Abstract
We demonstrate that the site-specific integrase encoded by phage TP901-1 of Lactococcus lactis subsp. cremoris has potential as a tool for engineering mammalian genomes. We constructed vectors that express this integrase in Escherichia coli and in mammalian cells and developed a simple plasmid assay to measure the frequency of intramolecular integration mediated by the integrase. We used the assay to document that the integrase functions efficiently in E. coli and determined that for complete reaction in E. coli, the minimal sizes of attB and attP are 31 and 50 bp, respectively. We carried out partial purification of TP901-1 integrase protein and demonstrated its functional activity in vitro in the absence of added cofactors, characterizing the time course and temperature optimum of the reaction. Finally, we showed that when expressed in human cells, the TP901-1 integrase carries out efficient intramolecular integration on a transfected plasmid substrate in the human cell environment. The TP901-1 phage integrase thus represents a new reagent for manipulating DNA in living mammalian cells.
Prokaryotic enzymes have supplied us with abundant tools for engineering DNA. For example, restriction enzymes and ligases, largely derived from bacterial and phage genomes, provided the tools for recombinant DNA. This technology has allowed construction of molecules at will in vitro, causing a wholesale transformation of biomedical science over the past 25 years. More recently, in vivo engineering of the genomes of living higher eukaryotic cells is becoming possible, often through the agency of prokaryotic enzymes such as Cre, an autonomous, site-specific, tyrosine-catalyzed recombinase from phage P1 (1). Recombinases such as Cre and FLP require no host-specific cofactors and perform well in higher eukaryotic cells, carrying out efficient site-specific recombination between two identical recognition sites (18, 22). These enzymes are useful for carrying out deletion and translocation-type recombination reactions in living cells (21).
Another useful reaction is integration for the purpose of creating knockin and knockout alterations of the genome, such as those desirable in gene therapy, creation of transgenic organisms, and modification of cells in culture. For integration, a unidirectional recombinase such as a phage integrase is ideal, because there is no reverse reaction that could depress net integration frequency (9). Phage integrases mediate recombination between nonidentical phage attP and bacterial attB recognition sites (13). The well-studied lambda integrase is, like Cre and FLP, a member of the tyrosine-catalyzed recombinase family (17). However, the integrases from lambda phage and the closely related phage HK022 have cofactor requirements that hamper their use in eukaryotic cells (11, 15).
Some phage integrases are members of the unrelated serine-catalyzed family of recombinases (24) and are autonomous with no cofactor requirements, which makes them potentially ideal for use in foreign host environments, such as mammalian cells. The integrase encoded by phage φC31 of Streptomyces spp. (12, 20) requires no cofactors (27). We have shown that the φC31 integrase works well in human and mouse cells (9, 30), mediating efficient intramolecular integration in transfected plasmid DNA and intermolecular integration into the chromosomes. Based on these results, we examined the related integrase from Streptomyces phage R4 (16) and found that it, too, works in human cells (19).
Although not highly similar to each other, the φC31 and R4 integrases are members of an identifiable subgroup of particularly long serine recombinases (16, 27) that contains another distantly related integrase, that of phage TP901-1 (6). We carried out this study to test the hypothesis that this integrase might also possess properties useful in engineering higher eukaryotic genomes. TP901-1 is a temperate bacteriophage that infects Lactococcus lactis subsp. cremoris and can be induced by UV light (2). After infection, the bacteriophage is able to lysogenize by integrating its 38.4-kb genome site specifically into the bacterial chromosome (7). It has been shown that the phage integrase encoded by the orf1 gene, a 425-bp region immediately upstream of orf1, and the attP sequence just downstream of orf1 are sufficient to catalyze integration into the chromosomal attB site in L. lactis (6). The goal of this investigation was to test whether the TP901-1 integrase could function outside its native host, in Escherichia coli and in human cells. We also defined minimal attP and attB sites recognized by the integrase and performed in vitro studies with the enzyme. This work introduces the TP901-1 integrase as a valuable tool for engineering DNA in the context of living mammalian cells.
MATERIALS AND METHODS
Intramolecular assay plasmids.
