Abstract
Four low-molecular-weight penicillin binding proteins (LMW PBPs) of Escherichia coli are closely related and have similar dd-carboxypeptidase activities (PBPs 4, 5, and 6 and DacD). However, only one, PBP 5, has a demonstrated physiological function. In its absence, certain mutants of E. coli have altered diameters and lose their uniform outer contour, resulting in morphologically aberrant cells. To determine what differentiates the activities of these LMW PBPs, we constructed fusion proteins combining portions of PBP 5 with fragments of other dd-carboxypeptidases to see which hybrids restored normal morphology to a strain lacking PBP 5. Functional complementation occurred when truncated PBP 5 was combined with the terminal membrane anchor sequences of PBP 6 or DacD. However, complementation was not restored by the putative carboxy-terminal anchor of PBP 4 or by a transmembrane region of the osmosensor protein ProW, even though these hybrids were membrane bound. Site-directed mutagenesis of the carboxy terminus of PBP 5 indicated that complementation required a generalized amphipathic membrane anchor but that no specific residues in this region seemed to be required. A functional fusion protein was produced by combining the N-terminal enzymatic domain of PBP 5 with the C-terminal β-sheet domain of PBP 6. In contrast, the opposite hybrid of PBP 6 to PBP 5 was not functional. The results suggest that the mode of PBP 5 membrane anchoring is important, that the mechanism entails more than a simple mechanical tethering of the enzyme to the outer face of the inner membrane, and that the physiological differences among the LMW PBPs arise from structural differences in the dd-carboxypeptidase enzymatic core.
Escherichia coli expresses four low-molecular-weight (LMW) dd-carboxypeptidase penicillin binding proteins (PBPs) that share considerable nucleic acid sequence identity (PBPs 4, 5, and 6 and DacD), suggesting that they diverged from a common primordial enzyme (16). The classic explanation for this apparent redundancy is that the dd-carboxypeptidases can modify peptidoglycan in similar ways so that they serve as auxiliaries of one another (3). However, arguing against this idea is the observation that PBP 5 plays a predominate role among the LMW PBPs in maintaining the normal morphology of E. coli, because the loss of this protein severely alters the diameter, contour, and topology of mutants lacking multiple PBPs (5, 12, 18, 19). Thus, among the LMW PBPs, PBP 5 must have unique properties that allow the protein to modify bacterial shape.
There are at least two structural differences among the dd-carboxypeptidases that might explain how PBP 5 contributes to uniform cell shape and why the homologous enzymes are not equivalent substitutes. First, differences may exist in the amphipathic carboxy terminus that is proposed to anchor each enzyme to the outer face of the cytoplasmic membrane (6, 13, 14, 21). Variations among the dd-carboxypeptidase anchors might affect protein localization, enzymatic activity, or interactions with other components of the murein biosynthetic machinery (6, 10, 19). Previously, we established that the PBP 5 anchoring sequence does have physiological significance: anchorless PBP 5 does not reverse the morphological defects of a dacA mutant and is lethal at approximately 1/10 the amount of wild-type PBP 5 (19). However, that work did not address the question of whether anchoring sequences from different LMW PBPs perform equivalent functions.
A second possible structural distinction among the dd-carboxypeptidases was revealed by the recent work of Davies et al., who reported the crystal structure of a soluble version of PBP 5 (4). The bulk of PBP 5 consists of two distinct domains oriented approximately 90° to one another. Comparison of the PBP 5 crystal structure with the amino acid sequences of PBP 6 and DacD revealed that the core dd-carboxypeptidase domain (domain I) is highly conserved and is similar to the class A β-lactamases (4). In contrast, the β-sheet-rich carboxy-terminal domain (domain II) is less well conserved and has no homologues in published databases outside of the LMW PBPs (4). Davies et al. speculated that domain II could mediate protein-protein interactions between PBP 5 and other components of the murein biosynthetic apparatus or, alternately, that the domain could serve as an inert linker to position the active site near its peptidoglycan substrate in the periplasm (4).
