Abstract
Sinorhizobium meliloti is a free-living soil bacterium which is capable of establishing a symbiotic relationship with the alfalfa plant (Medicago sativa). This symbiosis involves a network of bacterium-host signaling, as well as the potential for bacterium-bacterium communication, such as quorum sensing. In this study, we characterized the production of N-acyl homoserine lactones (AHLs) by two commonly used S. meliloti strains, AK631 and Rm1021. We found that AK631 produces at least nine different AHLs, while Rm1021 produces only a subset of these molecules. To address the difference in AHL patterns between the strains, we developed a novel screening method to identify the genes affecting AHL synthesis. With this screening method, chromosomal groEL (groELc) was shown to be required for synthesis of the AHLs that are unique to AK631 but not for synthesis of the AHLs that are made by both AK631 and Rm1021. We then used the screening procedure to identify a mutation in a gene homologous to traM of Agrobacterium tumefaciens, which was able to suppress the phenotype of the groELc mutation. A traR homolog was identified immediately upstream of traM, and we propose that its gene product requires a functional groELc for activity and is also responsible for inducing the synthesis of the AHLs that are unique to AK631. We show that the traR/traM locus is part of a quorum-sensing system unique to AK631 and propose that this locus is involved in regulating conjugal plasmid transfer. We also present evidence for the existence of a second quorum-sensing system, sinR/sinI, which is present in both AK631 and Rm1021.
Cell density-dependent gene expression, termed quorum sensing, is recognized as a widespread phenomenon in both gram-negative and gram-positive bacteria. The model organism for this form of gene regulation in gram-negative bacteria is Photobacterium fischeri, a luminescent marine bacterium. The process begins with the low-level production of an autoinducer signal at low cell densities, like those found in seawater. The autoinducers are thought to pass through the cell membrane by diffusion, so as the cell density increases during symbiotic association with the squid host, autoinducers accumulate in and around the cells (28). When a threshold level of autoinducers within the cell is reached, the LuxR regulator becomes activated by binding the autoinducer (23, 28). LuxR then induces expression of the autoinducer synthase gene, luxI, along with the genes necessary for luminescence (8, 9).
In addition to P. fischeri, quorum sensing has been identified in numerous other organisms, including Agrobacterium tumefaciens, Pseudomonas aeruginosa, Erwinia carotovora, and several others (for recent reviews see references 5, 15, and 36). Most of the quorum-sensing systems characterized so far occur in bacteria that are able to establish relationships, either pathogenic or symbiotic, with plant or animal hosts. The quorum-sensing mechanism in these cases usually regulates one or more genes that play a role in pathogenesis or symbiosis. For instance, in nature the P. fischeri lux genes for luminescence are activated only during association with a squid or fish host, in which the bacteria reach high cell densities (21, 24, 35). A. tumefaciens, which causes crown gall tumors in susceptible plants, uses quorum sensing to regulate transfer of the pathogenic Ti plasmid through luxR and luxI homologs, traR and traI (16, 26). Quorum sensing in this case is under an additional type of regulation since traR is induced by opines, which are released by the tumors. P. aeruginosa possesses a slightly more complex quorum-sensing system that is composed of two sets of luxR/luxI homologs, lasR/lasI and rhlR/rhlI, which function in a hierarchical manner to control the production of various enzymes and toxins that are required for virulence (29, 53).
Several Rhizobium species have recently been shown to produce N-acyl homoserine lactones (AHLs) (3, 22, 42, 45, 47). Rosemeyer et al. observed that in Rhizobium etli quorum sensing seemed to be involved in restricting the number of nodules formed during invasion of the common bean, Phaseolus vulgaris, although the mechanism has not been elucidated. Rhizobium leguminosarum also possesses a quorum-sensing system that is involved in restricting nodulation number, as well as in causing the early onset of the stationary phase and inducing the rhiABC operon (4, 22, 41). Regulation of quorum sensing in R. leguminosarum is not yet completely understood, but a recent report has shown that there is evidence of a complex network reminiscent of the lasR/rhlR hierarchy in P. aeruginosa (31).
Sinorhizobium meliloti (formerly Rhizobium meliloti) is a gram-negative soil bacterium which is capable of establishing a symbiotic relationship with the alfalfa plant (Medicago sativa). Shaw et al. (47) and Cha et al. (3) have shown that several strains of S. meliloti produced one or more AHLs. This suggests that quorum sensing exists in S. meliloti, as it does in R. etli and R. leguminosarum. However, the role, if any, of quorum sensing in the S. meliloti-alfalfa relationship remains unclear.
In this paper, we begin to characterize the quorum-sensing systems in two well-characterized S. meliloti strains, AK631 (a derivative of Rm41) and Rm1021, which are commonly used to study the symbiotic process in alfalfa. Despite their close relationship, these two strains have been shown to differ in several ways, including exopolysaccharide production, host restriction systems, and the number of plasmids which they carry (2, 10, 51, 52). Here we show that AK631 and Rm1021 also differ in their quorum-sensing systems. We identified two mutations that have an effect on AHL production in AK631. A mutation in the chromosomal groEL (groELc) results in decreased levels of AHLs, and a double mutant containing mutations in groELc and a traM homolog produces AHL levels slightly higher than those produced by the wild type. The phenotypes of these mutants are consistent with data from the P. fischeri and A. tumefaciens systems (7, 14, 26) and strongly suggest that a quorum-sensing system exists in S. meliloti. Moreover, we show here that TraM and the corresponding regulator, TraR, are part of a quorum-sensing system that is present in AK631 but not in Rm1021. We obtained sequencing data which shows the presence of putative trbH and trbI homologs immediately upstream of traR. This is consistent with the arrangement of the trb operon in Rhizobium strain NGR234 and R. leguminosarum (13) and suggests that the traR/traM locus is involved in regulating plasmid transfer, as it is in R. leguminosarum (J. A. Downie, personal communication) and A. tumefaciens (16, 39), or in regulating tra gene expression, as it is in Rhizobium strain NGR234 (C.W. Fuqua, personal communication). In addition, the recent release of the S. meliloti Rm1021 genome (17) shows the presence of a set of luxR/luxI homologs, which we term sinR and sinI, respectively. We show that these genes are common to both AK631 and Rm1021, and the locus is sufficient to support AHL production in Escherichia coli. Therefore, if quorum sensing is involved in symbiosis in S. meliloti, as it is in other rhizobia, these results may provide some insight into the relationship between alfalfa and S. meliloti, one of the most widely studied of the symbiotic nitrogen fixers.
MATERIALS AND METHODS
Bacterial strains and media.