The assay plasmids used for intramolecular integration were generated as follows. A 304-bp fragment containing the TP901-1 bacterial attachment site, attB, was PCR amplified from L. lactis (American Type Culture Collection) genomic DNA by using the primers 5′CTCAAGCTCGAGGGGATATCTCGTTACCCATTTATTCTAATATGG3′ and 5′GCTCAACGGATCCTCATGATCCAACTCATAAAGTTG3′. This fragment was ligated into pCR2.1 (Invitrogen, Carlsbad, Calif.) to create pTA-attB304, from which the 304-bp attB was subcloned as a BamHI-XhoI fragment into BamHI- and SalI-digested pBCβGal (9), generating pBB-attB304. The 333-bp attP fragment was obtained by PCR amplification from L. lactis subsp. cremoris strain 901-1, which is lysogenic for phage TP901-1 (a gift from Horst Neve, Bundesanstalt fur Milchforschung, Kiel, Germany). The 109-bp P-arm was generated by using the primers 5′GCGAGTTTTTATTTCGTTTATTTCAATTAAGGTAACTAAAAAACTCCTTTTAAGG3′ (fwd1A) and 5′GCAGGTCCCGGGCCTTCTATGCATGAGATAACTG3′ (rev1B). The 237-bp P′-arm was generated by using the primers 5′GCGATCCCGCGGGCTGCTTAAAGCTAAGATTAGCG3′ (fwd2A) and 5′GTTACCTTAATTGAAATAAACGAAATAAAAACTCG3′ (rev2B). These two PCR products were then combined, subjected to seven rounds of PCR amplification, and then further amplified with primers fwd2A and rev1B in order to reconstruct the 333-bp attP fragment, which was ligated into pCR2.1 to create plasmid pTA-attP333. The attP was liberated as an XmaI-SacII fragment and cloned into the XmaI and SacII sites of pBB-attB304 to create pBB-B304-P333. In this assay plasmid, the two att site cores are in the same orientation, flanking the lacZ gene (Fig. 1A).
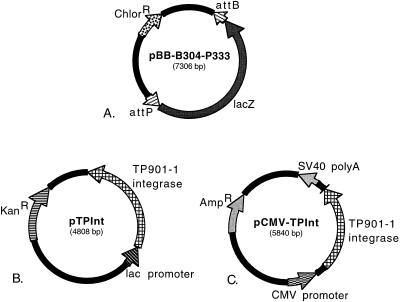
FIG. 1.
Plasmids for analyzing TP901-1 integrase-mediated recombination. (A) pBB-B304-P333 carries attB and attP recognition sites for the integrase in a direct orientation, flanking the lacZ gene, such that intramolecular integration mediated by the enzyme brings about excision of lacZ, which is detectable on X-Gal indicator plates as a change from blue colonies to white colonies. att sites of different sizes were cloned into pBB-B304-P333 in place of the full-length att sites to determine the minimal lengths of the recognition sites. (B) pTPInt expresses the TP901-1 integrase in E. coli by using the lac promoter. (C) pCMV-TPInt expresses the integrase in mammalian cells from the cytomegalovirus immediate early promoter. AmpR, ampicillin resistance; ChlorR, chloramphenicol resistance; KanR, kanamycin resistance; SV40 polyA, poly(A) addition site from simian virus 40.
A 53-bp attB was constructed by treating with kinase and annealing the oligonucleotides 5′TCGACGGGATATCGCAAAAAAAGCAAAAAGCATTTACCTTGATTGAGATGTTAATTGTGTTGGCATGAA3′ and 5′AGCTTTCATGCCAACACAATTAACATCTCAATCAAGGTAAATGCTTTTTGCTTTTTTTGCGATATCCCG3′, which were then ligated into SalI-HindIII-digested pBCβGal, generating plasmid pBB-B53. A 56-bp attP was constructed by treating with kinase and annealing the oligonucleotides 5′GGCCAAGCTTTCCAACTCGCTTAATTGCGAGTTTTTATTTCGTTTATTTCAATTAAGGTAACTAAAAAA3′ and 5′CTAGTTTTTTAGTTACCTTAATTGAAATAAACGAAATAAAAACTCGCAATTAAGCGAGTTGGAAAGCTTGGCCGC3′, which was followed by ligation into the SacII and SpeI sites of pBB-attB53 in order to create the assay plasmid pBB-B53-P56. For each additional att site reduction plasmid (Fig. 2), kinase-treated and annealed oligonucleotides of the reduced attB and attP sites were ligated into this vector, replacing attB53 and attP56, respectively.
FIG. 2.
