gene before cloning into the pET32a vector: A23G/A24C, A35G/G36C, A44G/G45C, A47G/A48G and A56G/A57C, where nucleotide 1 is the 5′ end of the NC gene. These changes resulted in the substitution of alanine residues for the lysines at positions 3, 11 and 14, and the arginines at positions 7 and 10. Mutagenesis was performed with the Quick Change Site Directed Mutagenesis Kit (Stratagene). The recombinant N-term ala protein was expressed and purified essentially as described previously (11). In some experiments, the zinc fingers of wild-type NC were disrupted by treating the protein with 5 mM EDTA in the presence of 100 µM TCEP. Experiments with SSHS NC were also performed in the presence of 5 mM EDTA. All protein concentrations were determined by quantitative amino acid analysis on a Beckman System 6300 amino acid analyzer (Beckman Coulter, Inc., Fullerton, CA) and/or spectrophotometrically. The ɛ280 was taken as 5.7 × 103 cm−1 M−1 for wild-type NC and was adjusted as appropriate for mutant NCs. Virtually all results presented have been reproduced with multiple batches of NC protein.
Oligonucleotides
All oligonucleotides were oligodeoxynucleotides and were synthesized by Leo Lee (SAIC-Frederick). They were purified after synthesis by high-performance liquid chromatography (HPLC) and their concentrations were determined from their absorbance at 260 nm. ɛ260 values were: for (TG)4, 75 400; for (A)30, 363 400; for (TG)10, 188 200; for (A)10(TG)5(A)10, 363 400; and for (TG)5(A)10(TG)5, 308 800. For attachment to the SPR surface, the oligonucleotides were synthesized with biotin at the 3′ end using biotin TED CPG (Glen Research, Sterling, VA). Free biotin was removed using a G25 Microspin column (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions. Oligonucleotides used in the FA studies were synthesized with a fluorescein CPG (Glen Research) at the 3′ end. These fluoresceinated oligonucleotides were then purified by HPLC using a Waters Xterra MS C18 column (4.6 × 50 mm, particle size 2.5 µm; Milford, MA). The oligonucleotides were eluted over 20 min with a linear gradient of 2–98% Buffer B in Buffer A. (Buffer A consisted of 95% 0.1 M triethylammonium acetate/5% acetonitrile and Buffer B consisted of 70% 0.1 M triethylammonium acetate/30% acetonitrile.) The molecular weights of the eluted oligonucleotides were verified by MALDI-MS.
SPR analysis
Binding experiments were performed on BIAcore 3000 and BIAcore S51 instruments (BIAcore Inc., Piscataway NJ). Biotinylated oligonucleotides were immobilized on streptavidin-coated sensor chips. These chips were prepared in the laboratory by amine coupling using the BIAcore amine coupling kit (#BR1000-50) according to the manufacturer's instructions. Approximately 3500–3800 RU's of streptavidin were attached to BIAcore 3000 chips. Chips for the BIAcore S51 instrument were coated with ∼500 RU's of streptavidin. Oligonucleotides were reconstituted in buffer consisting of 10 mM Tris (pH 7.5), 300 mM NaCl and 1 mM EDTA. The amount of oligonucleotide immobilized on each surface is listed in the relevant figure legends. Proteins were serially diluted in a running buffer, consisting of 1× HBS [10 mM HEPES, 150 mM NaCl (pH 7.5)], 5 mM β-mercaptoethanol, 100 µM TCEP, 0.005% Tween-20 and 1 µM ZnCl2. The concentrations described in each sensorgram were injected at 25°C. After each injection of analyte during a BIAcore experiment, bound analyte is allowed to dissociate freely for a specified period of time. Disruption of any complex that remained bound after dissociation was achieved using an injection of 0.1% SDS. Samples with different concentrations of protein were injected in non-sequential order. The BIAcore 3000 data were X- and Y-transformed using BIAevaluation 3.01 software and non-random noise was removed by subtracting a buffer injection from each curve. Small refractive index mismatches were removed using MATLAB software. The curves obtained from all four flow cells and all NC concentrations were globally fit using the complex model shown in Figure 3, as described in the Supplementary Data.
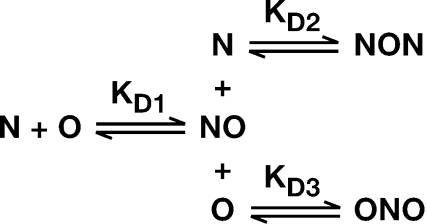
Figure 3.
Proposed reaction-scheme for interaction of NC with (TG)4, showing three possible binding reactions, each with its respective Kd. N, NC; O, (TG)4.
The data with very low densities of oligonucleotide were gathered on the BIAcore S51 using a 1 min injection of NC protein at 100 µl/min, followed by a 1 min dissociation and 30 s regeneration with 0.1% SDS. The NC injections were done in duplicate, and non-random noise was removed by subtracting a reference surface from each surface during a simultaneous measurement. The corrected data were globally fit to a 1:1 binding model as described in the Supplementary Data.
Tryptophan fluorescence assays
A Perkin Elmer (Wellesley, MA) luminescence spectrophotometer (LS50B) was used to measure fluorescence quenching. The instrument was set to an excitation maximum of 290 nm and an emission maximum of 350 nm. The excitation and emission slit widths were 2.5 and 10 nm, respectively. The filter cut-off was set at 350 nm and the integration time for sample readings at 10 s. The buffer was comprised of 1× HBS, 1 mM ZnCl2, 100 µM TCEP and 5 mM β-mercaptoethanol. A total of 3 ml buffer was added to two paired Hellma quartz cuvettes. NC was added to one cuvette for a final concentration of 400 nM, while an equivalent volume of buffer was added to the other cuvette as a control. Aliquots of the octanucleotide d(TG)4 (referred to as (TG)4 in the present work) were added to both cuvettes, and fluorescence readings were taken in triplicate. The average fluorescence readings of the control solution were subtracted from that of the NC solution to correct for dilution effects.
FA
Equilibrium binding isotherms for HIV-1 NC binding to (TG)4 were monitored by FA. Serial dilutions of wild-type or mutant NC were mixed in 96-well Costar polypropylene plates (Corning, NY) with 10 nM (TG)4 tagged at the 3′ end with fluorescein in 1× HBS, 5 mM β-mercaptoethanol, 100 µM TCEP, 1 µM ZnCl2 and 0.04% polyethylene glycol 20 000. After 10 min incubation, 2 aliquots of 50 µl were removed and transferred into 384-well Costar polypropylene plates (Corning, NY) and read using a Tecan Ultra plate reader (Durham, NC) with excitation and emission wavelengths of 485 and 535 nm, respectively.
ESI-FTMS
Sample solutions containing the desired concentrations of NC and oligonucleotides were prepared in 0.15 M ammonium acetate (pH 7.0) according to procedures described earlier (16). The NC protein employed in these experiments is identical to NCWT (Figure 1), except for M in place of I at the N-terminus. After mixing the different components, 5 µl aliquots were loaded on to quartz nanospray emitters (New Objective, Woburn, MA) and a platinum wire was inserted from the end to provide the necessary voltage. All determinations were performed on a Bruker (Billerica, MA) Apex III Fourier transform mass spectrometer equipped with a 7T shielded superconductive magnet and a thermally-assisted Apollo atmospheric pressure ion source. All data were acquired in negative ion mode and processed using XMASS 6.0.1 (Bruker, Billerica, MA). Typical analysis provided ∼150 000 resolution and <10 p.p.m. mass accuracy in broadband mode, using a three-points external calibration.