Table 1 lists the strains used in this work. All S. meliloti strains were cultured in Luria-Bertani (LB) broth supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4 (LB/MC). Antibiotics were added, as appropriate, at the following final concentrations: streptomycin, 500 μg/ml; neomycin, 200 μg/ml; tetracycline, 10 μg/ml; and gentamicin, 50 μg/ml. The concentrations were adjusted for the combination of tetracycline (5 μg/ml) and neomycin (100 μg/ml). A. tumefaciens NTL4(pZLR4) was grown on LB medium containing 50 μg of gentamicin per ml. Chromobacterium violaceum CV026 was grown on LB medium without drugs. E. coli strains were grown on LB medium containing 25 μg of kanamycin per ml, 5 μg of gentamicin per ml, 10 μg of tetracycline per ml, 100 μg of ampicillin per ml, 20 μg of chloramphenicol per ml, or 100 μg of spectinomycin per ml as needed. The concentrations were adjusted for the combination of tetracycline (5 μg/ml) and kanamycin (10 μg/ml). S. meliloti and A. tumefaciens strains were grown at 30°C, while C. violaceum and E. coli were grown at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| S. meliloti strains | ||
| AK631 | Rm41 exoB | 12 |
| Rm1021 | SU47 str-21 | 30 |
| Rm11500 | Spontaneous Smr mutant of AK631 | This study |
| Rm11501 (Tn5) | Rm11500 containing groELc1583::Tn5 | This study |
| Rm11501 (Tn5-233) | Rm11500 containing groELc1583::Tn5-233 | This study |
| Rm11502 | Rm11501 (Tn5-233) containing traM::Tn5 | This study |
| B4T1 | Rm1021 groELc350::Tn5 #B4 | 38 |
| Rm11507 | Rm11500 containing groELc350::Tn5 from B4T1 | This study |
| Rm11508 | Rm1021 containing groELc1583::Tn5 from Rm11501 | This study |
| Rm11509 | Rm11500 containing traM::Tn5 | This study |
| Rm11510 | Rm1021 containing pRme41a (traM::Tn5) | This study |
| Rm11513 | Rm11500 containing traR::GM | This study |
| Indicator strains | ||
| A. tumefaciens NTL4 (pZLR4) | NT1 derivative carrying a traG::lacZ reporter fusion | 32 |
| C. violaceum CV026 | CV017 derivative containing cviI::Tn5xylE | 34 |
| E. coli strains | ||
| MM294 A | pro-82 thi-I endA hsdR17 supE44 | 20 |
| Epicurian Coli XL10-Gold | —a | Stratagene |
| MT616 | MT607(pRK600) | 20 |
| HB101 | —a | Promega |
| DH5α | —a | Life Technologies |
| Plasmids | ||
| pRK600 | pRK2013 Nmr::Tn9 | 20 |
| pRK602 | pRK600 Ω Tn5 | 20 |
| pRK607 | pRK2013 Ω Tn5-233 | 20 |
| pPH1JI | GmrSpr, IncP plasmid | 20 |
| pLAFR1 | pRK290 containing cos site | 20 |
| pLAFR3 | pLAFR1 containing HaeII fragment of pUC8 | 48 |
| pUCGM | pUC derivative carrying a gentamicin cassette | 46 |
| pBlueKan | pBluescript containing mini-Tn5Km from pCP15 | L. Reitzer |
| pTraM | pLAFR1 library clone from Rm11500 containing traM | This study |
| pSinRI | pPCR-Script Amp SK (+) carrying the sinR/sinI locus | This study |
| pTraR | pLAFR1 library clone from Rm11500 containing traR | This study |
| pTraR::GM | pPCR-Script Amp SK (+) carrying traR::GM | This study |
| pTraRsub | traR-EcoRI fragment from pTraR in pPCR-Script Amp SK (+) | This study |
| pTraRsub::GM | pTraRsub carrying the traR::GM disruption | This study |
| pTraRsub::GM/sacB | pJQ200SmSp carrying traR::GM from pTraRsub::GM | This study |
| pJQ200SK | Shuttle vector carrying sacB | 40 |
| pUTminiTn5Sm/Sp | pUT derivative carrying a streptomycin-spectinomycin cassette | 6 |
—, see source for characteristics.
Screening for AHL-producing mutants.
Random transposon mutagenesis of an Smr derivative of AK631 termed Rm11500 was performed by using E. coli MM294(pRK602) as previously described (20). For transposon mutagenesis of strains already harboring a Tn5 insertion, the Tn5 was first replaced with Tn5-233 by using MM294(pRK607), as described previously (20), and this was followed by mutagenesis with MM294(pRK602). The mutants obtained from transposon mutagenesis were then plated on LB/MC containing antibiotics and incubated at 30°C until colonies were just visible. The colonies were replicated onto LB/MC lacking drugs and incubated at 30°C overnight. The replicated colonies were then overlaid with the indicator organism (preparation of the overlay is described below) and incubated at 30°C overnight to allow the indicator organism to grow and color to develop.
Inverse PCR, cloning, and sequencing of Tn5 mutants.
Chromosomal DNA from transposon-containing mutants was digested with XmnI or SmaI, which cut within Tn5, and with StuI and EcoRV, which cut outside the Tn5 region. The resulting fragments were circularized in a large volume to promote intramolecular ligation. PCR was then performed by using primers complementary to the transposon's IS50 region and 30 cycles of 30 s at 95°C, 30 s at 53°C, and 2 min at 72°C. The PCR fragments were purified, polished, and cloned by using a Stratagene PCR-Script Amp cloning kit. The cloned fragments were sequenced and then analyzed by using a BLAST search (1) and the Jotun Hein alignment method of the DNASTAR Megalign program.
Cloning of traM and traR.
The sequence data for Rm11502 and Rhizobium strain NGR234 were used to design primers for the open reading frames of traM (5′-CTAATCGCCATTGTCGAAAGGCATA-3′ and 5′-GCGAAAAGCAGTGTCACTACG-3′) and traR (5′-ACGCGTCGACATGTCCGTGAACGGAAACCTTCGC-3′ and 5′-CCGCTCGAGTCAGACCAGGCCACGGTCC-3′). These oligonucleotides were used to isolate the clones carrying traM and traR from an Rm11500 library. This library was constructed by ligating EcoRI-digested chromosomal DNA into the EcoRI site of pLAFR1 and then transforming the recombinant plasmids into HB101. The library was then divided into aliquots and transferred into a 96-well microtiter dish at a titer of about 30 clones per well. A PCR was performed to screen the wells for the presence of traM or traR by using 30 cycles of 30 s at 95°C, 30 s at 55°C, and 2 min at 72°C. The plasmid carrying traM (pTraM) was then introduced into strains for complementation by triparental mating (20).
Disruption of traR.
To disrupt traR, first the open reading frame was amplified by using the primers described above. The PCR product was polished by using Pfu polymerase and cloned into the EcoRV site of pPCR-Script Amp SK (+). The SmaI-gentamicin cassette from pUC-GM (46) was then ligated into the NruI site of the traR open reading frame, creating pTraR::GM. The disrupted open reading frame was then amplified and used as the primer in the modified PCR protocol described by Geiser et al. (18). The PCR conditions for the modified PCR were as follows: 5 min at 95°C, followed by 18 cycles of 30 s at 95°C, 30 s at 55°C, and 6 min at 72°C and then a final 10-min elongation step at 72°C. The template for the reaction was pTraRsub. pTraRsub was obtained by subcloning the traR EcoRI fragment from the library clone (pTraR) into the EcoRI site of pPCR-Script Amp SK (+). The modified PCR protocol resulted in incorporation of traR::GM into pTraRsub, creating pTraRsub::GM. Next, the shuttle vector pJQ200SmSp was created by inserting the BamHI-streptomycin-spectinomycin cassette from pUTminiTn5Sm/Sp (6) into the BglII site of pJQ200SK (40). The traR::GM disruption was then transferred from pTraRsub::GM as an ApaI-BstXI fragment into pJQ200SmSp, creating pTraRsub::GM/sacB, which was then used to transfer the mutation to Rm11500, and the S. meliloti clone (Rm11513) carrying the chromosomal disruption was selected by plating on media containing the appropriate antibiotics and 5% sucrose (40).
Genetic manipulations between AK631 and Rm1021.
The groELc350::Tn5 mutation in B4T1 (38), an Rm1021 derivative, was transduced into Rm11500 by using φM12h1 (52), creating Rm11507. For the groELc1583::Tn5 mutation in Rm11501, a library was made by ligating EcoRI-digested chromosomal DNA with EcoRI-digested pLAFR3. The library was transformed into DH5α, and the clone containing groELc1583::Tn5 was isolated by selecting for the npt cassette in Tn5. This plasmid was then transferred to Rm1021 by triparental mating. Allelic exchange of the wild-type groELc for the mutant copy was achieved by homogenotization (20), creating Rm11508. The traM::Tn5 mutation in Rm11502 was transduced into a fresh Rm11500 background by using φM12h1. Rm11510 was obtained by conducting biparental mating between Rm1021 and Rm11502, followed by selection for φM12-sensitive and neomycin-resistant colonies.