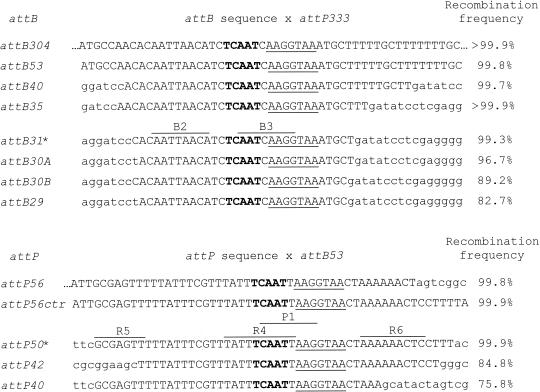
Efficiency of integrative recombination mediated by TP901-1 integrase between att sites of various sizes in E. coli. For each sequence shown, the uppercase letters represent att DNA sequences, while the lowercase letters represent flanking vector sequences. Boldface type indicates the 5-bp common core, and the underlined sequence is the 7-bp identical region shared by attB and attP. Previously identified repeats are indicated by lines above the attB31 and attP50 sequences, as discussed in the text. The asterisks next to these two att sites indicate that they are the smallest sites that were still fully active in this assay. The recombination frequency is the intramolecular integration frequency, calculated by determining the ratio of white colonies to total colonies and multiplying by 100. Each att site is followed by a number indicating the length of the att DNA sequence. In attB30A and attB30B there were deletions from the left and right sides of the sequence, respectively. attP56ctr is an attP sequence in which the att base pairs are centered precisely around the 5-bp core. Each attB was tested by integrative recombination against a 333-bp attP, while each attP was tested against a 53-bp attB. Frequency calculations were made by using bacterial strain DH-TPInt and are based on total numbers of bacterial colonies ranging from 500 to 8,800.
Integrase expression plasmids.
The TP901-1 integrase gene orf1 was PCR amplified from DNA extracted from L. lactis subsp. cremoris strain 901-1 by using the primers 5′GCCATTAGACTAGTGGTACAAAAACAATGACTAAG3′ and 5′CGAGTTGGGATCCCTCGCAATTAAGCGAGTTGG3′. The PCR product was ligated into vector pCR2.1, generating plasmid pTA-TPInt. Plasmid pInt (9) contains the φC31 integrase gene under the control of the bacterial lacZ promoter. The φC31 integrase gene was removed by digestion with BamHI and SpeI and was replaced by a BamHI-SpeI fragment from pTA-TPInt containing the TP901-1 integrase, generating the bacterial expression plasmid pTPInt (Fig. 1B). Plasmid pCMVInt (9) expresses the φC31 integrase in mammalian cells. The φC31 integrase gene was removed by digestion with BamHI and SpeI and replaced with a BamHI-SpeI fragment containing the TP901-1 integrase gene. This ligation generated the mammalian expression plasmid pCMV-TPInt, in which the cytomegalovirus immediate early promoter drives expression of the TP901-1 integrase (Fig. 1C). The TP901-1 integrase gene plus 422 bp of upstream sequence, including orfA, was amplified from L. lactis subsp. cremoris strain 901-1 by using the primers 5′GCCATTAGACTAGTGATATTCGGCAAAAAGTTTACCG3′ and 5′CGAGTTGGGATCCCTCGCAATTAAGCGAGTTGG3′. The PCR product was ligated into vector pCR2.1, generating plasmid pTA-TPInt+orfA. The TPInt+orfA BamHI-SpeI fragment was then ligated into pInt as described above for pTPInt to generate the bacterial expression plasmid pTPInt+orfA.
Plasmids for production of TP901-1 integrase.
The TP901-1 integrase gene was PCR amplified from pCMV-TPInt with the primers 5′GTCTAGAAATTAAGAAGGAGATAATGACTAAGAAAGTAGCAATCTAT3′ and 5′TGGATCCCAATTAAGCGAGTTGGAATTT3′. The PCR product was ligated into pCR2.1 (Invitrogen) to create plasmid pTA-TP901. The integrase gene was removed from pTA-TP901 by XbaI and BamHI digestion and ligated into XbaI- and BamHI-digested pET-11a (Novagen, Madison, Wis.), generating plasmid pET11-TP901. This plasmid contains the TP901-1 integrase gene located 24 bp downstream of the T7 promoter.
Bacterial intramolecular integration assay.
E. coli DH10B was transformed with pTPInt, grown under kanamycin selection conditions, and made electrocompetent by standard protocols. The resulting strain DH-TPInt cells were then used for the intramolecular integration bacterial assay. Twenty nanograms of assay plasmid DNA was electroporated into DH-TPInt cells, allowed to recover, and then plated on Luria-Bertani (LB) broth plates containing 25 μg of chloramphenicol per ml, 60 μg of kanamycin per ml, and 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml and grown at 37°C. Intramolecular integration resulted in excision of lacZ from the plasmid, producing a white colony. The frequency of intramolecular integration was calculated by dividing the number of white colonies by the total number of colonies and multiplying the quotient by 100.
Mammalian intramolecular integration assay.
