Abstract
Type III secretion systems (TTSS) are essential virulence determinants of many gram-negative bacteria and serve, upon physical contact with target cells, to translocate bacterial proteins directly across eukaryotic cell membranes. The Shigella TTSS is encoded by the mxi/spa loci located on its virulence plasmid. By electron microscopy secretons are visualized as tripartite with an external needle, a transmembrane domain, and a cytoplasmic bulb. In the present study, we generated a Shigella spa32 mutant and studied its phenotype. The spa32 gene shows low sequence homology to Salmonella TTSS1 invJ/spaN and to flagellar fliK. The spa32 mutant, like the wild-type strain, secreted the Ipas and IpgD, which are normally secreted via the TTSS, at low levels into the growth medium. However, unlike the wild-type strain, the spa32 mutant could neither be induced to secrete the Ipas and IpgD instantaneously upon addition of Congo red nor penetrate HeLa cells in vitro. Additionally, the Spa32 protein is secreted in large amounts by the TTSS during exponential growth but not upon Congo red induction. Interestingly, electron microscopy analysis of the spa32 mutant revealed that the needle of its secretons were up to 10 times longer than those of the wild type. In addition, in the absence of induction, the spa32 mutant secreted normal levels of MxiI but a large excess of MxiH. Taken together, our data indicate that the spa32 mutant presents a novel phenotype and that the primary defect of the mutant may be its inability to regulate or control secretion of MxiH.
The gram-negative Shigella bacterium is the cause of bacillary dysentery, an invasive disease of the human colonic epithelium (13, 42). The three essential steps for Shigella virulence are invasion of epithelial cells, intracellular multiplication, and the spread of the invading bacteria into adjacent cells. The capacity of Shigella to enter cells is governed by proteins encoded by a subset of genes within three contiguous operons (ipa, mxi, and spa) in a 30-kb region of the 230-kb pWR100 virulence plasmid (36). The Ipa proteins (invasins) are essential for the invasion of epithelial cells, and their secretion is mediated by the proteins encoded at the mxi and spa loci whose products constitute a type III secretion apparatus (TTSS) (or secreton) (6, 27).
The major function of TTSSs is to transport proteins from the bacterial cytoplasm into the host cell plasma membrane or cytoplasm upon contact with host cells (5, 11, 12). In Shigella flexneri, the mxi, the spa and the ipa operons are expressed at 37°C, but Ipa proteins remain in the bacterial cytoplasm until the secretion machinery is activated by host cell contact or by external, presumably surrogate, signals such as serum or asmall amphipathic Congo red (CR) dye molecule (4, 27, 37). Physical contact between the bacterium and the host cell induces insertion of two Ipas (IpaB and IpaC) into the host membrane to form a 25-Å pore that might be used to translocate the other invasins into target cells (6). The Ipas then catalyze the formation of a localized actin-rich, macropinocytic-like ruffle on the host cell surface which internalizes the bacterium (8, 48). Bacterial internalization initiates a cycle of intra- and intercellular spreading (34).
Th e Shigella type III secreton was found by electron microscopy of osmotically shocked and negatively stained cells to be composed of three parts: a cytoplasmic bulb, a transmembrane neck domain, and a 50- to 60-nm-long, extracellular and hollow needle through which secretion of Ipas might occur when bacteria contact epithelial cells (6, 7). This molecular machinery strongly resembles the Salmonella SPI1 TTSS1 (17, 20, 21) and flagellar basal bodies. The “needle complex” of Shigella is composed at least of MxiD, MxiG, MxiJ, MxiH, and MxiI (7, 47). The major needle component is MxiH, which is essential for the secretion of Ipa invasins (7).
The role of the Shigella Spa proteins is poorly understood. Yet, the spa region is highly conserved among all TTSS-encoding operons. Sasakawa and coworkers previously reported that spa32 mutant, which was able to bind CR at 37°C, suggesting a functional secretion apparatus (43, 49). These researchers also reported that cell surface-located Spa32 and contact between bacteria and HeLa cells were required for triggering the release of Ipa proteins from the Shigella outer membrane. Very recently, Schuch and Maurelli (45) reported that the spa33 gene is required for Ipa secretion and that its product is exported to the surface of the bacteria by the Mxi/Spa TTSS.
We performed here further studies on the spa32 gene product. We generated a nonpolar spa32 mutant, localized the Spa32 protein in Shigella, and studied the multiple aspects of its complex function.
MATERIALS AND METHODS
Bacterial strains and growth media.
S. flexneri strains are derivatives of the wild-type strain M90T (serotype 5) (40). The M90T-Sm (Smr) and SF401 (mxiD) strains have been described previously (1). Escherichia coli strains are derivatives of K-12 strain; the M15 strain harboring the pREP4 plasmid (Table 1) was transformed with two pQE30 derivatives (see below), and the Top10 strain (Table 1) was transformed with pBAD derivatives (see below); DH5α(λpir) was transformed with derivatives of the suicide vector pGP704 (29), and SM10-λpir was used to transfer derivatives of pGP704 to S. flexneri. Bacteria were grown in Luria-Bertani (LB) medium (Sigma) or tryptic casein soy broth at 37°C (Sigma). Antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; and streptomycin, 100 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant features and/or method for construction | Source or reference |
|---|---|---|
| Strains | ||
| S. flexneri | ||
| M90T-Sm | S. flexneri strain is a derivative of the wild-type strain M90T(serotype 5) (41) which is resistant to streptomycin (Smr) | 1 |
| SF401 | mxiD-deficient secretion mutant | 1 |
| MJ321 | spa32 mutant (pWR100-spa32::aphA-3), still producing truncated Spa32 protein (the first 91 N-terminal amino acid residues) (Spa32n) | This work |
| MJ322 | MJ321+pMJ7 (native Spa32) | This work |
| MJ323 | MJ321+pMJ8 (His6-Spa32) | This work |
| MJ005 | pWR100-mxiD-lacZ | This work |
| MJ325 | pWR100-spa32::aphA-3-mxiD-lacZ | This work |
| E. coli | ||
| DH5α(λpir) | Host for cloning suicide vector pGP704 | 29 |
| Sm10(λpir) | Host used for conjugation mating with S. flexneri | 29 |
| XL1-Blue | General host for cloning vectors | Pharmacia |
| M15 | Host for high expression of His6 recombinant proteins | Qiagen |
| Top10 | Host for high expression of His6 recombinant proteins | Invitrogen |
| Plasmids | ||
| pGP704 | Suicide vector, with R6K origin, used for the construction of spa32 mutant | 29 |
| pTZ18R | Cloning vector | Pharmacia |
| pUC19 | Cloning vector | New England Biolabs |
| pMJ1 | pUC19 carrying spa32 and its flanked regions | This work |
| pMJ2 | pMJ1 carrying internal spa32 in-frame deletion | This work |
| pMJ3 | pMJ2 carrying insertion of the aphA-3 cassette into spa32 | This work |
| pMJ5 | pGP704 suicide vector carrying spa32 inactivated by the aphA-3 cassette from pMJ2 | This work |
| pMJ7 | pTZ18R carrying the spa32 gene expressed from the p lac promoter | This work |
| pMJ8 | pQE30 carrying the entire spa32 gene; His6 N-terminal fusion construct | This work |
| pNJH8 | pQE30 carrying the mxiH gene; His6 N-terminal fusion construct | This work |
| pNJ54 | pBAD His mycA carrying the mxiI gene; His6 C-terminal fusion construct | This work |
| pQE30 | Expression vector | Qiagen |
| pBAD His-mycA | Expression vector | Invitrogen |
| pLAC8 | pGP704 suicide vector carrying mxiD-lacZ fusion | 1 |
Plasmids and strain construction.