Figure 1.
Sequences of NC and NC mutants. Changes from wild-type (WT) are shown in bold.
Mathematical treatment of results
All results were analyzed as described in detail in Supplementary Data.
RESULTS
SPR analysis
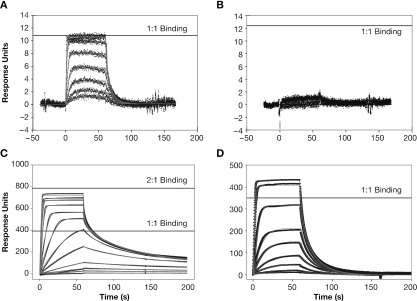
The sequences of HIV-1 NC protein and of the mutants used in the present work are shown in Figure 1. We had previously observed that HIV-1 NC exhibits sequence-specific binding to oligonucleotides containing the sequence d(TG)n (13), and that 5 bases of the repeating sequence d(TG)n are necessary and sufficient for stable NC binding. We first measured the binding of wild-type NC to the octanucleotide d(TG)4 by SPR, using a buffer containing 0.15 M NaCl. As in our previous studies (13), the oligonucleotide was immobilized on a BIAcore chip. Solutions of NC (at a series of different NC concentrations) were passed across the chip, and bound NC was then eluted with buffer. The instrument records the time-course of binding of NC to, and its subsequent dissociation from, the oligonucleotide on the chip. Our initial experiments were with a chip containing the oligonucleotide at an extremely low density: the average distance between the immobilized 3′ ends of the oligonucleotide molecules on this chip, as calculated from the concentration of oligonucleotide on the chip and the Poisson distribution, was 420 Å. Since NC contains only 55 amino acids, it would be only ∼200 Å long if it were fully extended. Thus, immobilization on the chip served to physically isolate oligonucleotide molecules from each other, so that NC bound to one (TG)4 molecule could not interact with other (TG)4 molecules. The resulting SPR profile is shown in Figure 2A, in which the horizontal line represents the response expected if each NC molecule bound to one (TG)4 molecule. It can be seen that a clear saturation was obtained at a level corresponding to one NC bound to each oligonucleotide molecule. This level shows that under these conditions, only a single NC molecule can bind to the octanucleotide (TG)4. The data were analyzed as described in Materials and Methods, and were consistent with a single binding system with a Kd of only 3.0 nM (Table 1). The grey lines in the Figure are the curves obtained from the fit to this model. Figure 2B shows that no binding to (A)8 can be detected under these conditions, as expected from our earlier analysis using SPR chips more densely loaded with oligonucleotide (13).
Figure 2.
SPR results: (A) Binding of NC to a 5.4 RU surface of (TG)4 in 150 mM NaCl. Solutions of 1.7, 3.13, 6.25, 12.5, 25, 50, 100 and 200 nM NC were passed over the surface at 100 µl/min. The horizontal line represents the response expected for a 1:1 complex of NC and (TG)4. The grey lines are the profiles expected from fitting the data to a simple 1:1 binding model as described in the Supplementary Data. (B) Binding of NC to (A)8. Solutions of NC as in (A) were passed over a 5.8 RU (A)8 surface. (C) Binding of NC to a 156.6 RU surface of (TG)4. Solutions of 1, 2.5, 5, 10, 25, 50, 100, 200, 400, 600, 800 and 1000 nM NC were passed over this surface. The two horizontal lines represent the signals expected if one (lower line) or two (upper line) NC molecules binds to each (TG)4 molecule. The grey lines are the profiles expected from fitting the data to the complex binding model shown in Figure 3, as described in the Supplementary Data. (D) Binding of NC to (TG)4 in buffer containing 250 mM rather than 150 mM NaCl. Solutions of 5, 10, 25, 50, 100, 200, 400, 800 and 1200 nM NC were passed over a surface containing 159.4 RU of (TG)4. The horizontal line represents the response expected for a 1:1 complex of NC and (TG)4. Grey lines are as in (C).
Table 1.
Binding of wild-type NC to (TG)4: summary of Kd determinations by different methods
| Method | [NaCl] (M) | Kd1a (nM)–NO | Kd2 (nM)–NON | Kd3 (nM)–ONO | ΔAICb |
|---|---|---|---|---|---|
| SPR, very low density | 0.15 | 2.95 ± 0.021 | – | – | – |
| SPR | 0.15 | 5.4 ± 0.08 | 25 ± 0.5 | 1600 ± 970 | + |
| TFQ | 0.15 | 4.70 ± 0.67 | 49.9 ± 10.23 | 239.9 ± 20.05 | + |
| FA | 0.15 | 2.7 ± 0.4 | 21 ± 6.6 | 5300 ± 10 700 | + |
| SPR | 0.25 | 200 ± 0.5 | 1300 ± 350 | – | + |
aKd values were determined by global analysis of the SPR, TFQ or FA data using the equations in Supplementary Data. The error terms were obtained using a linear approximation.
bΔAIC: Akaike's Information Criterion statistic (see Supplementary Data). ΔAIC > 0 implies that the complex model (see Figure 3) gives a better fit to the data than a 1:1 binding model.
We also repeated the SPR analysis using a chip displaying a higher density of (TG)4. As can be seen in Figure 2C, the results were quite different in this case. First, the binding did not reach saturation; rather, increasing the concentration of NC applied to the chip led to a rise in the amount of NC bound, exceeding that expected for equimolar binding to the (TG)4 (this level is indicated by the lower horizontal line in the Figure). Secondly, the kinetics of elution from the chip were sharply biphasic, with some NC washing off very rapidly and another portion eluting much more slowly: note that by 75–100 s, the binding curves in Figure 2A have fallen to zero, while in Figure 2C some NC remains bound to the oligonucleotide at 150–200 s, even in the most dilute NC solutions. The results suggest that at high (TG)4 densities, NC is ‘trapped’ on the chip. These data are in complete accord with our previous findings (13).
Taken together, the SPR results indicate that the interaction between NC and (TG)4 is potentially quite complex. The binding of NC at a level greater than equimolar (Figure 2C) implies that under these conditions, additional NC can bind to an NC:(TG)4 complex. In addition, the fact that the SPR profile is a function of the (TG)4 density on the chip (compare Figure 2C with A) suggests that when NC is bound to oligonucleotide, it can still interact with a second oligonucleotide molecule, provided that the latter is in close enough proximity. [Indeed, we previously presented SPR data showing that the stable binding of NC to the tetranucleotide (TG)2 is dependent upon the spacing of (TG)2 molecules on the chip (13)]. In other words, both NC and (TG)4 appear to be bivalent in NC:(TG)4 interactions.
These properties obviously have significant implications for the quantitative analysis of these interactions. For example, when (TG)4 is titrated into an NC solution, the latter will initially be in vast excess. While the first binding event will lead to a 1:1 complex (which we will refer to as ‘NO’, where ‘N’ and ‘O’ represent NC and oligonucleotide, respectively), the NO complexes are evidently capable of interacting with additional NC, generating ‘NON’ complexes (as indicated in the upper pathway in Figure 3). Conversely, when NC is added stepwise to a (TG)4 solution, the initial NO complexes will likely interact with a second (TG)4 molecule to form ‘ONO’ complexes (lower pathway in Figure 3). Moreover, if, as suggested by these considerations, both reactants are bivalent, then higher-order complexes can also be formed: e.g. if NC has two binding sites for oligonucleotides, then the two NC molecules in an NON complex will each have an unfilled site. Thus an NON complex can potentially bind additional (TG)4 molecules, giving rise to ONON or ONONO. In turn, since (TG)4 can evidently bind two NC molecules, ONON has the potential to form NONON, etc. The most probable result would appear to be a heterogeneous mixture of complexes between the protein and the oligonucleotide.