Because the dd-carboxypeptidase PBPs can be considered to be multiply mutated allelic products of PBP 5, we rearranged the structural components of four of these proteins to determine whether domain I, domain II, or the membrane anchor distinguished PBP 5 from its nearest relatives in producing morphologically normal E. coli. Using a PCR-based domain-swapping strategy, we constructed fusion proteins that combined portions of PBP 5 with homologous portions of other dd-carboxypeptidases or with the heterologous transmembrane domain of ProW (8) and tested the ability of each fusion protein to complement the morphological defects of a multiple PBP mutant. The results suggest that the functional uniqueness of PBP 5 resides in domain I, the dd-carboxypeptidase enzymatic core.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli XL-1 Blue (recA endA hsdR supE thi recA gyrA relA lac) (Stratagene, La Jolla, Calif.) and E. coli DH5α (deoR recA endA hsdR supE thi gyrA relA) were used as hosts for constructing recombinant plasmids. Strains used in the morphological experiments were derived from CS109 (W1485 rpoS rph) (C. Schnaitman), as follows: CS604-2 (CS109 Δ[mrcA-yrfE-yrfF] ΔdacB ΔdacC ΔpbpG ΔampC ΔampH); CS701-1 (CS109 Δ[mrcA-yrfE-yrfF] ΔdacB ΔdacA ΔdacC ΔpbpG ΔampC ΔampH); and CS703-1 (CS109 ΔmrcA ΔdacB ΔdacA ΔdacC ΔpbpG ΔampC ΔampH) (5, 17). PBP genes were expressed under the control of the arabinose promoter of pBAD18-CAM, provided by J. Beckwith (7). Strains were grown on Luria-Bertani (LB) broth or agar plates, with chloramphenicol (20 μg/ml) added as required to maintain selection of pBAD plasmids. Overnight broth cultures of E. coli strains were diluted 1:250 into fresh LB medium and were allowed to enter mid-logarithmic growth before complementation experiments were performed (five to six doublings). When necessary, glucose (0.2%, wt/vol) was added to the medium to inhibit gene expression from the arabinose promoter. To induce protein expression in complementation experiments, cells were grown in the absence of glucose or in the presence of arabinose (0.0005%) (19). Unless otherwise noted, all chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Molecular techniques.
Plasmids were isolated from E. coli by using QIAprep Spin Miniprep and Midiprep kits (Qiagen Corp., Valencia, Calif.) according to the manufacturer's instructions. Competent cells were prepared and transformed by electroporation, using a Gene Pulser apparatus from Bio-Rad (Hercules, Calif.) according to the manufacturer's instructions. CS109 chromosomal DNA for PCR amplifications was prepared by boiling 200 μl of overnight culture with 800 μl of distilled water for 10 min, followed by centrifugation at 14,000 × g for 1 min and collection of the supernatant (18). DNA agarose gel electrophoresis was performed as described previously (22). DNA purification from agarose gels was performed with QIAquick gel extraction kits (Qiagen Corp.) as described by the manufacturer. Restriction digests and ligations were performed with enzymes purchased from New England Biolabs (Beverly, Mass.). Expression of PBP fusion proteins from recombinant plasmids was confirmed by labeling cells with 125I-penicillin X, separating total cellular protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualizing the proteins by autoradiography as described elsewhere (11, 19). Except when used in complementation experiments, plasmids were constructed and maintained in the E. coli strains DH5α and XL1-Blue.
PCR.
PCR was performed in a model 2400 Gene Amp thermal cycler (Perkin-Elmer, Boston, Mass.). Oligonucleotide primers for PCR were from Gibco Life Sciences (Grand Island, NY). Stock solutions of individual deoxynucleoside triphosphates for PCR were from Promega (Madison, Wis.). Deep Vent DNA polymerase was from New England Biolabs.
Construction of PBP gene fusions.