Strains and plasmids used or constructed in this study are listed in Table 1.
DNA analysis, plasmid construction, and transformation of E. coli and S. flexneri were performed according to standard methods (39). A 2,475-bp DNA fragment containing the 879-bp spa32 gene flanked by upstream and downstream regions of 798-bp each was amplified by PCR with spa32-1 (5′-GCTCGCATGCCTTTTGGAGGATGAT-3′; sense) and spa32-2 (5′-GGCCGGATCCAAGAACCATTTACT-3′; antisense) primers and with the pWR100 virulence plasmid DNA as a template. The amplified fragment contains a BamHI site at the 5′ end and a SphI site at the 3′ end. Plasmid pMJ1 was constructed by inserting the BamHI/SphI-digested 2,475-bp PCR fragment (encompassing the spa32 gene and flanking regions) into the BamHI/SphI-digested pUC19 (Fig. 1B). Plasmid pMJ2 was constructed by an in-frame deletion within the spa32 gene by removing the 540-bp MscI -HincII fragment of pMJ1 (from codons 91 to 270 of Spa32) (Fig. 1B). Plasmid pMJ3 was constructed by inserting the 850-bp SmaI DNA fragment carrying the aphA-3 gene (26) into the unique SfuI site of pMJ2 (Fig. 1B). In this construct, the spa32 gene was interrupted at codon 92, which leads to the production of a truncated Spa32 protein (Spa32n) that contains the first 91 N-terminal amino acid residues, the codons 92 to 271 were deleted, and the last 21 codons were inserted in-frame with the translational start codon located at the 3′ end of the aphA-3 cassette (26). Plasmid pMJ5 was constructed by inserting the 2,795-bp SphI/KpnI fragment of pMJ3 (encompassing the inactivated spa32 gene and its flanking regions) into the corresponding sites of the suicide vector pGP704. PMJ5 was then transferred to S. flexneri M90T-Sm by conjugal mating, and clones in which the double recombination event had occurred were selected based on their resistance to streptomycin and kanamycin and their sensitivity to ampicillin. The structure of the resulting pWR100::spa32, carrying the inactivated spa32 was confirmed by PCR with the spa32-1 and spa32-2 primers, and the corresponding strain was designated MJ321 (and is referred to as the spa32 mutant).
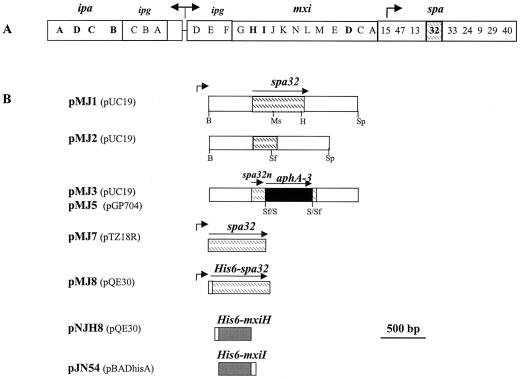
FIG. 1.
Plasmids used for the inactivation of the spa32 gene. (A) Schematic representation of the 30-kb DNA of the pWR100 plasmid required for the entry of S. flexneri into eukaryotic cells. Arrows indicate positions and orientation of previously described promoters. Open bars, except for the spa32 gene (indicated by a dashed box), show the relative positions of the icsB-ipgABC-ipaBCDA, ipgDEF-mxi, and spa loci. (B) Structure of plasmids used to inactivate the spa32 gene (pMJ2-5), to complement the spa32 mutant (pMJ7-8), and to produce and purify His-tagged recombinant proteins Spa32 (pMJ8), MxiH (pNJH8), and MxiI (pJN54). The black box represents the nonpolar aphA-3 gene that confers resistance to kanamycin. The horizontal arrows indicate the extent and the orientation of the corresponding genes, while small arrows indicate positions of the promoters known to control expression of these genes. A small open box represents the His6 tag motif. Constructs are drawn with the relevant restriction sites: B, BamHI; Ms, MscI; H, HincII; Sf, SfuI; and Sp, SphI.
Plasmid pMJ7, used for complementation experiments, was constructed by inserting the 1,540-bp EcoRI DNA fragment from pMJ1 into the corresponding site of pTZ18R (Pharmacia) (Fig. 1B). The introduction of this plasmid into MJ321 gave rise to strain MJ322.
mxiD-lacZ fusions in MJ321 and wild-type strains were constructed by using the pLAC8 (mxiD-lacZ) suicide plasmid according to the method of Allaoui et al. (1). Briefly, pLAC8 (mxiD-lacZ) was transferred by conjugal mating from E. coli SM10-λpir to S. flexneri M90T (wild type) and to MJ321 strains, respectively. The appropriate simple integration of the pLAC8 in the virulence plasmid pWR100 (wild type) or pWR100::spa32 (spa32 mutated strain) was ascertained by selection of the transconjugants on plates containing streptomycin and ampicillin. The resulting strains were designated MJ005 (M90T-mxiD-lacZ) and MJ325 (MJ321-mxiD-lacZ).
Construction of recombinant plasmids expressing His6 hybrid proteins.