As noted above, the results in Figure 2C are clearly incompatible with a simple model in which interactions are limited to NO formation as in the first pathway in Figure 3. We analyzed the data using the full scheme shown in Figure 3. Assuming the occurrence of the additional binding reactions, leading to NON and ONO complexes in addition to NO, gave an excellent fit to the data, as shown by the red lines in the Figure. The Kd's calculated for the three reactions are 5.4 nM for NO, 1.6 µM for formation of ONO from NO, and 25 nM for formation of NON from NO, as shown in Table 1. The utility of the model shown in Figure 3 for fitting the data is indicated in Table 1 by the positive value for the Akaike's Information Criterion (ΔAIC) statistic. This parameter includes a penalty for introduction of additional variables, and a positive ΔAIC thus implies that each variable makes a valuable contribution to the fit of the data (see Supplementary Data).
It was of interest to probe the contribution of electrostatic forces to the interaction between NC and (TG)4. Therefore, we also analyzed the binding of NC to the oligonucleotide in 250 (rather than 150) mM NaCl, again using an oligonucleotide density similar to that in Figure 2C. As seen in Figure 2D, no saturation was reached, even at 1 µM NC. However, the amount of NC bound to the chip was significantly lower than at 150 mM NaCl (Figure 2C), and the increase in binding above the equimolar level was relatively modest. These data imply that the formation of NON and ONO complexes detected at 150 mM NaCl (Figure 2C) was greatly reduced at 250 mM NaCl. We attempted to fit the results in Figure 2D with a simple 1:1 binding model, and found that the results were satisfactory at NC concentrations up to 100 nM (the 5th curve). However, the level of binding with more concentrated NC solutions was not consistent with this model. Assuming that NON and ONO complexes could also form under these conditions greatly improved the fit of the data at these concentrations, as indicated by red lines in Figure 2D and by the ΔAIC statistic (Table 1). The Kd's for the three reactions, leading to NO, NON and ONO, respectively, are shown in Table 1. The drastic reduction in affinities accompanying an increase in the ionic strength of the solution indicates that there is a major electrostatic component in the interaction of NC with (TG)4.
TFQ
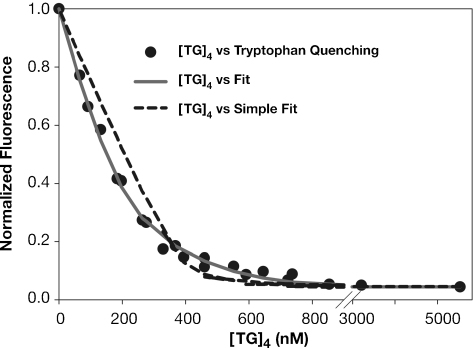
NC contains a single tryptophan residue. The fluorescence of the tryptophan is quenched when NC binds nucleic acids, and many analyses of NC–nucleic acid interaction have used this response as a reporter of the binding. Figure 4 shows the fluorescence at 350 nm as (TG)4 was titrated into a 0.4 µM NC solution in the standard buffer containing 150 mM NaCl. It can be seen that the fluorescence decreased nearly to zero over the course of the titration. The dashed line in Figure 4 shows an attempt to fit these data using the assumption that a single NC molecule interacts with a single (TG)4 molecule; this curve would extrapolate to zero fluorescence at 400 nM (TG)4, when one mole of (TG)4 had been added per mole of NC in the solution. It is evident that the curve (representing the fit) exhibits systematic deviations from the data; thus, the results are not precisely compatible with this model.
Figure 4.
TFQ results. (TG)4 was titrated into a 400 nM solution of NC and tryptophan fluorescence was monitored as described in Materials and Methods. The fluorescence readings were divided by the fluorescence of NC before addition of (TG)4 to obtain the normalized fluorescence shown here. Attempts were made to fit the data assuming a 1:1 binding system (dashed line) (giving a Kd of 3.4 ± 0.07 nM) or the more complex model shown in Figure 3 (solid grey line). The results shown here are normalized by dividing by the fluorescence value of NC alone. As indicated in Supplementary equation 5, the final fluorescence obtained after several equivalents of (TG)4 had been added [i.e. the residual fluorescence of NC bound to (TG)4] was subtracted from these values before attempting to fit the curves.
We attempted to fit the results to the scheme shown inFigure 3, in which NO can in turn participate in NON or ONO formation (and assuming that all NC molecules in NO, NON and ONO complexes completely lose fluorescence). As shown in the solid line, this model gave an excellent fit with the data. In particular, it is notable that at the lower (TG)4 concentrations, where NC is in excess, the fluorescence falls more steeply than in the dashed line representing the 1:1 binding model. This is presumably because under these conditions, a single (TG)4 molecule can quench the fluorescence of both of the NC molecules in a NON complex. In contrast, at higher (TG)4 concentrations (i.e. ∼450–650 nM), the fluorescence is quenched slightly more slowly than predicted by the 1:1 binding model; we would suggest that this is due to the binding of some (TG)4 molecules to NO complexes, forming ONO and having no effect on NC fluorescence. The three Kd's calculated from this fit are shown in Table 1, and are very similar to those obtained by SPR. Again, the superiority of the fit using this model is indicated by the positive ΔAIC value. Results of fits using only Kd1 and Kd2 or only Kd1 and Kd3 are shown in Supplementary Figure S2.
FA
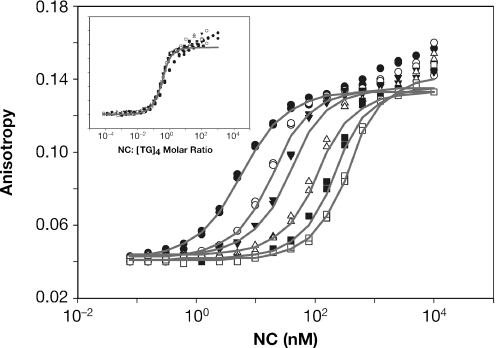
We also analyzed interactions between NC and (TG)4 by FA. NC was titrated into solutions of fluoresceinated (TG)4 and the anisotropy of the fluorescein reporter was monitored. Results of this experiment, for a series of different (TG)4 concentrations between 10 nM and 1 µM, are shown in Figure 5. It can be seen that addition of NC increased the anisotropy of the fluoresceinated oligonucleotide in a dose-dependent manner. As expected, each (TG)4 concentration gave a different titration curve; i.e. the concentration of NC required to shift the fluorescein anisotropy is dependent on the (TG)4 concentration. It is striking to note that there was no strict plateau in the anisotropy values; rather, the anisotropy continued to rise slowly, even at the highest NC concentrations. This observation supports the idea that complexes between NC and (TG)4 are still capable of interacting with additional NC to form NON-type complexes.
Figure 5.
FA results. NC was titrated into solutions containing 10, 50, 100, 300, 600 or 1000 nM fluoresceinated (TG)4 and the anisotropy of the oligonucleotide was measured. Grey lines are fits to the data using the model shown in Figure 3. FA of two aliquots was measured independently for each point in the titrations. Inset: the FA results in the figure were plotted vs. the NC:(TG)4 ratio in the solution.