Portions of the gene encoding PBP 5 were fused with segments of other PBP genes by using a PCR-based strategy. Oligonucleotide primers used to amplify each gene segment are listed in Table 1. Sequences of the individual oligonucleotides are listed in Table 2. The general strategy was as follows. In the first PCR (PCR 1) a 5′ segment of the gene encoding PBP 5 (dacA) or PBP 6 (dacC) was amplified from E. coli CS109 chromosomal DNA by using forward primer P1 and reverse primer P2. An NheI site and Shine-Dalgarno sequence were included in the design of the 5′ terminus of the P1 primer so that these appeared in the final PCR product. The 5′ end of the P2 primer included 18 to 21 nucleotides that were exactly complementary to the sequence at the 5′ end of the P3 primer. Therefore, the sequence of the P2 oligonucleotide defined the junction at which the two gene fragments were fused. In a parallel PCR (PCR 2) the 3′ segment of the genes encoding PBP 5, PBP 6, and DacD (dacD) was amplified with primers P3 and P4. The design of the 5′ end of the P4 primer included a HindIII site. Next, the product of PCR 1 was amplified (PCR 3) by using as primers P1 and the product of PCR 2, which could hybridize to 18 to 21 nucleotides at the 3′ end of the PCR 1 product. Because priming by the PCR 2 product was inefficient, this procedure yielded only a small amount of composite product in which the 3′ segment of the PBP 5 or PBP 6 gene was replaced by the 3′ terminus of a separate gene. Therefore, the product of PCR 3 was purified by agarose gel electrophoresis, and primers P1 and P4 were employed to reamplify the composite gene (PCR 4). This final PCR product was purified by agarose gel electrophoresis, digested with restriction enzymes NheI and HindIII, and ligated into the vector pBAD18-CAM, where each respective fusion protein could be expressed under the control of the arabinose promoter. The nucleotide sequences of all inserts were confirmed by the sequencing facility of the Department of Biochemistry, Colorado State University (Fort Collins).
TABLE 1.
Plasmids and fusion proteins
| Plasmida | Terminus encodedb
|
Primersc | Sequence at junctiond | |
|---|---|---|---|---|
| Amino | Carboxyl | |||
| pAG6 | Wild-type PBP 6 | Wild-type PBP 6 | ||
| pPJ5C | Wild-type PBP 5 | Wild-type PBP 5 | ||
| pPJ5D | aa 1-385 of PBP 5 | Anchorless PBP 5 | ||
| pPJ5/4 | aa 1-385 of PBP 5 | 17 C-terminal aa of PBP 4 | A, B, C, D | RPLVVLQEIPEGN|PLVRFESRLYKDIY |
| pPJ5/6 | aa 1-376 of PBP 5 | 31 C-terminal aa of PBP 6 | A, E, F, G | QLDGKTIEQRPLV|VMENVEEGGFFGRV |
| pPJ5/D | aa 1-376 of PBP 5 | 23 C-terminal aa of DacD | A, H, I, J | QLDGKTIEQRPLV|VTLESVGEGSMFSR |
| pPJ5/W | aa 1-385 of PBP 5 | 58 C-terminal aa of ProW | A, K, L, M | RPLVVLQEIPEGN|LRGIGRLDMGLATV |
| pPJ5I/6II | aa 1-294 of PBP 5 | 113 C-terminal aa of PBP 6 | A, N, O, G | KKLLTWGFRFFET|VTPIKPDATFVTQR |
| pPJ6I/5II | aa 1-287 of PBP 6 | 109 C-terminal aa of PBP 5 | P, Q, R, S | EKLLTWGFRFFET|VNPLKVGKEFASEP |
All plasmids were constructed by cloning DNA fragments into the NheI-HindIII sites of pBAD18-CAM (Cmr). Plasmid pAG6 replaces pPJ6 and was constructed according to the description for pPJ6 as described previously (19). Plasmids pPJ5C and pPJ5D were described previously (19).
The number of amino acids and source of the protein encoded by each cloned DNA fragment. Each plasmid expressed a single hybrid PBP consisting of the amino acid segments indicated. The amino acids are numbered from the initiating methionine of the complete open reading frame. Subtract 29 from this number to arrive at the residue position in the mature protein, which is the convention adopted for reporting residue locations in the crystal structure.
Oligonucleotide primers used to construct the two segments of each of the cloned DNA fragments. The sequence of each oligonucleotide is given in Table 2. The oligonucleotides are presented in the order P1, P2, P3, and P4, as described in Materials and Methods.
Partial amino acid sequence (one-letter code) just before and after the junction site (vertical line) of hybrid PBPs, as encoded by the plasmids listed. The underlined sequences in pPJ5I/6II and pPJ6I/5II represent the α 10 helix and the first half of the β12 β-barrel that define the junction between domains I and II in the crystal structure of PBP 5 (4).
TABLE 2.