Plasmid pMJ8, used for the purification of His6-Spa32 and for complementation experiments, was constructed by inserting the 915-bp BamHI/KpnI PCR DNA fragment, carrying the spa32 gene, into the corresponding sites of the expression vector pQE30 (Qiagen). The PCR DNA fragment was amplified by using the spa32-3 sense primer (5′-ACGCGGATCCATGGCATTAGATAATATAAACC-3′) and the spa32-4 antisense primer (5′-TGGCGGTACCTCTGTAGTTTTTCGTTAT-3′), which creates a BamHI at the 5′ end and a KpnI site at the 3′ end. Plasmid pNJH8, which encodes His6-MxiH was constructed as follows. The mxiH gene was amplified by using the mxiHI sense primer (5′-AGCGGATCCAGTGTTACAGTACCG-3′) and the mxiH2 antisense primer (5′-CGCGTCGACTGGATTATCTGAAGT-3′), which creates a BamHI site at one end and a HindIII site at the other. pNJH8 was constructed by inserting the 263-bp BamHI-HindIII mxiH PCR-digested PCR DNA fragment into the corresponding sites of expression vector pQE-30 (Qiagen) (Fig. 1B). Plasmid pNJ54 encoding MxiI-His6 was constructed as follows. The mxiI gene was amplified by using the mxiI1 sense primer (5′-CATGCCATGGTTTACATTTATCCAGTC-3′) and mxiI2, an antisense primer (5′-CCCAAGCTTAGACTTTAATAAAGTTTC-3′). The amplified DNA contains a NcoI site at the 5′ end and a HindIII site at the 3′ end. Plasmid pNJ54 was obtained by cloning the 305-bp NcoI/HindIII PCR-digested DNA fragment into the corresponding sites of the pBAD His-mycA vector.
Preparation of antibodies against Spa32, MxiH, and MxiI.
E. coli M15 was transformed with pMJ8 (His6-Spa32) and with pNJH8 (His6-MxiH), respectively (the two recombinant proteins contain N-terminal His6 tag fusions). The resulting strains were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 and then induced for 4 h with IPTG (isopropyl-β-d-thiogalactopyranoside) at a 1 mM final concentration. The Top10 strain harboring the pNJ54 plasmid (MxiI-His6), in which the six histidines were fused to the C terminus of the full-length MxiI protein, was grown at 37°C to an OD600 of 0.4 and then induced 4 h with arabinose at a final concentration of 0.2%. In all cases, the His6-tagged recombinant proteins were found in inclusion bodies and were solubilized in a 0.1 M NaH2 PO4-10 mM Tris (pH 8) buffer containing 8 M urea. Proteins were further purified according to the Qiagen expression protocol under denaturing conditions. The purified proteins were eluted from the nickel-nitrilotriacetic acid column with a 0.1 M NaH2 PO4-10 mM Tris (pH 8) buffer containing 6 M urea and imidazole at a final concentration of 200 mM. The eluted protein solutions were further submitted to dialysis in 1 M stepwise reductions in urea concentration until a concentration of 3 M urea was reached. For each His6-Spa32, His6-MxiH, or MxiI-His6, a rabbit was injected three times with 500 μg of the protein sample.
Protein analysis.
Proteins were analyzed by sodium dodecyl sulfate (SDS)-12 to 14% polyacrylamide gel electrophoresis (PAGE) (23) and/or Western blot. For some preparations, Tricine gels containing a 16% polyacrylamide concentration were used to separate the MxiH and MxiI proteins. The primary antibodies used in this study were the monoclonal antibodies anti-IpaC and anti-IpaB (1); anti-polyhistidine monoclonal antibodies (Sigma); and anti-IpaD (27), -Spa32, -MxiH, and -MxiI polyclonal antibodies. Goat anti-mouse immunoglobulin G (IgG)-peroxidase conjugate antibodies (Sigma) or donkey anti-rabbit IgG horseradish peroxidase-linked whole antibodies (Amersham Life Sciences) were used as secondary antibodies, and detected proteins were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Preparation of culture supernatants.
Two secretion modes are usually used to study Ipa proteins secretion in Shigella (37). After their production, the Ipa proteins are stored within the cytoplasm, and only 5% of them are secreted via the TTSS when bacteria are grown to mid log phase. The latter mode of secretion is called uninducible secretion, basal secretion, or leakage. The second mode of secretion is called inducible secretion, a condition during which bacteria are induced by the addition of CR. Under this condition, ca. 90% of the Ipa proteins are instantaneously secreted by the TTSS. This form of secretion is also called full or complete secretion.
Bacteria were grown with aeration on LB medium at 37°C by using a C25 incubator shaker at 175 rpm (New Brunswick Scientific). Proteins were analyzed from 50-ml log-phase cultures (OD600 = 0.3). Under inducing conditions, proteins from culture supernatants were prepared from 10-ml portions of these cultures. Bacteria were recovered by centrifugation (10,000 × g, 10 min), suspended in 500 μl of phosphate-buffered saline (PBS), and incubated with CR (at a final concentration of 10 μg/μl) for 15 min at 37°C (6, 33). Cells were removed by centrifugation (21,000 × g, 20 min), and 250 μl of supernatants was recovered. Then, 20 μl of these samples, corresponding to 0.8 ml of the original culture, was used for SDS-PAGE and Western blot analysis.
To analyze protein secretion under noninducing conditions, cells were removed from the remaining 40 ml of log-phase cultures (grown at 37°C on LB medium) by centrifugation (10,000 × g, 30 min). Portions (30 ml) of culture supernatants were filtered through a 0.2-μm-pore-size syringe filter and needle (18GA2 1.2×50; Becton Dickinson, Drogheda, Ireland) and precipitated by the addition of 40% (wt/vol) ammonium sulfate. The mixtures were incubated overnight at 4°C under mild agitation and then centrifuged (10,000 × g, 45 min). The pellets were washed in 800 μl of sterile water and centrifuged at high speed (21,000 × g, 45 min). The dried pellets were suspended in 40 μl of PBS. Then, 5-μl portions of these samples, corresponding to 3.5 ml of the original culture, were used for SDS-PAGE and Western blot analysis.
Amino acid sequencing.
Proteins were separated by SDS-PAGE, blotted onto polyvinylidene difluoride membrane, and stained with amido black. The bands of interest were applied to a protein sequencer (Eurogentec, Liège, Belgium).
Other tests.
Invasion assays, electron microscopy, and β-galactosidase assays were performed as described previously (6, 7, 28, 41).