The data shown in Figure 5 was analyzed in several ways. The inset shows the same titration data, but here each FA value is plotted against the molar ratio of NC to (TG)4 present in the solution. It is evident that when the results are graphed in this way, all of the titrations except that with 10 nM (TG)4 fall on the same line. (Presumably the curve obtained with 10 nM (TG)4 diverges from the others because this concentration of oligonucleotide is near the Kd1 of the reaction, so that the two reactants do not associate with each other completely until excess NC has been added.) The curve representing the higher NC concentrations was analyzed to determine the ‘valency’ of NC, i.e. the maximum number of (TG)4 that can be bound by an NC molecule, as described in Supplementary Data. In three separate experiments, this value was found to be 2.17 ± 0.25. The fact that the titration does not saturate until ∼2 equivalents of NC have been added shows clearly that the two reactants can interact further after formation of the expected 1:1 complex.
We also fit the data in Figure 5 using the complex model shown in Figure 3. As indicated by the lines in Figure 5, this treatment gave good fits to the data, yielding the three Kd values shown in Table 1. The values for the 3 Kd's are similar to those obtained by SPR and TFQ, and the superiority of the fit obtained from the model in Figure 3 is indicated by its positive ΔAIC value. (It will be evident that the fit is imperfect at very high molar ratios; this is presumably due to the formation of higher-order complexes such as NONON, which are not used in fitting the data.)
Dissection of the interaction of NC with (TG)4
NC is a highly basic protein, with a particularly basic N-terminal region; it also contains two zinc fingers (Figure 1). The zinc fingers are known to participate in binding to nucleic acids in vitro and are critical in some tests of nucleic acid chaperone activity in vitro (15,17–20). They are also essential for selective packaging of genomic RNA during virus assembly by the Gag polyprotein in vivo (2).
In order to explore the role of the basic N-terminal residues in binding of NC to (TG)4, we analyzed binding by the ‘N-term’ mutant, in which the five lysine and arginine residues within the first 14 residues had been replaced with alanines (Figure 1), by FA. We found (data not shown) that the anisotropy induced by the mutant NC rises to a clear plateau, so that there is no increase as the protein concentration is raised from 2 to 10 µM (a 1000-fold excess of protein over (TG)4). In contrast, the curve obtained with wt NC continues to rise slowly over this range, indicating the formation of NON-type complexes. The results with the mutant are consistent with a simple 1:1 binding system with a Kd of 62 nM (see Table 2). Thus, the basic residues in the N-terminus are critical both for the high-affinity of wt NC for (TG)4 and for its ability to interact with NO complexes.
Table 2.
Summary of Kd values for mutant NC proteins
| Protein | Method | Kd1a (nM)–NO | Kd2 (nM)–NON | Kd3 (nM)–ONO | ΔAICb |
|---|---|---|---|---|---|
| N-term | FA | 62 ± 8.8 | – | – | − |
| SSHS | FA | 30 500 ± 14 600 | – | – | – |
| F16A | SPR | 119 ± 0.5 | 14 000 ± 3000 | 987 ± 11 | + |
| 1:1 mutant | SPR | 10 ± 0.8 | – | – | − |
| 2:2 mutant | SPR | 48 ± 0.5 | – | 718 ± 40 | + |
| 2:1 mutant | SPR | 34 ± 0.6 | – | 2350 ± 72 | + |
aKd values in 0.15 M NaCl were determined by global analysis of the SPR or FA data using the equations noted in Supplementary Data.
bΔAIC: Akaike's Information Criterion statistic (see Supplementary Data).
We also tested the contribution of the zinc fingers to the interaction of NC with (TG)4. Again using FA, we analyzed binding of the ‘SSHS’ NC protein, in which the six zinc-coordinating cysteines were all replaced with serine. We found that the affinity of NC for the oligonucleotide was drastically reduced under these conditions. Only a single binding system with a Kd of 30.5 µM was detectable in this experiment (Table 2). Similar results were obtained with wt NC when the titration was performed in the presence of 5 mM EDTA (data not shown). These data imply that the zinc fingers, as well as the N-terminal basic residues, are crucial for the high-affinity interaction of NC with d(TG)4 and for its ability to form ONO- or NON-type complexes.
As an extension of our exploration of the role of the zinc fingers in the interaction with (TG)4, we also analyzed ‘finger-swap’ NC proteins, in which the sequence of one or both of the zinc fingers was replaced with that of the other (see Figure 1). Using SPR, we found that the ‘1:1’ mutant protein formed NO complexes with nearly the same Kd1 as wt NC, while the ‘2:2’ and ‘2:1’ proteins had ∼10-fold reduced affinity (Tables 1 and 2). Interestingly, the data indicated that only the latter two proteins retained the ability to form ONO complexes, and all three mutants were defective with respect to NON formation (see Table 2). We also investigated the binding of the F16A mutant of NC, in which an alanine residue is substituted for the phenylalanine within the N-terminal finger [known to engage in hydrophobic interactions with nucleic acid bases (21)]. In this case, the affinity of the protein for (TG)4 was reduced to ∼20–40-fold relative to wt NC, but it could still form both NON and ONO complexes at detectable levels.
ITC analysis
We also characterized the interactions between NC and (TG)4 by ITC. The results of this analysis are shown in Figure S3 and are discussed in Supplementary Data. Briefly, ∼18 kcal/mol was released when the oligonucleotide was titrated into a solution of 18 µM NC. The titration curve centered around a molar ratio of 0.5 and reached saturation at a molar ratio of ∼1.0, in agreement with the idea that a single oligonucleotide molecule can bind two NC molecules in a NON-type complex (22). We also performed a ‘reverse’ titration in which NC was titrated into a solution of 10 µM (TG)4. Approximately 12 kcal/mol was released over the course of the titration, but no clear saturation was observed. Thus the ITC data did not point to a precise stoichiometry for the interactions between NC and (TG)4, as might be expected from their apparent ability to form NO, ONO, NON and other, higher-order complexes (see above). The dependence of the total heat released upon the direction of the titration also underscores the complexity of the interactions between NC and (TG)4. In addition, we probed the role of electrostatic interactions in the binding, both by repeating the titration of NC into (TG)4 in 250 mM, rather than 150 mM, NaCl and by using the ‘N-term’ mutant protein, in which the five lysine and arginine residues between positions 3 and 14 had been replaced with alanine. The amount of heat released was drastically reduced in both of these experiments, suggesting that that coulombic attractions play a major role in the interactions between NC and (TG)4.
Competition experiments using SPR
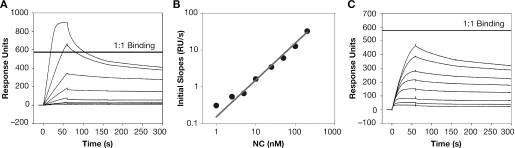
Although SPR technology is capable of measuring the binding of NC to high-affinity nucleic acid sequences such as the repeating oligonucleotide, (TG)n [see reference (13)], no binding to (A)n is detected under the same conditions, i.e. 150 mM NaCl (pH 7.5) (Figure 2B). Therefore, we devised an indirect SPR approach to detect the interaction of low-affinity ligands such as (A)n with NC. These experiments used an SPR chip coated with (TG)4 at a density of 344.2 response units (RU), similar to the conditions used in Figure 2C. In preliminary tests, a series of solutions containing different concentrations of NC was passed over the chip, and the initial binding rate of NC to the chip was measured. As expected, the initial binding rate under these conditions was directly proportional to the NC concentration (Figure 6A, B). This result shows that the initial binding rate can be used to measure the concentration of free NC in solution.