Human 293 cells (8) were grown in Dulbecco's modified Eagle medium supplemented with 9% fetal bovine serum and 1% penicillin/streptomycin to 60 to 80% confluence on 60-mm-diameter dishes. The cells were then transfected with 250 μg of pBB-B304-P333 assay plasmid or its reduced-size derivatives, 4 μg of pCMV-TPInt or salmon sperm DNA, and 12.75 μl of Lipofectamine (Gibco BRL). At 24 h after transfection, DNase I was added to the medium at a concentration of 50 U/ml in order to reduce the background of untransfected DNA. Plasmid DNA was isolated by the Hirt method (10) 72 h after transfection. A fraction of this DNA was then transformed into electrocompetent E. coli DH10B and plated on LB medium plates containing X-Gal and chloramphenicol, which selected for the assay plasmid but not the integrase plasmid. The intramolecular integration frequency was calculated by dividing the number of white colonies by the total number of colonies and multiplying the quotient by 100.
TP901-1 integrase production.
E. coli strain BL21-SI (Invitrogen) containing plasmid pET11-TP901 was grown at 30°C (to promote solubility) in 100 ml of LB broth without NaCl but with 100 μg of ampicillin per ml to an optical density at 600 nm of 0.727. The culture was induced with 0.3 M NaCl and 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) and grown for another 5 h. In this strain, salt and IPTG induce expression of the T7 polymerase, which in turn transcribes the TP901-1 integrase gene. The cells were resuspended in phosphate-buffered saline (pH 7.4) containing 10% glycerol and sonicated for 45 s on ice. The protein concentration was measured by using the Bio-Rad protein assay (Bio-Rad, Hercules, Calif.). The protein extract was tested for TP901-1 integrase function in vitro with the lacZ intramolecular integration assay.
In vitro assays with TP901-1 integrase protein extract.
To test the integrase function of the protein extract, various amounts of the extract were incubated with 500 ng of pBB-B304-P333 in binding buffer (2 mM Tris-HCl [pH 7.5], 10 mM NaCl, 0.1% glycerol, 10 μM EDTA) (27) in 20-μl reaction mixtures at 30°C for 16 h. The reaction mixtures were heat killed at 65°C for 20 min, and the DNA was purified by using a QIAquick PCR purification kit (Qiagen, Valencia, Calif.). Each reaction mixture was transformed into E. coli DH10B, and each transformation mixture was plated on LB medium with 100 μg of chloramphenicol per ml. The plates were incubated overnight at 37°C, and 24 h later the colonies were counted and the recombination frequency was calculated by dividing the number of white colonies by the total number of colonies and multiplying the quotient by 100. In both the time course and temperature optimization experiments we used 20-μl reaction mixtures containing 500 ng of pBB-B304-P333 in binding buffer with 29.6 μg of TP901-1 integrase protein extract. The reaction mixtures were incubated at 37°C, and the cells were heat killed by incubation at 65°C for 20 min at zero time and after 1, 2, 4, 8, and 15 h for the time course studies. Reaction mixtures were incubated at various temperatures for 16 h before the cells were heat killed in order to obtain a temperature curve.
RESULTS
Intramolecular integration is catalyzed by TP901-1 integrase in E. coli.
In order to assess the ability of the phage TP901-1 integrase to function outside the environment of its native gram-positive L. lactis host bacteria, we chose first to determine its functionality in the convenient laboratory gram-negative species E. coli. The attB and attP sites recognized by the TP901-1 integrase and the attL and attR sites that result after recombination contain a 5-bp TCAAT common core separated by a 1-bp mismatch from a 7-bp identical region (7) (Fig. 2). The sites are AT rich and contain several direct and inverted repeats that may be involved in integrase binding (4, 7). We constructed an assay plasmid, pBB-B304-P333, that carries the 304-bp TP901-1 bacterial attachment site attB and the 333-bp phage attachment site attP (5), in the same orientation, flanking the lacZ gene (Fig. 1A). Without integrative recombination, the pBB-B304-P333 plasmid gives rise to bacterial colonies that are blue on X-Gal indicator plates due to expression of lacZ. Intramolecular integration results in deletion of the lacZ gene, which is manifested by white colonies on X-Gal plates.
The only TP901-1 phage sequences required for integration into the L. lactis genome are attP, the orf1 gene, and 425 bp of sequence upstream of orf1 (6). orf1 encodes the 485-amino-acid TP901-1 integrase and is located just upstream of attP. The amino-terminal 150 to 180 amino acids of the integrase show ∼40% similarity to the amino-terminal catalytic domain of recombinases of the serine-catalyzed family and include the catalytic serine 12 residue (6). The extended carboxy-terminal region exhibits little identity with known proteins and presumably includes the DNA recognition domain. The 425 bp upstream of orf1 probably contains the native promoter for the gene but also encodes a 60-amino-acid orfA reading frame that is not likely to be expressed (6).