RESULTS
Spa32 is required for CR-induced secretion of the Ipa invasins.
The spa32 gene is encoded within the 30-kb DNA fragment of the Shigella virulence plasmid essential for entry into epithelial cells (Fig. 1A) (25, 44). In a previous work, Watarai et al. (49) constructed an in-frame deletion of spa32 in S. flexneri 2a by using the pCDV442 suicide vector and sacB for selection of sucrose-resistant clones. We found that this strain (CS25585) bound CR not only at 37°C but also at 30°C and displayed no secretons at 37°C (not shown). Therefore, we concluded that its CR-binding activity was unrelated to the expression of a functional TTSS. Indeed, the CS25585 strain had been screened for its ability to bind CR on sucrose plates (49), probably forcing the occurrence of an unrelated mutation that otherwise affected the CR-binding ability of this strain. Moreover, we failed, in the present study, to obtain a spa32 mutant by using a derivative of the pCDV442 that harbored an internal in-frame deletion of spa32. Indeed, for all of the clones tested, we found that the virulence plasmid pWR100 was partly deleted after sucrose selection (data not shown).
To characterize the role of spa32, we constructed a S. flexneri 5a strain, MJ321, in which the spa32 gene has been inactivated with a nonpolar cassette (Fig. 1B and Materials and Methods). The newly constructed strain was unable to bind CR and was thus white on agar plates at 37°C, suggesting that the spa32 gene product is required for the secretion of Ipa invasins. To test this, the proteins present in the culture supernatants of M90T (wild-type) and MJ321 strains induced by CR were analyzed by SDS-PAGE and immunoblotting. IpaA (70 kDa), IpaB (62 kDa), IpaC (41 kDa), and IpaD (39 kDa) were secreted by the wild-type strain but not by MJ321 (Fig. 2A). The lack of Ipa secretion was confirmed by using monoclonal antibodies directed against IpaC and IpaB and polyclonal anti-IpaD (Fig. 2B and data not shown). Significant amounts of IpaC were detected in whole-cell extracts of the wild type and the spa32 mutant, indicating that the lack of secretion was not due to the absence of synthesis or stability of the Ipa proteins (Fig. 2B).
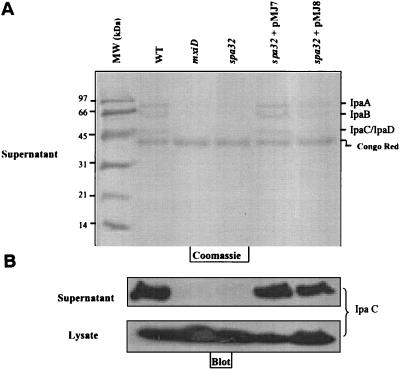
FIG. 2.
Spa32 is required for Ipa secretion upon CR induction. Strains used were: M90T (wild type), mxiD and MJ321 (spa32 mutant), MJ322 (MJ321+pMJ7), and MJ323 (MJ321+pMJ8). Cultures of S. flexneri strains (grown at 37°C) were harvested by centrifugation, suspended in PBS, and incubated in the presence of CR for 15 min at 37°C to induce Ipa secretion. Equal amounts of the culture supernatants and cell lysates were analyzed by SDS-12% PAGE stained with Coomassie blue (A) or were immunoblotted with a monoclonal antibody specific to the IpaC protein (B).
As expected, strain MJ321 was also unable to enter HeLa cells (data not shown). The mutant's inability to secrete Ipa proteins was restored to the wild-type phenotype by complementation of the mutant with expression vectors encoding the Spa32 protein from the p lac or the p tac promoters (Fig. 1B and Table 1). Indeed, the introduction of pMJ7 or pMJ8, two plasmids encoding the native Spa32 and a His6-Spa32, respectively, restored the ability of the MJ321 strain to secrete the Ipa proteins and to invade HeLa cells (Fig. 2 and data not shown).
Spa32 is not required for the initial secretion of Ipa invasins in the absence of CR induction.
It was previously reported that low-level secretion of the Ipa proteins via the TTSS in the absence of any induction could be observed in the wild-type strain (37). We therefore investigated whether this process was affected in the MJ321 spa32 mutant. We precipitated proteins from the culture supernatant of several strains grown until log phase and performed Coomassie blue staining of SDS-PAGE and Western blots. As shown in Fig. 3A and B, we found that IpaB, IpaC, and IpaD (data not shown) were present at wild-type levels in the culture supernatant of the MJ321 strain. As a control, we used the SF401 strain (mxiD mutant), which does not secrete any Ipa proteins under similar growth conditions, indicating that the Ipa proteins are released via the Mxi/Spa TTSS (Fig. 3A and B). This unambiguously demonstrates that the spa32 mutant, despite its apparent inability to bind CR on plates, retained its ability to secrete a small amount of the Ipa proteins prior to CR induction. Therefore, our data provides the first report of a Shigella mutant able to secrete a low-level the Ipa invasins in the medium but which has, however, lost its ability to penetrate HeLa cells in vitro.
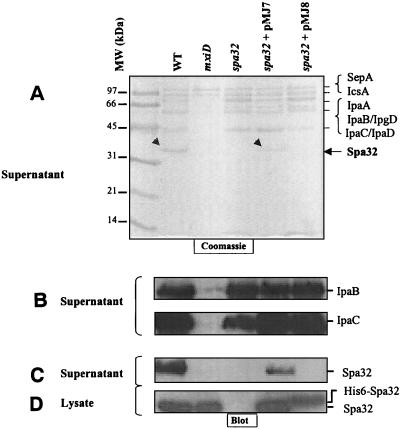
FIG. 3.
The spa32 mutant (MJ321) secretes Ipa proteins even in the absence of CR induction. Culture supernatant proteins were prepared from bacteria grown for 3 h at 37°C until the exponential growth phase was reached (OD600 = 0.3). Samples were centrifuged, and equal amounts of precipitated supernatants and cell lysates were analyzed by SDS-12% PAGE and Coomassie blue staining (A) or by immunoblotting with a monoclonal antibody specific for IpaB and IpaC (B) or a polyclonal antibody specific to Spa32 (C and D). The small arrowheads indicate the position of Spa32.
Electron microscopy analysis of the type III secretons of the spa32 mutant reveals secretons with ultralong needles.