Figure 6.
Analysis of competition between oligonucleotides in solution and immobilized (TG)4 for NC. (A) Binding of NC to (TG)4. One min injections of (from bottom to top) 0, 1, 2.5, 5, 10, 25, 50, 100 and 200 nM NC were passed over a surface of 344.2 RU's (TG)4 at 64 µl/min. (B) Rate of binding of NC to (TG)4 as a function of NC concentration. Initial slopes in (A) were measured using BIAevaluation 3.01, and the plot of initial slopes versus NC was fit using linear regression. (C) Effect of (TG)4 in solution on binding of NC to immobilized (TG)4. A total of 100 nM NC was incubated at 4°C with (from top to bottom) 0, 0.1, 0.25, 0.5, 1.0, 2.5, 5.0, 10.0, 25.0, 50.0 and 100.0 µM (TG)4 for at least 1 h before injecting a sample for 1 min at 64 µl/min.
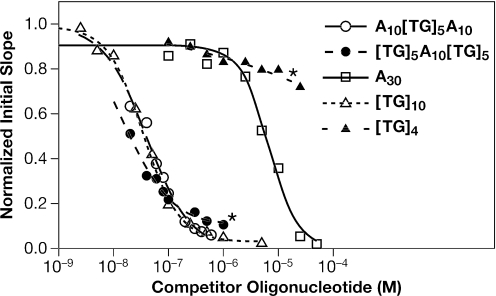
To exploit the ‘competition’ technique to detect binding of NC to (A)30, we mixed NC at 100 nM with a series of concentrations of (A)30. In all cases, the oligonucleotide was in excess over NC in the solution. The mixtures were incubated for 1 h and then passed over the (TG)4 chip surface. As expected, the initial binding rate of NC to immobilized (TG)4 decreased as the concentration of (A)30 increased (data not shown). This decrease reflects the removal of free NC from solution as it is bound by (A)30. The relationship between (A)30 concentration and the amount of NC removed provides a way of measuring the binding of NC to (A)30. As shown in Figure 7 (open squares), addition of sufficient (A)30 completely eliminated binding of NC to (TG)4, as expected. The results of these experiments were analyzed using the 4-parameter logistic equation as described in Supplementary Data, yielding an IC50 value (the concentration of oligonucleotide in solution required to depress the initial rate of binding to (TG)4 halfway to its final value) of 6.7 µM (see Table 3). The fact that the binding can be completely suppressed by addition of sufficient (A)30 is indicated in Table 3 by the value of zero for the ‘minimum initial slope’ (ISmin) (the normalized initial rate of binding to the immobilized (TG)4 that would still occur in an infinite concentration of oligonucleotide in solution).
Figure 7.
Competition by oligonucleotides in solution with the binding of NC to immobilized (TG)4. Solutions of 100 nM NC were incubated with varying amounts of different oligonucleotides for at least 60 min. These solutions were then passed over an SPR surface containing 327 RU's of (TG)4 and the initial slope of the SPR profile, representing the initial rate of binding to the immobilized (TG)4, was measured. The open squares represent preincubation with 0.1, 0.25, 0.5, 1.0, 2.5, 5, 10, 25 and 50 µM (A)30; open triangles: preincubation with 2.5, 5.0, 10, 25, 50, 100, 250, 500, 1000 and 5000 nM (TG)10; open circles: preincubation with 20, 40, 60, 80, 100, 300, 500 and 1000 nM (A)10(TG)5(A)10; closed circles: preincubation with 20, 40, 60, 80, 100, 300, 500 and 1000 nM (TG)5(A)10(TG)5; closed triangles: preincubation with 0.1, 0.25, 0.5, 1.0, 2.5, 5, 10 and 25 µM (TG)4. The lines are fits to a 4-parameter logistic equation, as described in Supplementary Data, yielding the values shown in Table 3. Asterisks highlight the two curves in which ISmin is greater than zero.
Table 3.
ISmin and IC50 values for competition by oligonucleotides
| Protein | Oligonucleotide | ISmina | IC50 (nM)a |
|---|---|---|---|
| wt NC | A10(TG)5A10 | 0.00 | 39.7 ± 3.67 |
| wt NC | (TG)5A10(TG)5 | 0.11 ± 0.023 | 15.96 ± 1.68 |
| wt NC | (A)30 | 0.00 | 6676 ± 785 |
| wt NC | (TG)10 | 0.00 | 36.96 ± 2.73 |
| N-term | (TG)4 | 0.00 | 602.6 ± 71.7 |
| EDTA–NC | (TG)4 | – | – |
| wt NC | (TG)4 | 0.77 ± 0.023 | 263.6 ± 100.8 |
aIC50 and ISmin values were determined from the SPR competition data using a 4-parameter logistic equation as described in Supplementary Data.
This competition protocol can be used to analyze the binding to high-affinity, as well as low-affinity substrates. We performed the same experiment using (TG)10 in place of (A)30. As shown in Figure 7 (open triangles), the IC50 for (TG)10 was ∼37 nM, and the ISmin in this interaction was zero (see Table 3).
Non-ideal behavior in competition experiments
We also tested the ability of (TG)4 in solution to compete with immobilized (TG)4 on the SPR chip surface. Since these oligonucleotides are identical, we expected the (TG)4 in solution to compete for NC with an IC50 in the low nanomolar range. However, the results were quite different from our predictions and from the results obtained with (A)30 or (TG)10. As shown in Figure 6C, the final height of the curves, i.e. the final amount of NC bound to immobilized (TG)4 at steady-state, was, as expected, quite sensitive to the concentration of (TG)4 in the solution. Thus, in all cases the (TG)4 in solution was able to depress the binding of NC to the immobilized (TG)4, and at high (TG)4 concentrations the SPR signal was reduced nearly to zero. However, the initial slopes (i.e. the initial rates of binding of NC to the chip) obtained with solutions of different (TG)4 concentrations were all very similar to each other, even when the (TG)4 in solution was in 1000-fold excess over NC (see Figure 6C). This observation implies that the mechanism of the competitive inhibition of NC binding is not that expected from a classical 1:1 binding event.
The results of a similar experiment are summarized by the closed triangles in Figure 7. It can be seen that the initial slope was reduced only ∼20% by preincubation with 10−5 M (TG)4 (a 100-fold excess of (TG)4 over NC), and responded only slightly to an additional 2.5-fold increase in the concentration of (TG)4 in the solution. These data are readily explained by the hypothesis that complexes formed in solution between NC and (TG)4 retain the ability to interact with (TG)4 on the SPR chip, forming ONO-type complexes, although the steady-state level of these complexes is low in the presence of excess (TG)4 in solution. The results are summarized in Table 3, showing that the ISmin is 77%. In other words, the initial rate of binding of NC to immobilized (TG)4 would only be reduced 23% by an infinite concentration of (TG)4 in solution. These results underscore the remarkable ability of NC to enter into higher-order complexes with nucleic acids containing the sequence (TG)n.
It was of interest to assess the contributions of the NC zinc fingers and basic residues to the interactions with (TG)4 observed here. We therefore tested the N-term ala mutant in an experiment analogous to that shown by the closed triangles in Figure 7. We found (Table 3) that the ISmin was zero for the mutant, with an IC50 of ∼600 nM. Thus, the basic residues in the N-terminal region of NC are required for the ONO-type complex formation detected in these experiments. NC from which the Zn+2 had been removed with 5 mM EDTA showed no detectable binding to (TG)4 in an SPR experiment.