Cloning of sinR and sinI.
The sinR/sinI locus was amplified from Rm1021 chromosomal DNA by using the following primers: 5′-TCAGGCGGCGCGTGCCGTTTCAAGCG-3′ and 5′-GGTGGCGCGCCTCCTCATTC-3′. The PCR product was polished with Pfu DNA polymerase and then ligated into the EcoRV site of pPCR-Script Amp SK (+). The resulting plasmid, pSinRI, was transformed into XL10-Gold.
Autoinducer bioassays.
For the NTL4(pZLR4) indicator, an overnight culture grown in LB medium with gentamicin was used to prepare a subculture (1:100) in MGM minimal medium (11 g of Na2HPO4 per liter, 3 g of KH2PO4 per liter, 0.5 g of NaCl per liter, 1 g of glutamate per liter, 10 g of mannitol per liter, 1 mg of biotin per liter, 27.8 mg of CaCl2 per liter, 246 mg of MgSO4 per liter) with gentamicin, which was grown for 6 to 8 h at 30°C with shaking. The culture was then mixed with an equal volume of 1.5% Bacto Agar (Difco), 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml was added, and the preparation was used as an overlay. For the CV026 indicator, a single colony was used to inoculate 25 ml of LB medium, and the culture was grown for 4 to 6 h at 30°C with shaking. An equal volume of LB top agar (0.75% Bacto Agar) was then mixed with the culture, which was then used as an overlay. The overlays described above were used to detect AHLs in both thin-layer chromatography (TLC) analyses and genetic screening analyses for AHL-producing mutants by pouring the indicator-containing top agar over a petri plate or a TLC plate and incubating the preparation at 30°C.
Preparation and TLC analysis of crude AHL extracts.
Five-milliliter S. meliloti or E. coli cultures were grown to saturation (optical density at 600 nm for AK631 derivatives, approximately 2.0; optical density at 600 nm for Rm1021 derivatives, 2.4; and optical density at 600 nm for DH5α carrying the sinR/sinI locus, 1.7). Whole cultures were extracted twice with equal volumes of ethyl acetate. The extracts were dried in a Brinkman SpeedVac centrifuge and resuspended in 500 μl of ethyl acetate. An appropriate amount of each crude AHL preparation was then spotted on a Whatman LKC18 analytical TLC plate. TLC plates were chromatographed in 70% methanol-30% water and dried thoroughly. Once dry, the plates were overlaid (see above) with top agar containing the indicator organism and incubated at 30°C overnight.
AHL standards were purchased from Sigma [N-(3-oxo-hexanoyl)-dl-homoserine lactone (oxo-C6 HSL)] and Fluka (N-butyryl-dl-homoserine lactone [C4 HSL], N-hexanoyl-dl-homoserine lactone [C6 HSL], and N-octanoyl-dl-homoserine lactone [C8 HSL]).
Growth curve analysis.
A 2-ml culture of each S. meliloti strain was inoculated with a single colony and grown in LB/MC in the presence of the appropriate antibiotics at 30°C until the culture was saturated. The strains were then subcultured (1:1,000) in LB/MC, and cell density was measured by monitoring the optical density at 600 nm. All growth curve determinations were repeated at least twice.
Nucleotide sequence accession number.
The traR/traM sequence from S. meliloti strain AK631 has been deposited in the GenBank database under accession number AF456804.
RESULTS
Identification of mutations affecting AHL synthesis in S. meliloti.
Previous survey studies have shown the production of AHLs by several different organisms when various indicator strains were used (3, 47). Cha et al. (3) demonstrated that crude supernatant preparations of two S. meliloti strains, Rm41 (parent strain of AK631) and Rm1021, have autoinducer activity. Interestingly, Rm1021 and Rm41 were observed to have different patterns of AHLs. When A. tumefaciens was used as an indicator organism, only one AHL was detected for Rm1021, while at least seven AHLs were detected for Rm41. The difference in AHL patterns suggested that the quorum-sensing systems in Rm1021 and Rm41 differ in one or more respects. Therefore, we set out to characterize quorum sensing in these two organisms.
In an effort to identify the genes involved in quorum sensing, we developed a novel screening procedure which detects deficiencies in AHL levels. In this procedure, mutants of Rm1021 and an AK631 Smr derivative, Rm11500, were generated by using random transposon mutagenesis. The mutants were plated to obtain isolated colonies, and the colonies were overlaid with top agar containing either A. tumefaciens NTL4(pZLR4) or C. violaceum CV026, two commonly used indicator strains. These two indicator organisms were chosen because they differ in the range of AHLs that they detect. A. tumefaciens is generally more sensitive to AHLs with medium to long chains, while C. violaceum is more sensitive to short-chain AHLs and is inhibited by long-chain AHLs (34, 47). Wild-type Rm11500 colonies normally activate both CV026 and NTL4(pZLR4), resulting in purple and blue haloes, respectively, surrounding the colonies, while Rm1021 is only able to activate NTL4(pZLR4). In this screening procedure, mutant colonies that were still able to produce AHLs activated the indicator organisms. Colonies that had acquired a mutation in a gene necessary for AHL production were not be able to activate the indicators and did not have a halo.
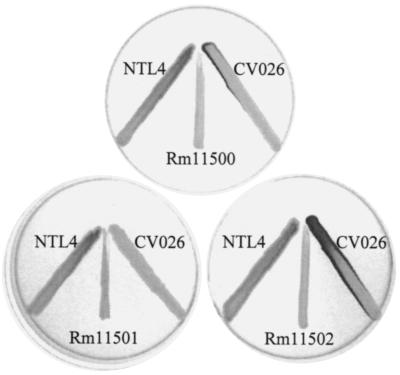
Using this screening procedure, we were able to identify a mutant, Rm11501, which was unable to activate the CV026 indicator (Fig. 1). Although this mutant did not activate CV026, it still had the ability to activate the A. tumefaciens indicator, suggesting that the mutation affected only a subset of the AHLs that are produced by AK631. Cloning and sequencing of the region surrounding the transposon insertion showed that the mutation resides in an open reading frame corresponding to the chromosomal groEL gene (groELc) of Rm1021. The mutation is referred to below as groELc1583::Tn5 to reflect the presence of the Tn5 insertion 1,583 nucleotides from the 5′ end of the gene (Fig. 2C). This is not a lethal mutation, possibly because S. meliloti contains at least two functional copies of groEL and groES (38, 43). Alternatively, the transposon insertion at the end of the gene may have resulted in a slightly truncated form of GroELc retaining partial activity. Interestingly, GroEL was shown to be required for the proper folding of the transcriptional activators LuxR in P. fischeri (7) and NodD in S. meliloti (38). It is, therefore, likely that a similar scenario occurs in S. meliloti, in which GroELc is responsible for the proper folding of a LuxR homolog.
FIG. 1.
Phenotypes of transposon mutants. The wild type (Rm11500) and mutant strains Rm11501 (groELc1583) and Rm11502 (groELc1583, traM) were streaked next to the two indicator strains [CV026 and NTL4(pZLR4)] to show the ability to activate the indicator.