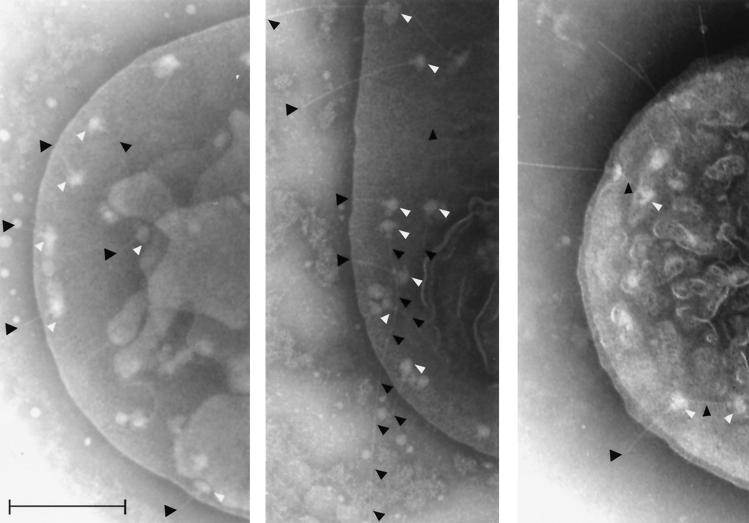
The MJ321 spa32 mutant's ability to secrete Ipa proteins suggested an at least partially functional TTSS, which we visualized by electron microscopy. Wild-type Shigella exhibits secretons with an average needle length of ca. 50 nm (Fig. 4, left panel). Electron microscopy analysis of the MJ321 strain indicated that the spa32 mutated strain showed extraordinary long secreton needles up to 400 to 900 nm in length (Fig. 4, middle panel). Complementation of the spa32 mutant with a plasmid expressing either the native Spa32 (MJ322 stain) (Fig. 4, right panel) or the His6-Spa32 (MJ323 strain) (data not shown) partly restored the wild-type needle substructures. We found that the two latter strains exhibited intermediate-length needles of up to 50 to 150 nm. Hence, the MJ321 is the first Shigella strain that contains the three parts of the secreton (bulb, neck, and an needle) but is no longer able to bind CR on plates, to secrete the Ipa invasins upon CR induction, or to invade HeLa cells in vitro.
FIG. 4.
The spa32 mutant strain (MJ321) exhibits an elongated needle structure. Electron micrographs of Shigella wild-type M90T (left panel), spa32 mutant (middle panel), and spa32 mutant complemented with the pMJ7 plasmid encoding the native Spa32 protein (MJ322) (right panel). Osmotically shocked and negatively stained bacteria (grown at 37°C) were visualized by electron microscopy as previously described by Blocker et al. (7). Bar, 200 nm. The white arrowheads indicate the transmembrane region and the cytoplasmic bulb of type III secretons; the needle (if particularly long) or needle tips are outlined or indicated by black arrowheads.
MxiH and MxiI are the components of the long needle of the type III secreton of the spa32 mutant.
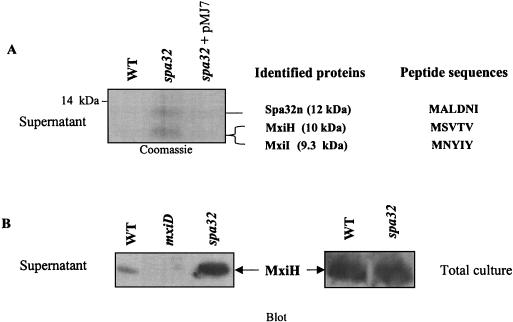
To analyze the components of the elongated needles observed on the spa32 mutant (MJ321), we isolated cell surface-associated structures from the MJ321 strain. Log-phase cultures were passed five times each through a 25-gauge needle prior to separation of the culture supernatant and the cell pellet fractions. Supernatant proteins were precipitated and separated by SDS-PAGE on a 16% polyacrylamide gel and then analyzed by Coomassie blue staining and immunoblotting (Fig. 5A). Two other strains—M90T (wild type) and MJ322 (MJ321+pMJ7)—were subjected as controls to the same analysis. As shown in Fig. 5A, the protein patterns show clear differences between strains in the low-molecular-weight range. A thick major band that migrated faster than the 14-kDa marker was detected for MJ321 but is missing in both the wild-type and the mxiD mutant strains. However, this protein band was present, although at a lower intensity, in the sample prepared from the MJ322 strain (MJ321+pMJ7), indicating a partial complementation of the spa32 mutant by pMJ7 (Fig. 5A). When longer 16% acrylamide gels with Tris-Tricine buffers were used, this protein band was revealed to be composed of at least two proteins of ca. 12 and 11 kDa. Upon sequencing the N terminus, the protein from the upper band contained the MALDNI amino acid residues. This sequence corresponds to the N-terminal end of Spa32 and was designated Spa32n (12 kDa); it is produced by the MJ321 strain (see Materials and Methods and Fig. 1B). In contrast, the lower protein band contained a mixture of two profiles of N-terminal amino acid residues: MSVTV and MNYIY (Fig. 5A), which were shown by sequence comparisons with the recently published sequence of the virulence plasmid pWR100 to correspond to the products of the mxiH (10 kDa) and mxiI (9.3 kDa) genes, respectively (2, 9).
FIG. 5.
The higher secreted amount of MxiH in the spa32 mutant is not due to its overproduction. (A) Wild-type strain (M90T), the spa32 mutant (MJ321), and its derivative complemented with pMJ7 (encoding the native Spa32) (MJ322) were grown to mid-log phase (OD600 = 2). Equal amounts of proteins from the culture supernatants were loaded on 16% Tris-Tricine SDS-PAGE gels, and then stained with Coomassie blue. The positions of the proteins, as well as the sequenced amino acid residues of the N terminus of Spa32n, MxiH, and MxiI are indicated on the right side of each panel. (B) Equal amounts of total culture proteins (supernatant and bacteria; the equivalent of 0.1 OD600) were separated by SDS-16% PAGE and then immunoblotted with the anti-MxiH polyclonal antibody. The strains used were the wild type (M90T), the spa32 mutant, and the mxiD mutant as indicated.