To test the ability of orf1 and orf1 plus orfA to mediate intramolecular integration in E. coli, we constructed pTPInt and pTPInt+orfA, bacterial expression plasmids in which the TP901-1 integrase gene is under control of the E. coli lacZ promoter. These expression plasmids are compatible with the pBB-B304-P333 assay plasmid. We established these integrase expression plasmids in E. coli DH10B, generating the TP901-1 integrase expression strains DH-TPInt and DH-TPInt+orfA, respectively. We assayed the frequency of intramolecular integration of the assay plasmid pBB-B304-P333 in the DH-TPInt+orfA and DH-TPInt bacterial strains, calculating the frequency of intramolecular integration by counting white bacterial colonies on X-Gal plates. In both strains, the integrative recombination frequency was >99% (Fig. 2). Restriction analysis of DNA purified from white bacterial colonies showed a digestion pattern consistent with a precise integration reaction between attB and attP, resulting in excision of the lacZ gene. This result indicated that the TP901-1 integrase was capable of functioning efficiently in E. coli and that the integrase protein was sufficient to carry out the integrative recombination reaction without Lactococcus-specific cofactors. The 425-bp upstream region was unnecessary, presumably because in our E. coli expression plasmid the lacZ promoter drove expression of the integrase and eliminated any dependence on the native promoter. Based on this result, the rest of our experiments were carried out with the TP901-1 integrase orf1 gene only, without the upstream sequences.
Minimal attB and attP sites required for efficient TP901-1 integrase activity.
After demonstrating that the TP901-1 integrase functioned efficiently in E. coli, we wanted to determine the minimal att sites recognized by the enzyme. We first analyzed the ability of the TP901-1 integrase to catalyze integrative recombination between symmetrically shortened attB sites and a 333-bp attP. As shown in Fig. 2, TP901-1 integrase efficiently recombined shortened attB sites ranging in length from 53 bp down to just 31 bp. The 31-bp attB still resulted in an integrative recombination frequency of >99% when it was paired with the full-length 333-bp attP (Fig. 2), as well as with a 56-bp attP (data not shown). However, reduction of the length of attB to 30 bp resulted in some loss of activity, and further shortening of attB to 29 bp resulted in reduction of the integrative recombination frequency to 82.7%.
Shortened attP sequences were tested in combination with attB53. attP sites that were 56 bp long, either asymmetrically disposed around the core (5) or centered on the core, retained full activity (Fig. 2). attP shortened to 50 bp still showed full integrative recombination activity, but shortening the attP site to 42 bp reduced the integrase activity to ∼85%. We concluded that at least in the context of intramolecular recombination in E. coli, an attB that was 31 bp long sufficed, while an attP that was approximately 50 bp long was needed for full activity.
In vitro studies.
A crude protein extract containing TP901-1 integrase was made by lysis of E. coli containing plasmid pET11-TP901 expressing the integrase from the T7 promoter. When samples of the culture taken at different times after induction of integrase gene expression were boiled in loading buffer and electrophoresed on a denaturing gel, a band at approximately the correct size, 55 kDa, appeared, and the intensity increased with time. The same band was also visible in the crude extract. The measured concentration of the extract was 7.4 mg/ml. The activity of the extract was tested in vitro by using the lacZ intramolecular integration assay, with scoring after transformation of in vitro-reacted DNA into E. coli lacking integrase. As expected, the integrative recombination frequency increased as the amount of protein extract increased. A maximum recombination frequency of 99% was obtained when 59.2 μg of protein was used. The postrecombination attR sites of four white colonies were sequenced to ensure that the TP901-1 integrase was completing the correct site-specific integration reaction, and all four colonies had the expected DNA sequence for a precise integrase-mediated event between attB and attP.
TP901-1 integrase showed good stability in vitro over time, with the integrative recombination efficiency increasing over 15 h (Fig. 3A). Between 2 and 4 h, the percentage of recombination in vitro increased from 6 to 36%; this was the interval with the largest recombination increase in the experiment. By 15 h, the percentage of in vitro recombination was 90%. The control reaction mixtures containing a crude protein extract made with the pET11 backbone in E. coli BL21-SI showed no integrative recombination in the in vitro assay.
FIG. 3.
Characteristics of TP901-1 integrase-mediated in vitro reactions. (A) Percentages of in vitro recombination over time. (B) Percentages of in vitro recombination in 16 h at various temperatures. Integrative recombination was assayed by transforming a reaction mixture containing pBB-B304-P333 into DH-TPInt and counting white colonies on X-Gal plates.
The TP901-1 integrase was relatively sensitive to temperature variations in vitro. The maximum integrative recombination frequency was seen at temperatures between 30 and 37°C. The average in vitro integrative recombination frequency for this temperature range was 86%. Outside this range, the integrative recombination frequency decreased sharply. The integrase did not work well at temperatures below 17°C, giving an average in vitro integrative recombination frequency of 11% at temperatures less than 17°C. The integrative recombination frequency fell to <0.1% at 50°C. Protein heated at 65°C for 20 min before incubation with substrate also gave an in vitro integrative recombination frequency of <0.1%, showing that the TP901-1 integrase could be heat killed. Control reaction mixtures with a protein extract lacking the integrase did not show any integrative recombination in this assay.