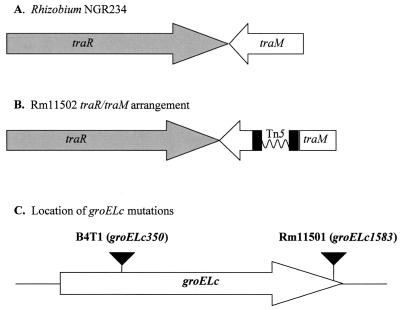
FIG. 2.
Organization of the traR/traM and groELc loci. (A) Arrangement of the traR and traM genes in Rhizobium strain NGR234. (B) Arrangement of the S. meliloti traR and traM genes, including the transposon insertion, in Rm11502, based on alignment of the cloned region with the NGR234 sequence. (C) Locations of the Tn5 insertions in the B4T1 and Rm11501 mutations.
AHL characterization of the groELc mutant, Rm11501.
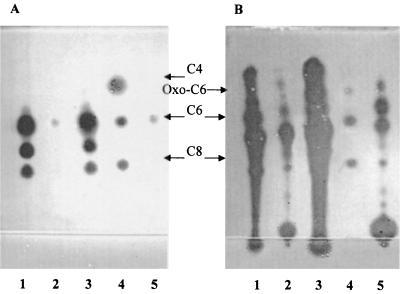
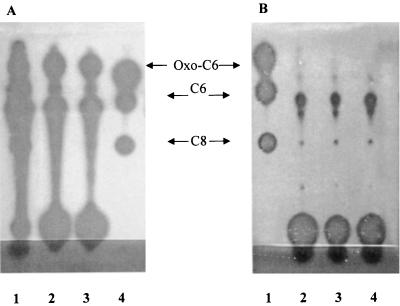
The pattern of AHLs from an Rm41 supernatant preparation has been characterized previously by Cha et al. (3), who used an A. tumefaciens indicator. Figure 3 shows the patterns of AHLs produced by the Rm41 derivative, AK631 (Fig. 3A and B, lanes 1), our other wild-type strain, Rm1021 (lanes 5), and the groELc1583 mutant, Rm11501 (lanes 2), as detected by the C. violaceum CV026 (Fig. 3A) and A. tumefaciens NTL4(pZLR4) (Fig. 3B) indicator strains. The AHL preparations were made by extracting 5-ml cultures with ethyl acetate, evaporating the ethyl acetate, and resuspending each preparation in 500 μl (final volume) of 70% methanol. Routine detection of AHLs in AK631 cultures by NTL4(pZLR4) required 5 to 10 μl of the crude extract, while detection by CV026 required all 500 μl. It should be noted that 100 μl of the Rm11501 groELc1583 mutant extract was used for detection by NTL4(pZLR4) (this was 10-fold more than the amount used for AK631). In addition to the decrease in the overall level of AHLs made by Rm11501, the pattern of spots that were detected by the two indicators was slightly different than the wild-type pattern (Fig. 3, compare lanes 2 with lanes 1). AK631 extracts typically produced at least nine spots detected by NTL4(pZLR4) and three spots detected by CV026. However, in Rm11501 extracts, only six spots were seen with NTL4(pZLR4) and only one spot was seen with CV026. The pattern of AHLs seen with Rm11501 was similar to the pattern produced by wild-type Rm1021; however, the Rm11501 AHL levels were still slightly higher than the Rm1021 AHL levels (routine detection of the AHL pattern also required 100 μl of Rm1021 extract [see below]).
FIG. 3.
(A) Plate overlaid with the CV026 indicator. Lane 1, AK631; lane 2, Rm11501 (groELc1583); lane 3, Rm11502 (groELc1583 traM); lane 4, C4 HSL, C6 HSL, and C8 HSL; lane 5, Rm1021. Each lane contained 500 μl of extract. (B) Plate overlaid with the NTL4(pZLR4) indicator. Lane 1, 10 μl of AK631; lane 2, 100 μl of Rm11501 (groELc1583); lane 3, 10 μl of Rm11502 (groELc1583 traM); lane 4, oxo-C6 HSL, C6 HSL, and C8 HSL; lane 5, 100 μl of Rm1021.
Mutation of a traM homolog suppresses the groELc1583 phenotype.
After identifying the groELc1583 mutant, Rm11501, we set out to determine what other loci may be involved in regulation or production of AHLs. To this end, we undertook a second round of transposon mutagenesis using Rm11501 to screen for mutations that suppress or compensate for the groELc1583 mutation. In doing so, we identified the double mutant Rm11502, which had regained the ability to activate both indicator strains (Fig. 1) despite the mutation in groELc1583. The region surrounding the second transposon insertion was cloned and sequenced. The sequencing data showed 82% homology to open reading frames in Rhizobium strain NGR234, which were designated traR and traM homologs (Fig. 2A and B) (13). In A. tumefaciens, TraM negatively regulates TraR (the LuxR homolog) activity by binding to it and forming an inactive complex (27, 33). Studies have shown that mutations in traM result in increased levels of active TraR (14, 26). This information is consistent with the phenotype of the Rm11502 mutant and suggests that due to mutation of the traM homolog, the levels of the TraR homolog increased enough to reestablish the positive feedback loop. Because of the extremely high sequence homology and conserved arrangement of the S. meliloti genes compared to the Rhizobium strain NGR234 genes, we use the same nomenclature and refer to the corresponding S. meliloti genes as traR and traM.
Figure 3 shows the pattern of AHLs detected from supernatant extracts of the groELc suppressor mutant, Rm11502 (lanes 3). The same amounts of extract that were used for wild-type strain AK631 were used for routine detection of AHLs in Rm11502. It is clear from comparing the pattern for Rm11502 with the pattern for AK631 that the number of spots was restored, and there was a slight increase in the level of total AHLs in the Rm11502 extract. The increase was due to the mutation in traM, which presumably eliminated the negative regulation of TraR.
Comparison of groELc mutant growth phenotypes.
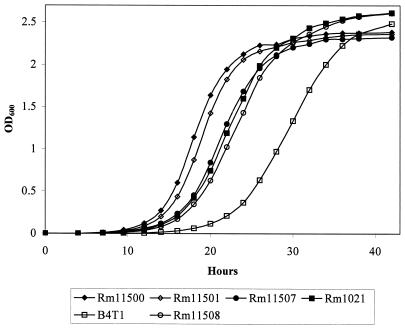
Sharon Long's lab has previously identified a transposon-induced mutation 350 nucleotides from the 5′ end in the chromosomal groEL gene of Rm1021, which is referred to in this paper as groELc350::Tn5 (Fig. 2C) (38). The Rm1021 groELc350 mutant, B4T1, was shown to have a slightly lower growth rate (∼25% longer doubling time) than Rm1021, but there were no other noticeable growth defects. We compared the growth rates of our groELc1583 mutant (Rm11500 derivative) and B4T1. Figure 4 shows the growth curves for all strains grown in LB/MC. The generation time for Rm1021 was about 15% longer than that for Rm11500. The B4T1 groELc350 mutant grew about 18% slower than its parent, Rm1021. The increase in doubling time for B4T1 is consistent with the previous observations (38). It is clear that growth of Rm11501 was not affected by the groELc1583 mutation, since its generation time was virtually identical to that of its parent strain, Rm11500.
FIG. 4.
Growth curve comparison of groEL mutants and wild-type S. meliloti strains. All strains were grown in LB/MC, and growth was monitored by measuring the optical density at 600 nm (OD600). Rm11500 and Rm1021 are the wild-type strains. Rm11501 (groELc1583) and Rm11507 (groELc350) are Rm11500 derivatives. B4T1 (groELc350) and Rm11508 (groELc1583) are Rm1021 derivatives.
Comparison of groELc mutant AHL patterns.