To monitor these proteins more easily, we purified recombinant MxiH, MxiI, and Spa32 proteins tagged with six histidines and generated antibodies against them (see Materials and Methods). The anti-Spa32 polyclonal antibody recognized Spa32n in the supernatant of the spa32 mutant (data not shown), and similar data were obtained for MxiH and MxiI (Fig. 5B and data not shown). This result indicates that the 91 N-terminal amino acid residues of Spa32 are sufficient to mediate its secretion via the TTSS. The amounts of secreted MxiI in both wild-type and spa32 mutant strains were similar (data not shown). However, the amount of MxiH detected in the culture supernatant prepared from the MJ321 strain was much higher than in the fractions prepared from for the wild-type strain (Fig. 5B). This difference was not due to the increased MxiH synthesis in the spa32 mutant strain since approximately equal amounts of MxiH were detected in the whole-cell culture of the wild-type and the spa32 mutant strains (bacteria and supernatant) from the two strains (Fig. 5B). These results suggest that the elongated needle substructures of MJ321 are due to the increased amount of MxiH secreted by the bacteria.
Spa32 is secreted by the TTSS in the absence of CR but not when secretion is induced.
We determined the localization of Spa32 during the bacterial growth cycle. We performed immunoblotting with the anti-Spa32 antibody on bacterial cell fractions prepared under various growth conditions. We found that native Spa32 was associated, in wild-type (M90T) and MJ322 strains, with the supernatant from log-phase cultures but not with the supernatant prepared after addition of CR (Fig. 3C and data not shown). Western blot analysis with antibodies directed against His6 (results not shown) or against Spa32 indicated that, in contrast to native Spa32, His6-Spa32 (expressed in MJ323 [MJ321+pMJ8]) was detected in the bacterial lysate fraction but was absent from the supernatant (Fig. 3C and D). In agreement with this result, a protein corresponding to the size of the spa32 gene product was detectable in a Coomassie blue-stained gel in the culture supernatant of the M90T and the MJ322 (MJ321+pMJ7) strains (Fig. 3A). However, this protein band was missing from the supernatant of the MJ323 (MJ321+pMJ8), thus confirming the lack of His6-Spa32 protein secretion (Fig. 3A). Moreover, Spa32 was not secreted by the mxiD mutant (Fig. 2C), which lacks visible secretons (6), indicating that Spa32 secretion occurs via the TTSS itself. The lack of Spa32 secretion by the mxiD mutant was not due to the absence of its synthesis, since Spa32 was detectable in the cell fraction in amounts equivalent to those seen in the wild-type strain (Fig. 3D). In further support of this, we confirmed by amino acid sequencing and Western blot that Spa32n (the N-terminal 91 amino acids of Spa32) is secreted without any cleavage, as would be expected for a protein secreted by the TTSS (Fig. 5A). In addition, since the His6 form of Spa32, which is not secreted, partially rescues Ipa secretion upon induction with CR (Fig. 2) and HeLa cell invasion in MJ321, we conclude that Spa32 need not be secreted to be functional. Similarly, we conclude that Spa32 need not be secreted to be functional, regulating the amount of MxiH secreted and thus needle length.
DISCUSSION
In the present study, we generated a spa32 mutant in S. flexneri 5a and showed that, although it can secrete a small amount of Ipa proteins into its growth medium, it is unable to secrete the Ipa proteins upon induction with CR. This mutant is also unable to invade HeLa cells in vitro, suggesting that its “contact-dependent” secretion is impaired. The wild-type phenotype was restored by complementation of the spa32 mutant with either native Spa32 or a His6-Spa32 protein. We characterized the spa32 mutant by electron microscopy of osmotically shocked cells and found that it exhibits a different TTSS (secreton) structure compared to the wild-type strain. Its secretons have transmembrane domains and cytoplasmic bulbs that appear identical to those of the wild-type strain, but the mutant strain has extraordinarily long needles, up to 10 times longer than those found in the wild-type strain. Furthermore, we identified the needle components of this spa32 mutant by preparing a fraction enriched in surface-associated structures. Three low-molecular-weight proteins were found in this fraction from the exponentially grown spa32-deficient strain, whereas they were not detectable in similar fractions from the wild-type strain. By N-terminal amino acid sequencing and Western blot, we showed that the three proteins corresponded to a truncated Spa32 (Spa32n) carrying the first N-terminal amino acid residues of Spa32, MxiH (10 kDa), and MxiI (9.3 kDa). The latter two proteins are required for Ipa secretion and are encoded by adjacent genes of the mxi locus (2, 7). These findings are in agreement with the previously proposed composition of the native needle complex of Shigella. Indeed, it was recently shown that both MxiH (7, 47) and MxiI are essential components of the Shigella TTSS needle (7).
MxiH is conserved in most of the bacteria species that are pathogenic for animals and have a TTSS (7), but MxiH homologues have not been reported in TTSSs of plant pathogens (15). In addition, the only MxiI homologue detected was the highly related Salmonella SPI1 TSSS protein PrgJ, which has been shown to be necessary for needle assembly in this organism (17). In Salmonella, PrgI is the major component of the needle complex and is secreted by the TTSS itself. Furthermore, PrgI is detected in the supernatant of the invJ/spaN mutant, which also displays abnormally long SPI1 TTSS needles (21). Interestingly, Spa32 shares 20% identity over its entire length with InvJ, suggesting a common ability to control the length of the corresponding TTSS needles. Additionally, both proteins share 14% identity with FliK of Salmonella (18), which is involved in regulating the hook-filament transition by switching to export of flagellar filament components (30, 32, 35).
One of the remarkable features of the flagellar structure of wild-type Salmonella is that the length of the hook, a hollow tubular structure composed of FlgE subunits, is tightly controlled at ca. 55 nm in wild-type Salmonella (14). The Shigella TTSS needle is of a similar and invariant length (ca. 50 nm [7, 47]). Very recently, Makishima et al. (24) examined hook length and found that it is determined not by molecular rulers (which FliK was initially thought to be) (16, 19) but from the inside of the bacterium by the C-ring proteins (FliG, FliM, and FliN), which appear to monitor the amount of hook protein secreted by the flagellar export apparatus.
Tamano et al. (47) recently reported that increased levels of MxiH in wild-type Shigella leads to much longer needles without affecting inducible Ipa protein secretion or invasion of HeLa cells. The elongated needle and the high amount of MxiH secreted in the spa32 mutated strain grown to mid log phase thus suggested that the deletion of spa32 might directly upregulate mxiH gene transcription or affect the stability of its mRNA or protein product. To test this, we introduced an mxiD-lacZ transcriptional fusion in both wild-type and MJ321 strains and found that the the level of β-galactosidase expressed from the mxi genes promoter in the spa32 mutant was 2.5 times higher than that of the wild-type strain (data not shown). This modest increase in mxiH gene expression cannot explain the >10-fold increase in the needle length observed in the spa32 mutant. However, amplification of protein production might also occur at the posttranscriptional or posttranslational level by increased mRNA stability, protein translation, and/or protein stability.