Primers and oligonucleotide sequences
| Primera | Oligonucleotide sequenceb | Genec |
|---|---|---|
| A | 5′-CTCTCTGCTAGCAGGAGGAATTCACCATGAATACCATTTTTTCCGC-3′ | dacA (F) |
| B | 5′-GCTTTCAAAACGCACTAACGG*GTTACCTTCCGGGATTTCTTG-3′ | dacA (R) |
| C | 5′-CCGTTAGTGCGTTTTGAAAGCCG-3′ | dacB (F) |
| D | 5′-CTCTCTCTCCAAGCTTCTAATTGTTCTGATAAATATCTTTATAC-3′ | dacB (R) |
| E | 5′-CTCTTCCACATTTTCCATCAC*AACCAGCGGGCGTTGCTCG-3′ | dacA (R) |
| F | 5′-GTGATGGAAAATGTGGAAGAGGGCGG-3′ | dacC (F) |
| G | 5′-CTCTCTAAGCTTTTAAGAGAACCAGCTGCC-3′ | dacC (R) |
| H | 5′-CCCGACAGATTCCAGGGT*AACCAGCGGGCGTTGCTCG-3′ | dacA (R) |
| I | 5′-ACCCTGGAATCTGTCGGGGAAGGCAG-3′ | dacD (F) |
| J | 5′-CTCTCTAAGCTTTCAGGCCTTATGGTGGAAATAATC-3′ | dacD (R) |
| K | 5′-CAGACGACCGATACCGCGAAG*GTTACCTTCCGGGATTTC-3′ | dacA (R) |
| L | 5′-CTTCGCGGTATCGGTCGTCTGG-3′ | proW (F) |
| M | 5′-CTCTCTCTCAAGCTTTTACTTAATGAATGGGCGGGTC-3′ | proW (R) |
| N | 5′-GGCATCAGGTTTAATTGGCGTCAC*GGTTTCAAAGAAACGGAAGCC-3′ | dacA (R) |
| O | 5′-GTGACGCCAATTAAACCTGATGCC-3′ | dacC (F) |
| P | 5′-CTCTTTGCTAGCAGGAGGAATTCACATGACGCAATACTCCTCTC-3′ | dacC (F) |
| Q | 5′-CTTTACCTACTTTCAGTGGGTTAAC*GGTTTCAAAGAAGCGGAAACCCCAGGT-3′ | dacC (R) |
| R | 5′-GTTAACCCACTGAAAGTAGG-3′ | dacA (F) |
| S | 5′-GCATGCAAGCTTCTAGATTTTTAACCAAACCAGTGATG-3′ | dacA (R) |
Primer designation in Table 1.
Underlined sequences are complementary to the 5′ (forward primer) or 3′ (reverse primer) ends of the gene fragments in each fusion construct. Italicized sequences are the portions of primer P2 that are complementary to the 5′ end of the gene fragment (primer P3) that will be fused to the 3′ end of the hybrid gene. An asterisk within the sequences of oligonucleotides used as P2 primers designates the fusion site between two coding sequences. Sequences in bold designate HindIII (AAGCTT) and NheI (GCTAGC) sites.
Gene to which each oligonucleotide anneals in PCR amplification. F, forward primer (P1 or P3 in the text); R, reverse primer (P2 or P4 in the text).
The fusion sites chosen to replace the carboxy terminus of PBP 5 with that of other PBPs were determined by aligning the protein sequences. The fusion site selected for replacing domains I and II of PBPs 5 and 6 was located in the sequence encoding α-helix 10, as represented in the crystal structure of PBP 5 (4). The amino acid sequences at each fusion junction are reported in Table 1.
Site-directed mutagenesis of PBP 5.
Site-directed mutagenesis of the carboxy-terminal amphipathic region of PBP 5 was performed with the QuikChange mutagenesis kit from Stratagene. Mutagenesis on supercoiled double-stranded pPJ5 DNA was carried out exactly according to the manufacturer's instructions, using oligonucleotide primer pairs (MWG Biotech Inc., High Points, N.C.) to create individual mutations in the dacA gene. The primers ranged from 34 to 61 bases, depending on the number of individual nucleotides to be altered (one to three). For each primer pair, one codon at the center was altered as described in Table 3. The exact primer sequences are available on request. The number of PCR cycles varied with the number of bases altered. Mutated pPJ5 DNA was transformed by heat shock into Epicurian XL1-Blue supercompetent cells (Stratagene) and plated on LB-chloramphenicol plates. After selection and purification of candidate plasmids, the existence of the correct mutations was confirmed by DNA sequencing (MWG Biotech Inc.).