We also compared the patterns of AHL production by the different groELc mutants. Figure 5 shows the AHL patterns for the groELc1583 mutant (Rm11501) (Fig. 5A, lane 2), the groELc350 mutant (B4T1) (Fig. 5B, lane 3), AK631 (Fig. 5A, lane 1), and Rm1021 (Fig. 5B, lane 2). Surprisingly, the groELc350 mutation in B4T1 does not affect AHL production since the pattern of AHLs for this strain is identical to the pattern for the parent strain, Rm1021. Both strains required about 100 μl of extract for detection of AHLs by the NTL4(pZLR4) indicator organism. In Rm11501, however, the mutation has a significant effect, which results in an approximately 10-fold decrease in AHL levels compared to the wild-type strain AK631 levels. Interestingly, the AHL pattern and the overall amount of AHLs of Rm11501 are very similar to those of Rm1021. Either groELc plays a different role in AK631 than in Rm1021 or the position of the mutation is responsible for the differences in the phenotypes. In B4T1, the transposon insertion is 350 bases from the 5′ end, while in Rm11501 the transposon is 53 bases from the 3′ end (Fig. 2C).
FIG. 5.
Comparison of groELc mutations in AK631 and Rm1021 backgrounds. TLC plates were overlaid with the NTL4(pZLR4) indicator. (A) Lane 1, 10 μl of AK631 extract; lane 2, 100 μl of Rm11501 (groELc1583) extract; lane 3, 100 μl of Rm11507 (groELc350) extract; lane 4, oxo-C6 HSL, C6 HSL, and C8 HSL. (B) Lane 1, oxo-C6 HSL, C6 HSL, and C8 HSL; lane 2, 100 μl of Rm1021 extract; lane 3, 100 μl of B4T1 (groELc350) extract; lane 4, 100 μl of Rm11508 (groELc1583) extract.
Exchange of groELc mutations.
To address the difference in transposon insertions, we transferred the Rm11501 groELc1583 to Rm1021 and transferred the B4T1 groELc350 to Rm11500. Figure 5 shows the effects of the resulting mutations on AHL production. As expected, the groELc1583 mutation in an Rm1021 background had no effect on the autoinducer pattern (Rm11508) (Fig. 5B, lane 4), while the groELc350 mutation in an AK631 background resulted in the same phenotype as the groELc1583 mutant (Rm11507) (Fig. 5A, lane 3). We also compared the growth rates of the mutants and found that the groELc1583 mutation has no effect on growth in either parent strain (AK631 or Rm1021) (Fig. 4). The groELc350 mutation, however, resulted in a slightly longer (about 18%) doubling time in both backgrounds. Therefore, the position of the mutation in the groELc locus does not seem to play a role in AHL production, despite its effect on growth. Both mutations presumably eliminate the ability of groELc to properly fold TraR. On the other hand, a mutation at the end of the gene (groELc1583) does not seem to have such a dramatic effect on GroELc's interaction with other proteins, since the growth rate remained unaltered in both backgrounds.
Further characterization of traR and traM.
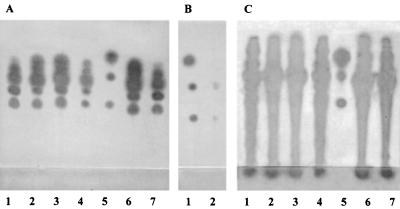
After identifying the traR and traM junction in the AK631 derivative, Rm11502, we wanted to characterize these genes and to determine if they are also present in Rm1021. We therefore aligned the cloned region obtained from the Rm11502 sequencing results with the traR and traM open reading frames in Rhizobium strain NGR234. We used the sequence data from Rm11502 to design primers complementary to the end of each gene and used the Rhizobium strain NGR234 data to design primers for the beginning of each open reading frame, for which we did not have S. meliloti data. These primers were used to screen the AK631 library for a clone carrying wild-type traM. Then, to investigate the role of traM in autoinducer production, complementation experiments were performed by using the plasmid from this clone, pTraM. First, the traM::Tn5 mutation in Rm11502 was transduced to Rm11500 in order to confirm the effect of the mutation in a wild-type background. Figure 6 shows the AHL pattern of the transductant (Rm11509) (Fig. 6A and C, lanes 3). The level of AHLs was slightly increased (only 2.5 μl of Rm11509 extract was used, compared to 5 μl of AK631 extract), and this effect could be complemented with the traM library clone [Rm11509(pTraM)] (Fig. 6A and C, lanes 4). We also found that pTraM complemented the double mutant, Rm11502, restoring the original groEL phenotype (data not shown). Therefore, it seems that TraM acts as a modulator of TraR activity and that TraR in turn regulates AHL production by a putative autoinducer synthase, referred to here as TraI. The slight, but consistent, increase in AHL production in a traM mutant is similar to the increase in AHL levels observed for Agrobacterium traM mutants (26). The role of TraM, however, is proposed to be in preventing the premature activation of TraR, and traM mutants result in plasmid transfer at low cell densities (14, 26). We have preliminary evidence that S. meliloti traM mutants show high levels of AHLs at lower cell densities, which is consistent with the proposed role of traM in preventing early activation of TraR (data not shown).
FIG. 6.
Effect of traM and traR mutations on AHL production. Plates were overlaid with the CV026 indicator (A and B) or the NTL4(pZLR4) indicator (C). (A) Lane 1, 500 μl of AK631 extract; lane 2, 500 μl of Rm11502 (groELc1583 traM) extract; lane 3, 500 μl of Rm11509 (traM) extract; lane 4, 500 μl of Rm11509(pTraM) extract; lane 5, C4 HSL, C6 HSL, and C8 HSL; lane 6, 500 μl of Rm11510 (Rm1021 carrying pRme41a::Tn5) extract; lane 7, 500 μl of Rm11510(pTraM) extract. (B) Lane 1, C4 HSL, C6 HSL, and C8 HSL; lane 2, 500 μl of Rm11513 (AK631 traR) extract. (C) Lane 1, 5 μl of AK631 extract; lane 2, 5 μl of Rm11502 extract; lane 3, 2.5 μl of Rm11509 extract; lane 4, 5 μl of Rm11509(pTraM) extract; lane 5, oxo-C6 HSL, C6 HSL, and C8 HSL; lane 6, 2.5 μl of Rm11510 extract; lane 7, 5 μl of Rm11510(pTraM) extract.
The traR/traM locus is present in AK631 but not in Rm1021.
To determine if the traR and traM genes are also present in Rm1021, PCRs were performed with Rm1021 and AK631 chromosomal DNA by using primers for the corresponding open reading frames. Bands of the expected sizes were obtained for traR and traM with AK631 but not with Rm1021 (data not shown). The PCR products obtained from AK631 were cloned and sequenced. BLAST search results showed that the traM open reading frame from AK631 is 77% identical to traM of Rhizobium strain NGR234 and 37% identical to traM of A. tumefaciens. The traR open reading frame showed 84 and 48% identity to Rhizobium strain NGR234 and A. tumefaciens traR open reading frames, respectively. However, no matches were found when we used the sequence data to search the S. meliloti Rm1021 genome database. This suggests that Rm1021 does not contain the traR and traM genes. The lack of this set of genes explains the differences in the observed patterns of AHLs between the two strains. In addition, introducing traM into Rm1021 did not affect AHL production (data not shown), which again suggests that the corresponding TraR protein does not exist in Rm1021.