To understand the role of Spa32 in releasing the Ipa proteins in an inducible manner, its cellular location was examined. Spa32 was detected in the supernatant of the wild-type strain growing exponentially (CR-independent secretion). The recent finding that Spa32 is secreted via the TTSS in the Δipa mutant, which constitutively secretes at least 15 other proteins (9), is supported by our data. In addition, the fact that Spa32 is secreted is in accordance with previous reports on spa32 homologues, InvJ and YscP, secreted molecules required for the secretion of Sip and Yop virulence proteins of Salmonella and Yersinia, respectively (10, 38, 46). Finally, the FliK protein has also been shown to be secreted (30), even though its biological action was pinpointed to occur inside the bacterium or as it exits the flagellar export apparatus (19). Interestingly, His6-tagged Spa32, which is not secreted and is detected in whole-cell protein extracts, restores almost normal needle length, inducible secretion of Ipa proteins, and invasion to the nonpolar spa32 mutant strain. Of note, FliK and Spa32 might similarly play a crucial role when present inside the bacteria and be secreted concomitantly or after action.
Therefore, this raises the question of why Spa32 is secreted at all? Perhaps, its secretion during type III secreton assembly is an artifact unrelated to its function. However, a body of evidence shows that, upon the completion of the hook assembly, FliK is very efficiently secreted (30). Moreover, any decrease in the level of FliK export has been shown to result in polyhook stub-filament structures with an inversely proportional higher number of repeated hooks, each 55 nm in length (19, 30). Thus, we propose that Spa32 might perform two biological functions: the first function, we propose, is exerted inside the bacterium, perhaps at the cytoplasmic face of the C-ring, to mediate the switch in substrate specificity from needle components to Ipa proteins. We propose that the second function, exerted near the entrance of the secreton channel, is to allow inducible Ipa secretion. This second step would be equivalent to the initiation of the hook-filament transition by FliK.
The detailed molecular nature of the biological functions of Spa32 remains to be determined. Yet, there are good reasons to believe that needle unit repeat control in TTSSs operates similarly (but perhaps not identically) to hook unit repeat control in the bacterial flagellum. Indeed, FliK was shown to function together with a membrane protein, FlhB, a component of the flagellar export apparatus, to mediate the switching of export substrate specificity upon completion of the hook assembly (22, 31). FliK was shown to interact with the carboxy-terminal cytoplasmic domain of FlhB. Interestingly, in our laboratory, we have preliminary unpublished data that indicate that the Shigella Spa40 protein, which shares 45% sequence identity with FlhB, interacts with Spa32 (3). Since the FlhB/FliK and Spa40/Spa32 protein pairs have counterparts in all bacteria with flagella and/or TTSSs, one can speculate that the mechanisms involved in regulation of hook repeat control and in switching to filament assembly or inducible secretion form common regulatory pathways.
The spa32 mutant secretes Ipa proteins at a low level but not in an inducible manner. This is the first report of a Shigella mutant that has lost its ability to invade HeLa cells, although it can still secrete all of the Ipa invasins. The novel phenotype of the spa32 mutant regarding Ipa secretion leads us to suggest that the TTSS of S. flexneri can exist in at least two secretion modes: (i) a noninducible one that occurs, for instance, in culture media in vitro during type III secreton assembly, and (ii) an inducible one that is activated upon addition of CR or contact with target cells. Furthermore, there are qualitative differences in the levels of secretion of different substrates by the two secretion modes. For example, while the secreton is being assembled, the early P-rod and needle proteins are secreted solely constitutively, and later their secretion is fully turned off, probably because there are no P-rod or needle components left. In a similar manner, Ipa proteins can “leak” at a low level during the later stages of log phase growth when secretons are fully assembled but mostly remain stored inside the bacterium awaiting the signal derived from host cell contact for complete secretion.
The findings of Tamano et al. (47) with a Shigella strain that overproduces MxiH and displays highly elongated needles indicate that the length of the TTSS needles is not crucial for correct adherence to and entry of the bacteria into eukaryotic host cells. The question then arises as to why a strain that produces a large amount of MxiH (47) and one deleted for spa32 (the present study), both of which display highly elongated needles, are differentially responsive to CR on plates and to induction of secretion and also differ in invasion ability. The MxiH-overproducing strain binds CR on plates, is capable of secretion upon CR induction in solution, and is invasive. In contrast, the spa32 mutant is white on CR plates, is unresponsive to CR induction in solution, and is noninvasive. This may simply reflect the inability of the bulbs in type III secretons of the spa32 mutant to bind and secrete Ipas efficiently upon addition of CR or host cell contact. Thus, instead of secreting predominantly Ipa proteins at this stage, it can only secrete more MxiH and MxiI (and Ipas at low level). On the other hand, the MxiH-overproducing strain, due to the presence of a wild-type Spa32, is able to switch its substrate specificity despite an elongated needle and is thus ready to respond to CR and host cell contact.
Thus, the primary defect of the spa32 mutant may be its inability to arrest MxiH export. Spa32 is probably also needed to switch from secretion of needle components to secretion of Ipas. Together with the C-ring components, it might act to monitor the level of MxiH and MxiI secretion and sense when TTSS needles of wild-type length have been assembled. Then, it may transmit a signal to the C-ring to arrest MxiH export and switch the substrate specificity of the export machinery to that of the effectors: Ipas and IpgD, via an interaction with the cytoplasmic domain of Spa40, before it is itself exported. Additional work is required to determine the mechanism by which the TTSS's needle length is controlled, as well as to understand the basis of the inducibility of secretion of the Ipa proteins.
Acknowledgments
J.M. and A.H. contributed equally to this study.
We thank C. Parsot for extremely helpful discussions during the course of this work. We thank M. B. Goldberg for careful reading of the manuscript.
This work was supported in part by grants from the Belgian FRSM (Fonds National de la Recherche Scientifique Médicale, Convention 3.4603.02), the Actions de Recherche Concertées (ARC; convention 98/03-224) de la Direction Générale de La Recherche Scientifique-Communauté Française de Belgique, and by a CEE fund (QLK2-CT-1999-00938). J.M. and N.J were supported by grants from the ARC convention 98/03-224, and A.H. is a recipient of a Ph.D. fellowship from the Fonds National de Recherches Industrielles et Agronomiques, Belgique. M.C. was supported by a Postdoctoral fellowship from the CEE. A.B. was supported by an Émile Roux Fellowship from the Pasteur Institute while in Paris and, since then, by the Guy G. F. Newton Senior Research Fellowship from the Guy Newton Trust, based at the Sir W. Dunn School of Pathology in Oxford, and K22 grant AI01847 from the National Institute of Allergy and Infectious Disease.