TP901-1 integrase catalyzes intramolecular integration in human cells.
The ability of TP901-1 integrase to function in E. coli and in vitro in the absence of added cofactors created the possibility that the enzyme could also function in mammalian cells. We constructed pCMV-TPInt (Fig. 1C) to place the integrase gene under expression of a promoter active in mammalian cells. To analyze TP901-1 integrase function in mammalian cells, the intramolecular integration assay plasmid pBB-B304-P333 (Fig. 1A) and plasmid pCMV-TPInt were cotransfected into human 293 cells. Seventy-two hours after transfection, plasmid DNA was extracted, transformed into E. coli DH10B lacking integrase activity, and spread on X-Gal indicator plates to determine whether intramolecular integration had occurred in the mammalian environment. The frequency of intramolecular integration in the mammalian cells was determined by counting white bacterial colonies.
As shown in Table 1, in the absence of the integrase plasmid, a background of 1.9% white colonies was obtained for a plasmid carrying full-length attB and attP sites. These colonies harbored plasmids that contained nonspecific deletions and other mutations engendered by the transfection process (14). Cotransfection of pBB-B304-P333 and the integrase expression plasmid yielded white colonies at a frequency of 23.2% (Table 1). PCR analysis of plasmid DNA extracted from 66 white colonies showed that >95% of the samples represented correct site-specific integration events, as shown by amplification of an expected 623-bp fragment containing attR. DNA sequencing of four junctions confirmed that the attB-attP recombination reaction was site specific and perfect to the base. Restriction analysis of the 66 plasmid samples showed that the 95% that underwent integrative recombination had no concurrent rearrangements. The remaining 5% of the events represented rearrangements of the assay plasmid, corresponding to the transfectional mutation rate observed when pBBBP-type plasmids were transfected without pCMV-TPInt. Controls were included to provide assurance that the integrative recombination events occurred in the human cells, not in E. coli. Direct transformation of pBB-B304-P333 into DH10B failed to produce white colonies. Likewise, transformation of pBB-B304-P333 plus pCMV-TPInt directly into DH10B produced negligible white colonies. In addition, a PCR was performed with the plasmid DNA extracted from the human cells before transformation into E. coli, and the 623-bp fragment diagnostic of site-specific integration was readily detected.
TABLE 1.
Intramolecular integration frequencies in human cells
| Vector(s) transfecteda | No. of white colonies | Total no. of colonies | Intramolecular integration frequency (%)b |
|---|---|---|---|
| pCMV-TPInt (4) | 0 | 0 | NAc |
| pBB-B304-P333 (5) | 197 | 10,462 | 1.9 ± 1.0 |
| pBB-B304-P333 + pCMV-TPInt (3) | 1,420 | 6,124 | 23.2 ± 5.4 |
| pBB-B53-P56 (2) | 94 | 3,470 | 2.7 ± 0.2 |
| pBB-B53-P56 + pCMV-TPInt (3) | 2,845 | 9,165 | 31.0 ± 1.6 |
| pBB-B31-P56 (3) | 68 | 1,607 | 4.2 ± 1.3 |
| pBB-B31-P56 + pCMV-TPInt (3) | 550 | 2,040 | 27.0 ± 2.3 |
The numbers in parentheses indicate the numbers of independent transfections analyzed. The numbers in the plasmid designations for pBBBP derivatives correspond to the sizes of the attB and attP sites in the plasmids.
Mean ± standard deviation.
NA, not applicable.
The frequency of TP901-1 integrase-mediated intramolecular recombination between smaller attB and attP sites in mammalian cells was also measured. Reduction of the size of attB to 53 bp and the size of attP to 56 bp still resulted in an intramolecular integration frequency of 31.0% (Table 1). Further shortening of attB to 31 bp also did not affect the ability of the integrase to recombine in the human cell environment with a 56-bp attP. This integrative recombination reaction occurred with a frequency of 27.0% (Table 1), which is similar to the frequency of recombination between two full-length att sites (Table 1). These recombination values are probably underestimates, because it is likely that not all of the plasmid DNA extracted from the human cells and scored in E. coli was exposed to the integrase.
DISCUSSION
This study illustrates that the integrase from phage TP901-1 is a robust and autonomous enzyme that can mediate site-specific integrative recombination between compact attB and attP sites in environments remote from its native host, including human cells.