It has been shown that Rm41 carries a large plasmid (pRme41a) which is not present in Rm1021 (2, 10). The role of this plasmid in symbiosis is unclear; however, it was shown to be self-transmissible (25). These findings, along with the evidence that the traR/traM locus is not present in Rm1021, suggest that traR and traM may reside on pRme41a. To test this hypothesis, we conducted mating between the double mutant (Rm11502) and Rm1021. We used the neomycin resistance of the traM::Tn5 mutation to select for Rm1021 transconjugants that had received the pRme41a plasmid. Figure 6 shows the AHL pattern for the resulting clone (Rm11510) (Fig. 6A and C, lanes 6). The presence of pRme41a::Tn5 in Rm1021 allowed production of the AHLs unique to AK631. The levels of these AHLs were high, as they were in Rm11509, owing to the traM mutation. This phenotype was complemented by pTraM, giving Rm1021 an AHL pattern similar to that of AK631 [Rm11510(pTraM)] (Fig. 6A and C, lanes 7). In addition, PCR results confirmed the presence of traR and traM in Rm11510 (data not shown). These results confirmed that traR and traM reside on pRme41a. Presumably, one or more AHL synthases (i.e., the putative TraI) encoded by the plasmid are responsible for the production of the AHLs unique to AK631.
traR disruption eliminates the pRme41a-encoded AHLs.
To verify that traR plays a role in production of the AHLs that are specific to the pRme41a plasmid, we constructed a traR mutation and analyzed its effect in different backgrounds. Rm11513 (AK631 traR) showed a dramatic decrease in AHL production (compare Fig. 6A, lane 1, and Fig. 6B, lane 2); however, two faint spots remained. These remaining AHLs could have been due to basal levels of AHL production by the pRme41a plasmid, or they could have been the result of an additional synthase independent of the plasmid. These data suggest that traR is responsible for inducing the pRme41a-specific AHLs. The fact that the pattern of AHLs present in the AK631 traR extract is similar to the pattern seen for groELc1583 (Fig. 6B, lane 2, and Fig. 3A, lane 2) suggests, but does not prove, that the effect of groELc on AHL production is through traR.
A second locus, sinR/sinI, is common to AK631 and Rm1021.
A search of the recently published Rm1021 genome revealed the presence of multiple genes with homology to luxR, but only one of these genes was positioned next to a putative autoinducer synthase. We therefore chose this locus to study more closely. To indicate that the genes at this locus belong to S. meliloti, we designated them sinR and sinI. sinR shows 34, 33, and 24% identity to cerR of Rhodobacter sphaeroides, cinR of R. leguminosarum, and luxR of P. fischeri, respectively, while sinI shows 39, 35, and 26% identity to the cerI, cinI, and luxI genes, respectively. A PCR was done to clone this sinR/sinI locus into E. coli XL10-Gold cells and to assay for autoinducer activity. Figure 7 shows that the sinR/sinI locus produced a pattern of AHLs similar to that produced by Rm1021, although the levels were significantly lower [100 μl of Rm1021 extract and 500 μl of XL10-Gold(pSinRI) extract were required for detection of AHLs]. A noticeable difference between the two AHL patterns is the absence from the E. coli extract of an AHL with fast mobility. It is possible that Rm1021 contains an additional synthase that is responsible for this AHL. Alternatively, E. coli may not be capable of synthesizing all of the AHLs for which the sinRI locus is responsible. Since a similar pattern of AHLs is also present in the AK631 groELc mutant (Rm11501), we conducted a PCR analysis to determine if the sinR and sinI genes are also present in AK631. PCR products of the expected sizes were obtained for the two genes, and the products were sequenced and determined to be identical to sinR and sinI of Rm1021 (data not shown). These results indicate that the sinR/sinI locus is sufficient for AHL production and represents a quorum-sensing system present in both Rm1021 and AK631.
FIG. 7.
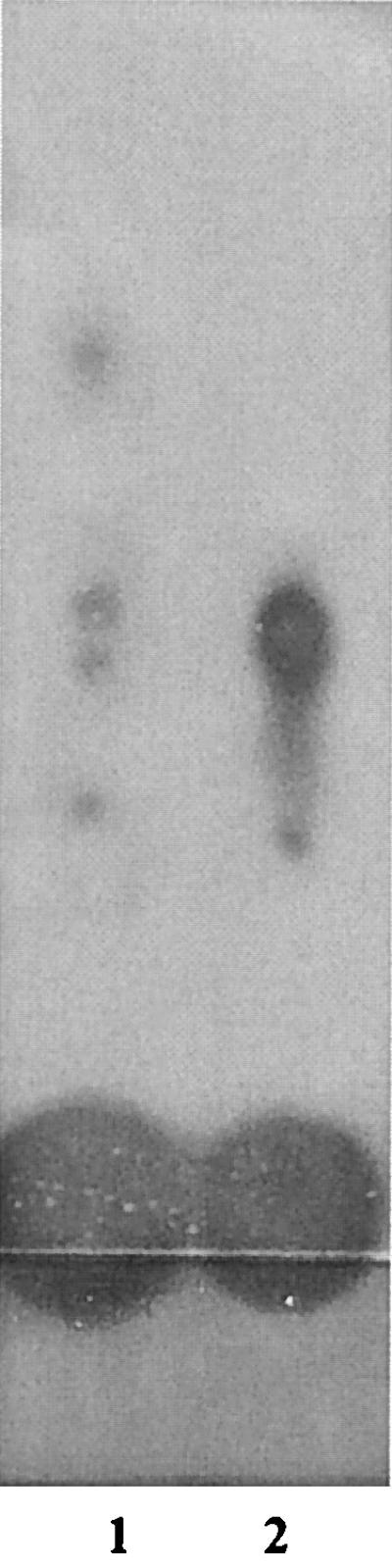
Role of sinRI in AHL production. The TLC plate was overlaid with the NTL4(pZLR4) indicator. Lane 1, 100 μl of Rm1021 extract; lane 2, 500 μl of XL10-Gold(pSinRI) extract.
DISCUSSION
Rm1021 and Rm41 are two independently isolated strains of S. meliloti, and they both are commonly used to study aspects of symbiosis with alfalfa (M. sativa). In the survey study of Cha et al. (3), these two strains were shown to produce autoinducers, which were able to activate the A. tumefaciens indicator organism. However, even though the two strains are closely related and are capable of establishing symbiosis with the same host, the patterns of putative AHLs that they produce are very different. In this study we addressed the observed difference in AHL production in an attempt to determine whether quorum sensing exists in S. meliloti and if it plays a role in symbiosis. We found that there is indeed a difference between the Rm41 derivative, AK631, and Rm1021 in terms of AHL production (Fig. 3). We observed at least nine different AHLs for AK631 using an A. tumefaciens indicator, a pattern similar to the one observed by Cha et al. for Rm41. Using the C. violaceum indicator, we also observed three spots for AK631. However, for Rm1021 we observed an approximately 10-fold decrease in the overall amount of AHLs compared to the amount for AK631, in addition to a different AHL pattern. In contrast to the findings of Cha et al., we observed six different AHLs for Rm1021 when A. tumefaciens was used and one spot when C. violaceum was used. The difference in the pattern observed for Rm1021 is most likely due to the amount of extract that was used in the analysis. As mentioned above, we had to use approximately 10 times more extract from Rm1021 than from AK631 for routine detection of AHLs.
To try to identify genes involved in AHL production by AK631 and Rm1021, we developed a novel screening procedure designed to detect mutants that have a deficiency in AHL production and/or transport. This procedure has revealed at least two interesting mutants that define loci involved in the generation of AHLs. One of these mutants, Rm11501, has a Tn5 insertion in the groELc locus (groELc1583). We have shown that in an AK631 background, a mutation in groELc has a dramatic effect on autoinducer production. Mutant Rm11501 produced only six AHLs detected by A. tumefaciens, and the overall amount of autoinducers was about 10-fold less than the amount observed with the parent strain (Fig. 3A and B, lanes 1 and 2). Interestingly, the result was a phenotype similar to that of Rm1021 (Fig. 3A and B, lanes 5). The effect on autoinducer production could be explained by a direct or indirect effect on TraR, due to the lack of a functional GroELc. This hypothesis is further supported by the fact that a traR mutation in either a wild-type or groELc background results in an AHL pattern similar to that observed for the groELc mutant alone (Fig. 3A, lane 2; Fig. 6B, lane 2; and data not shown).