Binding of NC to chimeric oligonucleotides containing both low-affinity and high-affinity sequences
As another approach to understanding the structure and mode of formation of ONO complexes, we also analyzed the binding of NC to a pair of oligonucleotides which contained both low-affinity [i.e. (A)n] and high-affinity [(TG)n] sequences. In one of these oligonucleotides, the (TG)n sequences were at the ends and the (A)n sequence was internal; in the other, terminal (A)n sequences surrounded a (TG)n stretch.
NC was mixed with different concentrations of these oligonucleotides, and the mixtures were then analyzed in a competition experiment by passing them over an SPR chip containing (TG)4, as in Figure 6. We found (Figure 7, open circles) that the 30-base oligonucleotide with terminal low-affinity sequences, (A)10(TG)5(A)10, like (A)30 (open squares) or (TG)10 (open triangles), was able to completely prevent the binding of NC to (TG)4 on the SPR surface (i.e. the ISmin was 0, as indicated in Table 3). The IC50 with which this oligonucleotide inhibited binding was 40 nM.
The addition of the other chimeric oligonucleotide to the NC solution produced a somewhat different result. As shown in Figure 7 (closed circles) and Table 3, (TG)5(A)10(TG)5 inhibited the binding of NC to immobilized (TG)4 with an IC50 of 16 nM. However, this oligonucleotide could not completely prevent the binding of NC to immobilized (TG)4: the initial rate of binding to the chip was only reduced to ∼87% even when this oligonucleotide was at concentrations as high as 1 µM. The non-zero ISmin, 0.11, obtained for this oligonucleotide (Table 3) implies that when NC is bound to it in solution, the protein is still capable of interacting with (TG)4 on the SPR surface to a significant degree. In other words, when (TG)n is at the ends of an oligonucleotide, the oligonucleotide can enter into an ONO complex under our experimental conditions, but if the oligonucleotide has internal (TG)n and terminal (A)n sequences, it cannot.
Direct detection of NON complexes
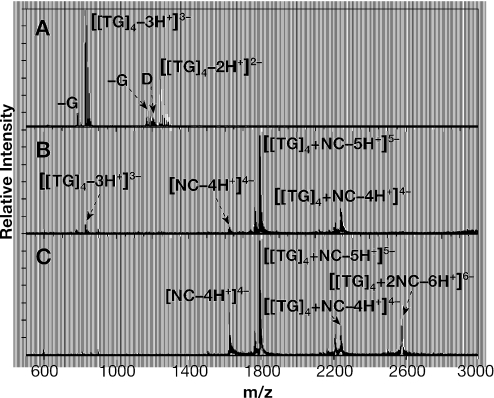
Finally, we characterized the composition of NC:oligonucleotide mixtures by electrospray ionization Fourier transform mass spectrometry (23). This ionization technique has proven capable of preserving relatively weak non-covalent interactions, including those involved in protein–nucleic acid complexes (24). More specifically, we recently employed ESI-FTMS to determine the stoichiometry and binding affinity of the complexes of NC with RNA stem–loops of the HIV-1 packaging signal (16). Under similar conditions, a 5 µM solution of (TG)4 provided intense signals with mass over charge ratio (m/z) of 822.48 and 1234.22, corresponding to the −3 and −2 charge states, respectively (see Figure 8A). Small amounts of (TG)4 in which a guanine-base has undergone hydrolysis [(TG)4-G] were also detected together with dimeric forms of intact and hydrolyzed (TG)4, which could be explained by possible ‘wobble’ base-pairing between G and T.No other nucleic acid species were detected in this analysis.
Figure 8.
ESI-FTMS spectra of (TG)4 and (TG)4-NC complexes. (A) Control 5 µM (TG)4, providing a 2470.46 Da experimental mass (2470.44 Da monoisotopic mass calculated from sequence, for a ∼8 p.p.m. mass accuracy). The signal labeled −G corresponds to a product in which a deoxyguanine is formally replaced by deoxyribose (observed decremental mass of ∼133 Da). D indicates (TG)4/(TG)4-G dimeric species. (B) Mixture containing 5 µM each of (TG)4 and NC. NC provided a mass of 6488.89 Da (6488.91 Da monoisotopic from sequence), while the 1:1 NO complex was 8959.33 Da (8959.35 Da monoisotopic from sequence). (C) Sample containing 5 µM (TG)4 and 15 µM NC. The 1:2 NON complex provided a mass of 15 447.93 Da (15 448.25 Da monoisotopic from sequence).
The addition of an equimolar amount of NC produced a mixture in which a 1:1 complex is the predominant species (Figure 8B). Consistent with an equilibrium situation, very small amounts of free NC and free (TG)4 were still detectable in the solution. Most interestingly, when the ratio of NC to (TG)4 was increased to a final 3:1, an additional species was observed in the corresponding ESI-FTMS spectrum (Figure 8C) consistent with a complex of one (TG)4 molecule with two NC molecules (referred to as NON above). The fact that free NC is still present in the solution suggests that the affinity of NC for an NC:(TG)4 complex (i.e. Kd2) is considerably weaker than its affinity for naked (TG)4. In other words, while NON could be detected here, there is no evidence for cooperativity in the binding of NC to the oligonucleotide. These results are in excellent agreement with those reported in Table 1. It is interesting to note that no signals could be found for dimeric (TG)4 species upon addition of NC (Figure 8B and C), suggesting that the affinity of (TG)4 for NC is higher than its affinity for a second (TG)4 molecule. Further addition of NC to achieve a 5-fold excess over (TG)4 did not result in the formation of detectable levels of higher-order stoichiometries (data not shown), thus indicating that NON is the largest complex observed under these experimental conditions.
When 5 µM (TG)4 was mixed with ∼2 µM NC, we only observed the 1:1 NO complex in addition to free (TG)4. This would suggest that the tendency to form ONO complexes under conditions of (TG)4 excess is relatively weak, again in good agreement with the Kd3 results shown in Table 1. We also detected virtually no binding of NC to (A)8 in these experiments, as expected (13). In analogy with the SPR competition experiments presented in Figures 6 and 7 and Table 3, ∼6 µM NC was added to a solution containing 5 µM each of (TG)4 and (A)8. The resulting solution was found to contain both NO and NON complexes with (TG)4, but no evidence for binding to (A)8 was obtained (data not shown). Thus, the affinity of NC for a NC:(TG)4 complex (again, Kd2), while weaker than Kd1, is still stronger than its affinity for (A)8.
DISCUSSION
In the present work, we have used a variety of techniques, including SPR, FA, ITC, TFQ and ESI-FTMS to detect and characterize the binding of NC to the octanucleotide (TG)4. These results led to the model presented in Figure 3, in which a 1:1 complex between NC and (TG)4 can be augmented either by an additional NC molecule (leading to an ‘NON’ complex) or an additional (TG)4 molecule (forming ‘ONO’). Direct evidence was obtained for the formation of these higher-order complexes, since (i) the results with SPR were a function of the density of (TG)4 on the SPR chip (Figure 2); (ii) in both SPR and FA, we did not observe a complete saturation when excess NC was added to (TG)4 (Figures 2C and 5); (iii) addition of some oligonucleotides to NC in solution caused only a partial reduction in the rate of binding of NC to (TG)4 on an SPR chip (Figures 6C and 7, Table 3); and (iv) NON complexes were directly observed by ESI-FTMS (Figure 8C). In fact, the only experimental situation in which the results were compatible with simple 1:1 binding of NC to (TG)4 was on SPR chips containing a very low density of (TG)4; this configuration evidently precluded interaction of the NC: (TG)4 complex with additional (TG)4 molecules. The model in Figure 3 was applied in the quantitative analysis of the results obtained by the other techniques used in the present work.