By using an intramolecular integration assay in E. coli, we characterized minimal recognition sites for the enzyme that were 50 bp long for attP and just 31 bp long for attB. These sites are somewhat smaller than the minimal 56-bp attP site (5) and 43-bp attB site (4) previously reported. In the previous cases, the minimal att sites were asymmetrically disposed about the 5-bp core, with most of the sequence upstream of the core, whereas the minimal att sites found in this study are centered on the 5-bp core. Numerous paired direct and inverted repeats have been identified within attP (5, 7) and attB (4) that may be involved in the integrase binding and recombination function. Our minimal attB sequence includes the B2 and B3 repeats (4), although only a single copy of each repeat is present (Fig. 2). Our minimal attP site includes the P1, R4, R5, and R6 repeats (5) (Fig. 2), although again only one copy of each repeat, not the pair, is present. These reduced sites function as well as full-length sites that are over 300 bp long in both E. coli (Fig. 2) and human cells (Table 1). The significance of the repeats is therefore unclear. The second copy present in the full-size att sites might provide enhancement of recombination that is undetectable with our assay because it is fully saturated.
The minimal attB31 sequence contains the C17 and A25 bases that have been shown to both reduce in vitro binding of the integrase and reduce recombination in E. coli (4). The TP901-1 integrase binds with similar affinity in vitro to attB and attP (4), as is also the case for the φC31 integrase (28), another member of the extended serine recombinase group. By mutating each base in the attB 5-bp core, it was revealed that only the first two bases are important (4). This result suggested that there is a 2-bp overlap region between attB and attP during recombination, as has been observed for other members of the serine recombinase family (24).
Our major interest in the TP901-1 integrase is in its potential as a tool for making directed genomic rearrangements in eukaryotic cells. Our in vitro studies with this enzyme reinforce the hypothesis that it possesses good reaction kinetics and stability and works well at 37°C (Fig. 3), making it eligible for use in mammalian cells. The small size of the att sites is consistent with a lack of cofactor requirements, which is also a highly desirable feature because it simplifies use of the enzyme in foreign hosts. The efficient function of the enzyme in vitro and in E. coli documented here and recently by other workers (4) and the positive results for human cells reported here suggest that the TP901-1 integrase should function in most cellular environments in a wide range of species. The enzyme joins the other two integrases of the extended serine recombinase family, φC31 (9, 26, 30) and R4 (19), in this respect. The intramolecular integration activity of the TP901-1 integrase in human cells is within twofold of the activities measured for the φC31 (9) and R4 (19) integrases in similar assays.
This study demonstrated efficient function of the enzyme in an intramolecular integration reaction in human cells (Table 1). This type of reaction is useful for creating chromosome rearrangements, such as deletions. We are currently investigating the ability of the enzyme to carry out such reactions in a mammalian chromosomal context. Another major area of interest is the enzyme's ability to carry out intermolecular integration reactions. The enzyme was designed to carry out such reactions in the normal phage life cycle, mediating integration between an incoming phage genome and the host bacterial genome. Plasmid integration vectors carrying attP mediate intermolecular integration into the native host attB site at a frequency of ∼20% (6). This integration reaction is largely unidirectional. A second phage protein, an excisionase encoded by orf7, is required to bring about an efficient reaction between attR and attL, which is required for phage excision (3). To maximize the integration reaction, the excisionase is simply left out.
It may be possible to use the TP901-1 integrase as an integration tool targeted to inserted wild-type att sites. We have obtained evidence for integration of a plasmid containing the TP901-1 attB site into a TP901-1 attP site that was placed into the genome of human 293 cells (Stoll and Calos, unpublished results). The TP901-1 integrase is therefore capable of intermolecular integration into chromosomes in the human cell environment. As well, the short length of the att sites makes feasible the idea of using native chromosomal sequences resembling the att sites as integration targets. Such pseudo-recombination binding sites exist in the case of the Cre recombinase (29) and the φC31 and R4 integrases (19, 30). The sequences of the att sites recognized by the TP901-1 integrase differ from those recognized by these other two integrases and offer additional points of entry into the genome, especially in AT-rich regions. It may be possible to enhance the reactivity of the enzyme with chromosomal sequences and to improve its affinity for particular native sequences by DNA shuffling (25) of the integrase gene, as has been shown for the φC31 integrase (23). If so, the TP901-1 integrase may become an important tool for engineering the genomes of higher living cells.
Acknowledgments
We thank Horst Neve for supplying L. lactis subsp. cremoris strain 901-1.
S.M.S. and D.S.G. were supported by a graduate training grant from NIH. This work was supported by NIH grant DK58187 to M.P.C.