GroEL is known as a molecular chaperone because of its role in helping proteins fold into their active conformations, oligomerization, and stabilization of unfolded polypeptides (19). It has been shown that in an E. coli strain carrying the P. fischeri lux operon, a groEL mutation resulted in a severe decrease in luminescence (7). Addition of autoinducers to an rpoH mutant, which could no longer induce expression of groEL, could compensate for the defect, but only slightly. These observations led to the conclusion that GroEL was required for the folding of LuxR into a conformation that is capable of binding autoinducers and becoming active. The fact that some autoinducers are produced by the groELc1583 mutant (Rm11501) implies that (i) more than one LuxR and/or LuxI homolog exists in AK631 and (ii) the different LuxR homologs either may not require a GroEL protein or may require a different GroEL protein to fold properly. It has been suggested that in Bradyrhizobium japonicum, in which there are multiple groEL loci, different GroEL proteins may have distinct functions (11), since they are expressed differentially. However, the different GroEL proteins in Bradyrhizobium may also have overlapping roles because a mutation in any individual gene did not affect nodulation and nitrogen fixation during symbiosis. It is not currently known how many functional groEL loci exist in AK631, but Rm1021 has been shown to harbor up to five different loci (38, 43). AK631 probably also has multiple groEL loci since a PCR product matching the predicted size of a wild-type locus is observed in the Rm11501 groEL mutant (data not shown). Thus, it is possible that the groELc gene product interacts with TraR and that other GroEL proteins are not able to compensate for a mutation in this gene. The observation that mutations in traR and/or groELc result in similar phenotypes (Fig. 3A and 6B and data not shown) supports this hypothesis. The interaction of GroELc with other proteins may, however, be redundant, enabling the cell to ensure that essential proteins are able to fold properly in the absence of one particular GroEL.
In a previous study Ogawa and Long (38) characterized a groELc350 mutant in Rm1021 (B4T1) which showed defects in the growth rate and also in nodulation. Interestingly, our groELc1583 mutation in an AK631 background (Rm11501) did not show any such defects (Fig. 4 and data not shown). Therefore, we wanted to determine if the differences were due to the different parent strains (AK631 versus Rm1021) or if they were due to the different positions of the transposon insertions (Fig. 2). When we compared the groELc mutations in Rm11501 and B4T1, we found that only the B4T1 mutation (groELc350) had an effect on growth (Fig. 4). Both groELc350 mutants (Rm11507 and B4T1) resulted in an approximately 18% decrease in the growth rate, as well as nodulation defects (data not shown). The groELc1583 mutants (Rm11501 and Rm11508), however, had no significant effect on growth in either an AK631 or Rm1021 background. When we compared autoinducer production from the different groELc mutants, we found that only AK631 derivatives were affected by the groELc mutations, regardless of the position of the transposon insertion (Fig. 5A). Mutating groELc in an AK631 background significantly decreased autoinducer production, while in Rm1021 no differences were observed (Fig. 5B). Moreover, the groELc mutations in the AK631 derivatives resulted in an AHL phenotype nearly identical to that of Rm1021. These results, in combination with the lack of a traR/traM locus in Rm1021, suggested to us that traR, traM, and the corresponding putative autoinducer synthase-encoding gene traI are unique to AK631.
After identifying the groELc1583 mutant, Rm11501, we used it in a second round of mutagenesis to screen for mutations that could suppress the AHL phenotype. This approach yielded a mutation in a gene homologous to traM of A. tumefaciens, which is a negative regulator of traR (the luxR homolog) (14, 26). We showed that a mutation in this gene, also designated traM, could compensate for the groELc1583 mutation (Fig. 3A and B, lanes 3). In A. tumefaciens mutating traM resulted in increased expression of tra genes, as well as increased AHL production. In addition, in an E. coli groEL mutant carrying the P. fischeri lux genes, overexpression of LuxR resulted in nearly wild-type levels of luminescence (7). These findings support our observations and suggest that TraM also acts as a negative regulator of TraR. When traM is disrupted, TraR levels increase enough to overcome the groELc mutation and establish the positive feedback loop necessary for autoinducer production. Complementation studies suggest that the traM mutation specifically interacts with the traR gene product, since no effect was seen when traM was introduced into Rm1021 (data not shown). This suggests that the luxR homolog in Rm1021, sinR, is significantly different from traR in AK631 and is also subject to different control mechanisms.
The difference between strains was further confirmed when the traM open reading frame was cloned and sequenced from AK631, but no homolog could be found in Rm1021, either by PCR or by searching the S. meliloti genome database. However, when pRme41a::Tn5 was transferred to Rm1021, the result was a phenotype similar to that of the AK631 traM mutant (Rm11509) (Fig. 6A and C, lanes 3 and 6). As in Rm11502 and Rm11509, the traM mutation in Rm11510 could be complemented by the traM library clone (Fig. 6A and B, lanes 7). In addition, the presence of traR and traM in Rm11510 could be detected by PCR (data not shown). These results suggest that the traR/traM locus resides on pRme41a and is involved in the production of the AHLs unique to AK631. It is important to note that the proposed role of TraM in Agrobacterium is to prevent premature activation of TraR and subsequent plasmid transfer, and mutations in traM result in plasmid transfer even at low cell densities (14, 26). We have obtained preliminary evidence that shows that the pRme41a-specific AHLs are not produced until AK631 cultures reach the stationary phase, but the traM mutant Rm11509 produces these AHLs even at lower cell densities (data not shown). These observations are consistent with the proposed role of traM in Agrobacterium and suggest that this gene is responsible for the timing of AHL synthesis, not the overall level of AHLs.
To identify the genes responsible for AHL production in Rm1021, we searched the recently published S. meliloti genome (17). We found a set of luxR and luxI homologs, which we call sinR and sinI, respectively. Cloning this locus into E. coli showed that these genes are sufficient for production of the AHLs present in Rm1021 (Fig. 7). PCR and sequencing results show that the sinR/sinI locus is also present in AK631 (data not shown). This observation correlates with the AHL phenotype of the groEL mutant, Rm11501, and suggests that the sinR/sinI locus is responsible for producing the AHLs that are common to AK631 and Rm1021. Thus, two quorum-sensing systems exist in AK631, and these systems may represent another example of a complex network like those seen in P. aeruginosa and R. leguminosarum (29, 31, 53).
It is not yet clear what role quorum sensing plays in S. meliloti symbiosis. It has been shown that in other rhizobia quorum sensing regulates phenomena such as induction of the stationary phase and restriction of the number of nodules of host plants (4, 22, 41, 42), but the precise mechanisms are not known. Preliminary studies have shown that both Rm11501 and Rm11502 have no apparent nodulation defects (data not shown). However, if a complex hierarchy exists in AK631, a combination of mutations affecting the quorum-sensing system may be necessary to see an effect on symbiosis. In A. tumefaciens, quorum sensing regulates transfer of the Ti plasmid, which involves induction of the tra and trb genes. Interestingly, Rhizobium strain NGR234 and R. leguminosarum have also been shown to carry the trb operon on their symbiotic plasmids between the traR/traM and traI loci (13). In addition, there is evidence that in R. leguminosarum quorum sensing is involved in regulating plasmid transfer through the TraR regulator (J. A. Downie, personal communication), and in Rhizobium strain NGR234 the trb genes are also activated by TraR (C. W. Fuqua, personal communication). We have obtained results from sequencing of a library clone carrying traR which show the presence of the trbI and trbH genes upstream of traR (data not shown). These results are in agreement with the arrangement of the corresponding genes in Rhizobium strain NGR234. Given the extremely high homology and identical arrangement of the traR/traM genes, as well as the arrangement of the trbI and trbH genes of NGR234 and AK631, it is possible that the entire trb operon also exists in AK631 and is involved in transfer of pRme41a. Further support for this idea was provided by the early studies of Huguet et al. (25). These authors identified regions of pRme41a that were homologous to regions B and C of the Ti plasmid, which correspond to the tra and trb gene clusters.