The binding results using all of the techniques are summarized in Table 1. It can be seen that the conclusions derived from SPR, FA and TFQ measurements were generally in very good agreement with each other. We conclude that under our standard conditions [0.15 M NaCl (pH 7.5)], NC binds with remarkably high-affinity (Kd of ∼5 nM) to the simple repeating sequence, (TG)n. It can then proceed to form either NON or ONO; the ability to form both types of ternary complex also raises the possibility of further permutations, e.g. NONO or ONON, etc. The data also indicate that the high affinity of NC for (TG)4 depends upon both the positively charged groups in the protein, which presumably engage in electrostatic interactions with the nucleic acid, and the zinc fingers, which are known to be important in binding of NC to other nucleic acids (21,25). Evidence for the electrostatic contribution included the reduction in affinity observed when some of the positive charges were removed from the protein (Table 2) or when additional counterions were added (Figure 2D), and the reduced enthalpy of binding in the presence of added counterions (Supplementary Figure S3).
One important question which is left unanswered by our results is why proximity of (TG)4 molecules, as in Figure 2C, is required for formation of NON as well as ONO complexes. It might be proposed that a high (TG)4 concentration leads to the formation of (TG)4 dimers or oligomers, and that these preformed oligomers then participate in NON formation. Indeed, (TG)4 dimers were detected by ESI-MS (Figure 8A). However, addition of one or more equivalents of NC reduced their concentration to below detectable levels (Figure 8B and C), suggesting that they are dissociated by interaction with NC. This result appears to argue against a role for these dimers in the formation of ternary NC–(TG)4 complexes. Furthermore, if dimer formation were a major contributor to Kd3, then the affinity would be expected to increase with ionic strength. However, raising the salt concentration was found to decrease the affinities for all three binding reactions (Table 1, Figure 2C), thus casting further doubt on the possible contribution of preformed dimers.
We showed previously that a 5-base oligonucleotide is long enough to support binding of NC (13). Thus, the octanucleotide (TG)4 potentially contains as many as four overlapping 5-base binding sites. However, SPR results with the two pentanucleotides, (TGTGT) and (GTGTG), displayed on the chip at densities similar to that used in Figure 2C, were very similar to those shown in Figure 2C (13). Thus the ‘redundancy’ of (TG)4 is not responsible for the high-affinity or complexity of its interactions with NC. It should be noted that an earlier study (26) measured the affinity of NC for (TG)4 at low ionic strength and the dependence of the affinity upon the Na+ concentration. Extrapolation of these values to the ionic strength used here gives a significantly lower affinity for the interaction than we observed for Kd1. We are not certain why this extrapolation method gave a different Kd from the mutually consistent results obtained by three independent methods, i.e. SPR, FA and direct TFQ measurements, in the present work.
The ability of NC to form ONO-type complexes indicates that it contains several domains or sites that bind nucleic acids. We cannot say precisely how many such domains are present in NC, since we do not know whether the two zinc fingers can function independently, nor whether basic regions other than the N-terminus can bind nucleic acids. As an approach to this question, we analyzed ‘finger-swap’ mutant NC proteins, in which one or both of the zinc fingers was placed in the position of the other. We found that the 2:1 and 2:2 proteins had significantly lower affinity for (TG)4 than the wild-type protein, although this affinity was far higher than when the ability to coordinate zinc was completely eliminated (Table 2). Results with F16A NC, lacking an aromatic residue in the first finger that is known to participate in tight binding to RNA stem–loops (21), were somewhat similar to those with 2:1 and 2:2. The reduced binding of the finger-swap mutants might indicate that the fingers normally function in concert with each other. An alternative explanation, however, is that a single zinc finger can function as an autonomous nucleic acid-binding domain, but its functional boundaries extend beyond the N- and C-terminal cysteines of the finger. The ratio 1:1 NC, in which only the C-terminal finger has been changed, formed NO complexes with nearly the same affinity as wt NC. It is interesting to note that HIV-1 containing 1:1 NC is more replication-competent than those with the other finger-swaps (27).
We had previously reported that the tetranucleotide, (TG)2, cannot support the binding of NC in SPR experiments unless the (TG)2 molecules are in close proximity to each other (13). These experiments suggested that NC could form a relatively stable complex with two tetranucleotide molecules, resembling the ONO-type complexes detected in the present work. We do not know how these complexes are formed, but it is significant that NC bound to a chimeric oligonucleotide, consisting of internal (TG)n sequences flanked by low-affinity (A)n sequences, did not engage in detectable ONO-type complex formation (Figure 7, Table 3). This observation raises the possibility that NC must be at the end of one nucleic acid molecule in order to interact with a second. [This may also be the reason that competition by (TG)10 gave results in accord with a simple binding model (Figure 7, Table 3): as in the (A)10(TG)5(A)10 oligonucleotide, the internal (TG) sequences provide binding sites for NC which are distant from the ends of the oligonucleotide.]
The possible requirement that NC be near the end of one oligonucleotide in order to interact with a second suggests that it may slide between them in an ONO-type complex. In many ways, this behavior of NC on single-stranded oligonucleotides is analogous to that of other proteins on double-stranded DNA. For example, the ability of lac repressor to rapidly ‘find’ a high-affinity base sequence in a large double-stranded DNA molecule appears to result from its ability to slide along DNA (28–33). In fact, Winter et al. (30) point out that the ability of lac repressor to bind DNA both specifically through sequence-dependent interactions and nonspecifically, i.e. electrostatically, is essential for its facility in finding the operator by a sliding mechanism. It seems possible that the zinc fingers and basic regions of NC play a similar respective roles in the interactions of NC with single-stranded nucleic acid.
NC has been reported to induce aggregation of single-stranded nucleic acids (34–36). These aggregates presumably represent higher-order complexes, which are expected to form if both NC and the nucleic acid are at high concentration and each is able to bind more than one molecule of the other.
What is the biological significance of these remarkable properties of NC protein? The functional importance of the high-affinity of NC for the repeating sequence, (TG)n, or its RNA counterpart, (UG)n, (13) is not clear, but it is noteworthy that alternating (UG)n sequences are found at several sites in HIV-1 genomic RNA. One (UG)-rich region is nucleotides 556–577 in the U5 region of the NL4-3 isolate of HIV-1. The function of this highly conserved sequence is not known, but it must be crucial for the virus, since subtle changes lead to a profound diminution in replicative capacity (37). These sequences appear to be significant in dimerization of genomic RNA (38), and may also contribute to efficient polyadenylation at the 3′ end of viral RNA (39). Similar sequences are also found in stem–loop 3, a region in the leader that appears to be important for encapsidation of the genome.