TABLE 3.
Complementation by membrane anchor mutants of PBP 5
| Plasmid | Mutationa | Carboxy terminusb | Primerc | Codon change | Complementationd |
|---|---|---|---|---|---|
| pPJ5 | None | FFGKIIDYIKLMFHHWFG | NA | NA | Yes |
| pAG2A | Phe2 to Ala | FAGKIIDYIKLMFHHWFG | 2 | TTC to GCC | Yes |
| pAG1A | Phe2 to Leu | FLGKIIDYIKLMFHHWFG | 1 | TTC to GAC | No |
| pAG3A | Phe2 to Asp | FDGKIIDYIKLMFHHWFG | 3 | TTC to CTG | No |
| pAG7A | Gly3 to Ala | FFAKIIDYIKLMFHHWFG | 7 | GGC to GCC | Yes |
| pAG8A | Lys4 to Ala | FFGAIIDYIKLMFHHWFG | 8 | AAA to GCA | Yes |
| pAG9A | Lys4 to His | FFGHIIDYIKLMFHHWFG | 9 | AAA to CAT | Yes |
| pAG4A | Asp7 to Ala | FFGKIIAYIKLMFHHWFG | 4 | GAT to GCG | Yes |
| pAG10A | Asp7 to Glu | FFGKIIEYIKLMFHHWFG | 10 | GAT to GAA | Yes |
| pAG6A | Ile9 to Asp | FFGKIIDYDKLMFHHWFG | 6 | ATT to GAT | No |
| pAG5A | His14 to Ala | FFGKIIDYIKLMFAHWFG | 5 | CAT to GCG | Yes |
| pAG11A | His15 to Ala | FFGKIIDYIKLMFHAWFG | 11 | CAC to GCC | No |
Numbering is for the last 18 amino acids of wild-type PBP 5.
Changed amino acids are in bold.
Oligonucleotide sequences of mutagenic primer pairs are available on request. NA, not applicable.
Complete or virtually complete restoration of normal cellular morphology to E. coli CS701-1 or CS703-1 by mutant protein.
Preparation of membrane and soluble fractions.
Strains carrying recombinant clones were grown overnight at 37°C in LB plus chloramphenicol (20 μg/ml) and 0.2% glucose to inhibit expression of proteins cloned under the arabinose promoter. The cultures were diluted 1:250 into LB plus chloramphenicol (in the absence of glucose) and were incubated at 37°C until the culture reached an A600 of 0.2. At this point arabinose was added (0.01% final concentration) and incubation was continued for 1 h. The cells were harvested by centrifugation at 6,000 × g for 15 min, the pellets were resuspended in 2 ml of 0.1 M phosphate buffer, pH 7.5, containing 0.2 mg of Pefabloc protease inhibitor (Roche Diagnostics Corp., Indianapolis, Ind.) per ml, and the cells were disrupted by three passages through an Aminco French pressure cell (Aminco, Urbana, Ill.) at 16,000 lb/in2. Unbroken cells and debris were removed by centrifugation at 4,000 × g for 5 min. Membranes were pelleted from this clarified supernatant by centrifugation at 175,000 × g for 45 min at 4°C in a Beckman Optima TLX ultracentrifuge. The supernatant containing the soluble fraction of the bacterial lysate was recovered and concentrated to 50 μl with a Biomax 10K Ultrafree-4 centrifugal filter unit (Millipore Corp., Bedford, Mass.). The pellet, containing membranes, was washed once and resuspended in 50 μl of 0.1 M phosphate buffer, pH 7.5, plus 0.2 mg of Pefabloc per ml and stored at −70°C. Samples representing equal numbers of bacterial cells were labeled with 125I-penicillin X for SDS-PAGE analysis.
Photography and sequence analysis.
Photography was performed and interpreted as described previously (18, 19). Homologous protein sequences were identified and compared with the BLASTP 2.1.3 program (1) as supplied on the National Institutes of Health Entrez web site (http://www3.ncbi.nlm.nih.gov/Entrez/) and with the Clustal W program (version 1.81) (23) as supplied on the European Bioinformatics Institute website (http://www2.ebi.ac.uk/clustalw/). Helical wheel representations were produced with a program maintained at the University of Virginia (http://cti.itc.Virginia.EDU/∼cmg/Demo/wheel/wheelApp.html).