It is interesting to speculate about the possibility that plasmids may be transferred upon reaching a quorum, as in A. tumefaciens, to ensure the prosperity of certain rhizobia in the rhizosphere or within nodules. It has been shown that certain species of rhizobia, including S. meliloti, produce substances known as rhizopines (37, 44, 50), which are made by bacteroids within the nodules and then catabolized by their vegetative counterparts in the rhizosphere. Although these rhizopines have not been detected with AK631 or Rm1021 (37, 50), other compounds or properties may be involved. It has been shown that the pRme41a plasmid encodes genes involved in catabolism of plant-produced compounds called calystegins (49). The significance of this property is unclear, since only three plants were shown to be capable of synthesizing calystegins and AK631 is not known to establish a symbiotic relationship with them. However, the ability to utilize compounds produced by plants or bacteroids could provide a competitive advantage to bacteria in the rhizosphere. The traR/traM quorum-sensing regulators may also have a role in other functions, such as host range specificity, which would be unique to AK631, while the sinRI locus, common to Rm1021 and AK631, may regulate components of symbiosis or the free-living state common to these two strains.
Acknowledgments
We thank Steve Farrand, Sharon Long, Lawrence Reitzer, and Graham Walker for providing strains. We also thank Allan Downie, Clay Fuqua, Lawrence Reitzer, Ron Yasbin, and the members of our laboratory for their helpful discussions and insights during this project.
This work was supported by National Science Foundation grant MCB-9733532 to J.E.G. The results were also based in part on work supported by the Texas Advanced Research Program under grant 009741-0022-2001.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Banfalvi, Z., E. Kondorosi, and A. Kondorosi. 1985. Rhizobium meliloti carries two megaplasmids. Plasmid 13:129-138. [DOI] [PubMed] [Google Scholar]
- 3.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 4.Cubo, M. T., A. Economou, G. Murphy, A. W. Johnston, and J. A. Downie. 1992. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J. Bacteriol. 174:4026-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolan, K. M., and E. P. Greenberg. 1992. Evidence that GroEL, not sigma 32, is involved in transcriptional regulation of the Vibrio fischeri luminescence genes in Escherichia coli. J. Bacteriol. 174:5132-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 9.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, H. M., M. Babst, T. Kaspar, G. Acuna, F. Arigoni, and H. Hennecke. 1993. One member of a gro-ESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO J. 12:2901-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrai, T., E. Vincze, Z. Banfalvi, G. B. Kiss, G. S. Randhawa, and A. Kondorosi. 1983. Localization of symbiotic mutations in Rhizobium meliloti. J. Bacteriol. 153:635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, C., M. Burbea, and S. C. Winans. 1995. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J. Bacteriol. 177:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 16.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 18.Geiser, M., R. Cebe, D. Drewello, and R. Schmitz. 2001. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. BioTechniques 31:88-92. [DOI] [PubMed] [Google Scholar]
- 19.Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 355:33-45. [DOI] [PubMed] [Google Scholar]
- 20.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 21.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray, K. M., J. P. Pearson, J. A. Downie, B. E. Boboye, and E. P. Greenberg. 1996. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 178:372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haygood, M. G., and D. L. Distel. 1993. Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes form unique lineages related to the genus Vibrio. Nature 363:154-156. [DOI] [PubMed] [Google Scholar]
- 25.Huguet, T., C. Rosenberg, F. Casse-Delbart, P. De Lajudie, L. Jouanin, J. Batut, P. Boistard, J.-S. Julliot, and J. Denarie. 1983. Studies on Rhizobium meliloti plasmids and on their role in the control of nodule formation and nitrogen fixation: the pSym megaplasmids and the other large plasmids, p. 35-45. In A. Puhler (ed.), Molecular genetics of the bacteria-plant interaction. Springer-Verlag, Berlin, Germany.
- 26.Hwang, I., D. M. Cook, and S. K. Farrand. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, I., A. J. Smyth, Z. Q. Luo, and S. K. Farrand. 1999. Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34:282-294. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 30.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lithgow, J. K., A. Wilkinson, A. Hardman, B. Rodelas, F. Wisniewski-Dye, P. Williams, and J. A. Downie. 2000. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 37:81-97. [DOI] [PubMed] [Google Scholar]
- 32.Luo, Z. Q., T. E. Clemente, and S. K. Farrand. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant-Microbe Interact. 14:98-103. [DOI] [PubMed] [Google Scholar]
- 33.Luo, Z. Q., Y. Qin, and S. K. Farrand. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275:7713-7722. [DOI] [PubMed] [Google Scholar]
- 34.McLean, R. J., M. Whiteley, D. J. Stickler, and W. C. Fuqua. 1997. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154:259-263. [DOI] [PubMed] [Google Scholar]
- 35.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55:123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, P. J., N. Heycke, Z. Banfalvi, M. E. Tate, F. de Bruijn, A. Kondorosi, J. Tempe, and J. Schell. 1987. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc. Natl. Acad. Sci. USA 84:493-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa, J., and S. R. Long. 1995. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 9:714-729. [DOI] [PubMed] [Google Scholar]
- 39.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 40.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 41.Rodelas, B., J. K. Lithgow, F. Wisniewski-Dye, A. Hardman, A. Wilkinson, A. Economou, P. Williams, and J. A. Downie. 1999. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 181:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosemeyer, V., J. Michiels, C. Verreth, and J. Vanderleyden. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 180:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusanganwa, E., and R. S. Gupta. 1993. Cloning and characterization of multiple groEL chaperonin-encoding genes in Rhizobium meliloti. Gene 126:67-75. [DOI] [PubMed] [Google Scholar]
- 44.Saint, C. P., M. Wexler, P. J. Murphy, J. Tempe, and M. E. Tate. 1993. Characterization of genes for synthesis and catabolism of a new rhizopine induced in nodules by Rhizobium meliloti Rm220-3: extension of the rhizopine concept. J. Bacteriol. 175:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schripsema, J., K. E. de Rudder, T. B. van Vliet, P. P. Lankhorst, E. de Vroom, J. W. Kijne, and A. A. van Brussel. 1996. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J. Bacteriol. 178:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 47.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tepfer, D., A. Goldmann, N. Pamboukdjian, M. Maille, A. Lepingle, D. Chevalier, J. Denarie, and C. Rosenberg. 1988. A plasmid of Rhizobium meliloti 41 encodes catabolism of two compounds from root exudate of Calystegium sepium. J. Bacteriol. 170:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wexler, M., D. Gordon, and P. J. Murphy. 1995. The distribution of inositol rhizopine genes in Rhizobium populations. Soil Biol. Biochem. 27:531-537. [Google Scholar]
- 51.Williams, M. N. V., R. I. Hollingsworth, S. Klein, and E. R. Signer. 1990. The symbiotic defect of Rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J. Bacteriol. 172:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams, M. N. V., S. Klein, and E. R. Signer. 1989. Host restriction and transduction in Rhizobium meliloti. Appl. Environ. Microbiol. 55:3229-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft, et al. 1995. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]