As noted above, NC and the NC domain of Gag polyprotein participate in an amazing variety of protein–nucleic acid interactions during the viral life cycle. Many, although not all, of the functions of NC involve its nucleic acid chaperone activity. The molecular mechanisms underlying this activity are not fully understood, but recent work lends strong support to the hypothesis that a major component of chaperone activity is the ability of NC to induce short-range attraction between nucleic acid molecules (10,20,40). Hargittai et al. (20) point out that this property is to be expected from the polycationic nature of NC. The results presented here provide the first quantitation of this activity, which we termed ‘Kd3’, the affinity of a complex between NC and one nucleic acid molecule for a second nucleic acid molecule. It is also notable that this affinity was significantly weaker than Kd1; thus NC does not bind to the short oligonucleotide studied here with positive cooperativity, in contrast to the suggestion that NC:NC interactions contribute to its biological activities (41). However, we cannot exclude the possibility that NC binds cooperatively to longer nucleic acid molecules.
In conclusion, we have used SPR, TFQ, FA, ITC and ESI-FTMS to characterize the binding of HIV-1 NC protein to the simple oligodeoxynucleotide, (TG)4. Our data show that this high-affinity interaction results from the combined action of multiple nucleic acid-binding domains in NC. The protein is capable of binding simultaneously to more than one nucleic acid molecule, and can also bind to complexes between an oligonucleotide and another NC molecule. It seems likely that these attributes contribute to the complex functional properties of NC and of the NC domain of the Gag protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are very grateful to Irina Orlov and Evan Behre at BIAcore Inc. for their work on the BIAcore S51. We also thank Dr Linda Kenney for helpful suggestions on fluorescence anisotropy and Drs. Ioulia Rouzina and Yun-Xing Wang for careful readings of the manuscript. In addition, we thank Leslie Hickman for her helpful analyses of SPR signals and Dominic Scudiero for permission to use the Tecan Ultra fluorescence anisotropy plate reader. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and was funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contracts NO1-CO12400 and NO1-CO12401. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organization imply endorsement by the U.S. Government. The participation of D.F. and N.A.H. was supported by the National Institutes of Health (R01-GM643208 and 1T32-GM066706). Funding to pay the Open Access publication charges for this article was provided by NO1-CO12401.
Conflict of interest statement. None declared.
REFERENCES
- 1.Swanstrom R., Wills J.W. Synthesis, assembly, and processing of viral proteins. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 2.Berkowitz R., Fisher J., Goff S.P. RNA packaging. Curr. Top Microbiol. Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Muriaux D., Mirro J., Harvin D., Rein A. RNA is a structural element in retrovirus particles. Proc. Natl Acad. Sci. USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khorchid A., Halwani R., Wainberg M.A., Kleiman L. Role of RNA in facilitating Gag/Gag-Pol interaction. J. Virol. 2002;76:4131–4137. doi: 10.1128/JVI.76.8.4131-4137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y.X., Campbell S., Harvin D., Ehresmann B., Ehresmann C., Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cen S., Huang Y., Khorchid A., Darlix J.L., Wainberg M.A., Kleiman L. The role of Pr55(gag) in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu W., Gorelick R.J., Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y.X., Copeland T.D., Henderson L.E., Gorelick R.J., Bosche W.J., Levin J.G., Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl Acad. Sci. USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rein A., Henderson L.E., Levin J.G. Nucleic acid chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 10.Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 11.Carteau S., Gorelick R.J., Bushman F.D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckman J.S., Bosche W.J., Gorelick R.J. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher R.J., Rein A., Fivash M., Urbaneja M.A., Casas-Finet J.R., Medaglia M., Henderson L.E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuilleumier C., Bombarda E., Morellet N., Gerard D., Roques B.P., Mely Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 15.Guo J., Wu T., Anderson J., Kane B.F., Johnson D.G., Gorelick R.J., Henderson L.E., Levin J.G. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagan N., Fabris D. Direct mass spectrometric determination of the stoichiometry and binding affinity of the complexes between nucleocapsid protein and RNA stem-loop hairpins of the HIV-1Psi-recognition element. Biochemistry. 2003;42:10736–10745. doi: 10.1021/bi0348922. [DOI] [PubMed] [Google Scholar]
- 17.Guo J., Wu T., Kane B.F., Johnson D.G., Henderson L.E., Gorelick R.J., Levin J.G. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 2002;76:4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams M.C., Rouzina I., Wenner J.R., Gorelick R.J., Musier-Forsyth K., Bloomfield V.A. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams M.C., Gorelick R.J., Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc. Natl Acad. Sci. USA. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hargittai M.R., Gorelick R.J., Rouzina I., Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 21.De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 22.Wiseman T., Williston S., Brandts J.F., Lin L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 23.Comisarow M.B., Marshall A.G. Fourier transform ion cyclotron resonance. Chem. Phys. Lett. 1974;25:282–283. [Google Scholar]
- 24.Hofstadler S.A., Sannes-Lowery K.A., Hannis J.C. Analysis of nucleic acids by FTICR MS. Mass Spectrom Rev. 2005;24:265–285. doi: 10.1002/mas.20016. [DOI] [PubMed] [Google Scholar]
- 25.Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K.J., Wu Z.R., Summers M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 26.Wu J.Q., Ozarowski A., Maki A.H., Urbaneja M.A., Henderson L.E., Casas-Finet J.R. Binding of the nucleocapsid protein of type 1 human immunodeficiency virus to nucleic acids studied using phosphorescence and optically detected magnetic resonance. Biochemistry. 1997;36:12506–12518. doi: 10.1021/bi970676f. [DOI] [PubMed] [Google Scholar]
- 27.Gorelick R.J., Chabot D.J., Rein A., Henderson L.E., Arthur L.O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 1993;64:3207–3211. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 29.Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia colirepressor–operator interaction: equilibrium measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 30.Winter R.B., Berg O.G., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 31.Barkley M.D., Lewis P.A., Sullivan G.E. Ion effects on the lac repressor–operator equilibrium. Biochemistry. 1981;20:3842–3851. doi: 10.1021/bi00516a027. [DOI] [PubMed] [Google Scholar]
- 32.Barkley M.D. Salt dependence of the kinetics of the lacrepressor-operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry. 1981;20:3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- 33.Berg O.G., von Hippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 34.Stoylov S.P., Vuilleumier C., Stoylova E., De Rocquigny H., Roques B.P., Gerard D., Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 35.Le Cam E., Coulaud D., Delain E., Petitjean P., Roques B.P., Gerard D., Stoylova E., Vuilleumier C., Stoylov S.P., Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Remy E., de Rocquigny H., Petitjean P., Muriaux D., Theilleux V., Paoletti J., Roques B.P. The annealing of tRNA3Lys to human immunodeficiency virus type 1 primer binding site is critically dependent on the NCp7 zinc fingers structure. J. Biol. Chem. 1998;273:4819–4822. doi: 10.1074/jbc.273.9.4819. [DOI] [PubMed] [Google Scholar]
- 37.Yamada O., Kraus G., Sargueil B., Yu Q., Burke J.M., Wong-Staal F. Conservation of a hairpin ribozyme sequence in HIV-1 is required for efficient viral replication. Virology. 1996;220:361–366. doi: 10.1006/viro.1996.0324. [DOI] [PubMed] [Google Scholar]
- 38.Russell R.S., Hu J., Laughrea M., Wainberg M.A., Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology. 2002;303:152–163. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 39.Bohnlein S., Hauber J., Cullen B.R. Identification of a U5-specific sequence required for efficient polyadenylation within the human immunodeficiency virus long terminal repeat. J. Virol. 1989;63:421–424. doi: 10.1128/jvi.63.1.421-424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urbaneja M.A., Wu M., Casas-Finet J.R., Karpel R.L. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 41.Tanchou V., Gabus C., Rogemond V., Darlix J.L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.