REFERENCES
- 1.Abremski, K., and R. Hoess. 1984. Bacteriophage P1 site-specific recombination, purification and properties of the Cre recombinase protein. J. Biol. Chem. 259:1509-1514. [PubMed] [Google Scholar]
- 2.Braun, V., H. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 3.Breuner, A., L. Bronsted, and K. Hammer. 1999. Novel organization of genes involved in prophage excision identified in the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7291-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breuner, A., L. Bronsted, and K. Hammer. 2001. Resolvase-like recombination perfomed by the TP901-1 integrase. Microbiology 147:2051-2063. [DOI] [PubMed] [Google Scholar]
- 5.Brondsted, L., and K. Hammer. 1999. Use of the integration elements encoded by the temperate lactococcal bacteriophage TP901-1 to obtain chromosomal single-copy transcriptional fusions in Lactococcus lactis. Appl. Environ. Microbiol. 65:752-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen, B., L. Brondsted, F. K. Vogensen, and K. Hammer. 1996. A resolvase-like protein is required for the site-specific integration of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 178:5164-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen, B., M. G. Johnsen, A. Stenby, F. K. Vogensen, and K. Hammer. 1994. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J. Bacteriol. 176:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-72. [DOI] [PubMed] [Google Scholar]
- 9.Groth, A. C., E. C. Olivares, B. Thyagarajan, and M. P. Calos. 2000. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. USA 97:5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 11.Kolot, M., N. Silberstein, and E. Yagil. 1999. Site-specific recombination in mammalian cells expressing the Int recombinase of bacteriophage HK022. Mol. Biol. Rep. 26:207-213. [DOI] [PubMed] [Google Scholar]
- 12.Kuhstoss, S., and R. N. Rao. 1991. Analysis of the integration function of the streptomycete bacteriophage φC31. J. Mol. Biol. 222:897-908. [DOI] [PubMed] [Google Scholar]
- 13.Landy, A. 1989. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 58:913-949. [DOI] [PubMed] [Google Scholar]
- 14.Lebkowski, J. S., R. B. DuBridge, E. A. Antell, K. S. Greisen, and M. P. Calos. 1984. Transfected DNA is mutated in monkey, mouse, and human cells. Mol. Cell. Biol. 4:1951-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorbach, E. N., M. Christ, M. Schwikardi, and P. Droge. 2000. Site-specific recombination in human cells catalyzed by phage lambda integrase mutants. J. Mol. Biol. 296:1175-1181. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura, M., T. Noguchi, D. Yamaguchi, T. Aida, M. Asayama, H. Takahashi, and M. Shirai. 1996. The sre gene (ORF469) encodes a site-specific recombinase responsible for integration of the R4 phage genome. J. Bacteriol. 178:3374-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Gorman, S., D. T. Fox, and G. M. Wahl. 1991. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251:1351-1355. [DOI] [PubMed] [Google Scholar]
- 19.Olivares, E. C., R. P. Hollis, and M. P. Calos. 2001. Phage R4 integrase mediates efficient integration in mammalian cells. Gene 278:167-176. [DOI] [PubMed] [Google Scholar]
- 20.Rausch, H., and M. Lehmann. 1991. Structural analysis of the actinophage φC31 attachment site. Nucleic Acids Res. 19:5187-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauer, B. 1994. Site-specific recombination: developments and applications. Curr. Opin. Biotechnol. 5:521-527. [DOI] [PubMed] [Google Scholar]
- 22.Sauer, B., and N. Henderson. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85:5166-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sclimenti, C. R., B. Thyagarajan, and M. P. Calos. 2001. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 29:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark, W. M., M. R. Boocock, and D. J. Sherratt. 1992. Catalysis by site-specific recombinases. Trends Genet. 8:432-439. [PubMed] [Google Scholar]
- 25.Stemmer, W. P. C. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomason, L. C., R. Calendar, and D. W. Ow. 2001. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage φC31 site-specific recombination system. Mol. Genet. Genomics 265:1031-1038. [DOI] [PubMed] [Google Scholar]
- 27.Thorpe, H. M., and M. C. M. Smith. 1998. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. USA 95:5505-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorpe, H. M., S. E. Wilson, and M. C. M. Smith. 2000. Control of directionality in the site-specific recombination system of the Streptomyces phage φC31. Mol. Microbiol. 38:232-241. [DOI] [PubMed] [Google Scholar]
- 29.Thyagarajan, B., M. J. Guimaraes, A. C. Groth, and M. P. Calos. 2000. Mammalian genomes contain active recombinase recognition sites. Gene 244:47-54. [DOI] [PubMed] [Google Scholar]
- 30.Thyagarajan, B., E. C. Olivares, R. P. Hollis, D. S. Ginsburg, and M. P. Calos. 2001. Site-specific genomic integration in mammalian cells mediated by phage φC31 integrase. Mol. Cell. Biol. 21:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]