REFERENCES
- 1.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol. Microbiol. 7:59-68. [DOI] [PubMed] [Google Scholar]
- 2.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1993. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J. Bacteriol. 174:7661-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allaoui, A., S. Woestyn, C. Sluiters, and G. R. Cornelis. 1994. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J. Bacteriol. 176:4534-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahrani, F. K., P. J. Sansonetti, and C. Parsot. 1997. Secretion of Ipa proteins by Shigella flexneri: inducing molecules and kinetics of activation. Infect. Immun. 65:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleves, S., and G. R. Cornelis. 2000. How to survive in the host: the Yersinia lesson. Microbes Infect. 2:1451-1460. [DOI] [PubMed] [Google Scholar]
- 6.Blocker, A., P. Gounon, E. Larquet, K. Niebuhr, V. Cabiaux, C. Parsot, and P. J. Sansonetti. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 8.Bourdet-Sicard, R., M. Rudiger, B. M. Jockusch, P. Gounon, P. J. Sansonetti, and G. Tran Van Nhieu. 1999. Binding of the Shigella protein IpaA to vinculin induces F-actin depolymerization. EMBO J. 18:5853-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. J. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 10.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis, G. R., and G. Denecker. 2001. Yersinia leads SUMO attack. Nat. Med. 7:21-23. [DOI] [PubMed] [Google Scholar]
- 12.Galan, J. E., and J. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 13.Hale, T. L. 1998. Bacillary dysentery, p. 479-493. In W. R. Hansler and M. Shuman (ed.), Topley and Wilson's microbiology and microbial infections, vol. 3. Arnolds, London, United Kingdom
- 14.Hirano, T., S. Yamaguchi, K. Oosawa, and S.-I. Aizawa. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176:5439-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawagishi, I., A. W. Homma, A. W. Williams, and R. B. Macnab. 1996. Characterization of the hook length control protein FliK of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 178:2954-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Nat. Acad. Sci. USA 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komoriya, K., N. Shibano, T. Higano, N. Azuma, S. Yamaguchi, and S.-I. Aizawa. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767-779. [DOI] [PubMed] [Google Scholar]
- 19.Koroyasu, S., M. Yamazato, T. Hirano, and S.-I. Aizawa. 1998. Kinetic analysis of the growth rate of the flagellar hook in Salmonella typhimurium by the population balance method. Biophys. J. 74:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S.-I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 21.Kubori, T., A. Sukhan, S.-I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 18:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutsukake, K., T. Minamino, and T. Yokoseki. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176:7625-7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Makishima, S., K. Komoriya, S. Yamaguchi, and S.-I. Aizawa. 2001. Length of the flegellar hook and the capacity of the type III export apparatus. Science 291:2411-2413. [DOI] [PubMed] [Google Scholar]
- 25.Maurelli, A. T., B. Baudry, H. d'Hauteville, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ménard, R., P. Sansonetti, and C. Parsot. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamino, T., B. Gonzalez-Pedrajo, K. Yamaguchi, S.-I. Aizawa, and R. M. Macnab. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295-304. [DOI] [PubMed] [Google Scholar]
- 31.Minamino, T., and R. M. Macnab. 2000. FliK, Interactions among components of the Salmonella flagellar export and its substrates. Mol. Microbiol. 35:1052-1064. [DOI] [PubMed] [Google Scholar]
- 32.Muramoto, K., S. Makishima, S. Aizawa, and R. M. Macnab. 1999. Effect of hook subunit concentration on assembly and control of length of the flagellar hook of Salmonella. J. Bacteriol. 181:5808-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niebuhr, K., N. Jouihri, A. Allaoui, P. Gounon, P. Sansonetti, and C. Parsot. 2000. IpgD a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol. Microbiol. 38:8-19. [DOI] [PubMed] [Google Scholar]
- 34.Niebuhr, K., and P. J. Sansonetti. 2000. Invasion of epithelial cells by bacterial pathogens the paradigm of Shigella. Subcell. Biochem. 33:251-287. [DOI] [PubMed] [Google Scholar]
- 35.Ohnishi, K., Y. Ohto, S.-I. Aizawa, R. M. Macnab, and T. Iino. 1994. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsot, C. 1994. Shigella flexneri: genetics of entry and intercellular dissemination in epithelial cells. Curr. Top. Microbiol. Immunol. 192:217-241. [DOI] [PubMed] [Google Scholar]
- 37.Parsot, C., R. Ménard, P. Gounon, and P. J. Sansonetti. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291-300. [DOI] [PubMed] [Google Scholar]
- 38.Payne, P. L., and S. C. Straley. 1999. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J. Bacteriol. 181:2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sansonetti, P. J. 2001. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross talks. FEMS Microbiol. Rev. 25:3-14. [DOI] [PubMed] [Google Scholar]
- 43.Sasakawa, C., K. Kamata, K. T. Sakai, S. Y. Murayama, S. Makino, and M. Yoshikawa. 1986. Molecular alteration of the 140-megadalton plasmid associated with loss of virulence and Congo red binding activity in Shigella flexneri. Infect. Immun. 151:470-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasakawa, C., K. Komatsu, T. Tobe, T. Suzuki, and M. Yoshikawa. 1993. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J. Bacteriol. 175:2334-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuch, R., and A. T. Maurelli. 2001. Spa33, a cell surface-associated subunit of the Mxi-Spa type III secretory of Shigella flexneri, regulates Ip protein traffic. Infect. Immun. 69:2180-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stainier, I., S. Bleves, C. Josenhans, L. Karmani, C. Kerbourch, I. Lambermont, S. Totemeyer, A. Boyd, and G. R. Cornelis. 2000. YscP, a Yersinia protein required for Yop secretion that is surface exposed and released in low Ca2+. Mol. Microbiol. 37:1005-1018. [DOI] [PubMed] [Google Scholar]
- 47.Tamano, K., S.-I. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran Van Nhieu, G., E. Caron, A. Hall, and P. J. Sansonetti. 1999. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 18:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J. 14:2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]