RESULTS
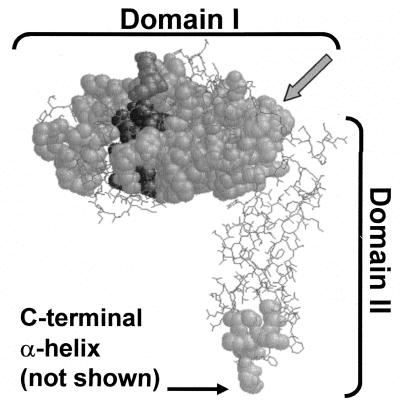
The nine proteins most closely related to E. coli PBP 5 were identified by BLAST searching, and their sequences were aligned with Clustal W (data not shown). We identified the amino acid sequences most highly conserved among the group and superimposed the positions of these segments onto the known crystal structure of PBP 5 (Fig. 1). The enzymes were highly similar to one another in domain I, which contains the dd-carboxypeptidase active site, but much less so in domain II, which forms a “stalk” of β-sheets between domain I and the carboxy-terminal membrane-anchoring sequence (not visible in the crystal structure).
FIG. 1.
Positions of highly conserved sequences among PBP 5 homologues. The most highly conserved sequences of nine dd-carboxypeptidase PBPs were mapped onto the crystal structure of PBP 5 (4). Conserved segments are shown as space-filled residues superimposed onto nonconserved segments, which are represented in wireframe. Darker residues are in or near the active site. The carboxy-terminal α-helix was absent from the crystal structure, but its point of attachment to the base of domain II is indicated. The gray arrow indicates the residues in the α10 helix that form the boundary between domains I and II. Sequences used in the alignments are listed in the legend to Fig. 5.
The carboxy-terminal membrane anchors of PBP 6 and DacD restore function to anchorless PBP 5.
To test the specificity of PBP 5 membrane anchoring, we constructed two plasmids, pPJ5/6 and pPJ5/DacD, in which the 27 carboxy-terminal amino acids of PBP 5 were replaced with the homologous membrane anchors of PBP 6 and DacD, respectively (Table 1). The plasmids were transformed into E. coli CS701-1 and CS703-1, and each construct expressed a PBP of the expected size (Fig. 2A, lanes 3 and 8, respectively). In addition, overexpression of the PBP 5/6 and PBP 5/DacD protein fusions was as lethal as wild-type PBP 5 (data not shown), indicating that the hybrids retained dd-carboxypeptidase activity.
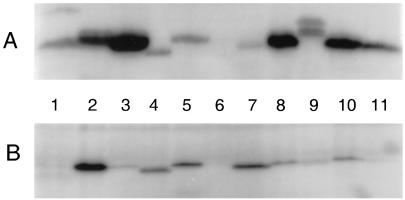
FIG. 2.
Membrane binding of wild-type and hybrid PBPs. Cloned PBP genes were induced for 1 h, after which membrane and soluble fractions were collected for labeling with 125I-penicillin X and visualization by SDS-PAGE (see Materials and Methods). (A) Membrane fractions; (B) soluble fractions. Lanes: 1, CS109 parental strain (pBAD18-Cam); 2, PBP 5 (pPJ5C); 3, PBP 5/6 (pPJ5/6); 4, PBP 6 (pAG6); 5, PBP 5/4 (pPJ5/4); 6, CS701-1 (pBAD18-Cam); 7, anchorless PBP 5 (pPJ5D); 8, PBP 5/D (pPJ5/D); 9, PBP5/ProW (pPJ5/W); 10, PBP 5I/6II (pPJ5I/6II); 11, PBP 6I/5II (pPJ6I/5II). CS701-1 was the host strain for samples in every lane except lane 1.
As predicted, each hybrid protein associated tightly to bacterial membranes (Fig. 2). Whereas anchorless PBP 5 fractionated almost completely into the soluble fraction (Fig. 2, lanes 7), the PBP5/6 and PBP5/DacD hybrids were retained on the membranes (Fig. 2, lanes 3 and 8). The appearance of some of each protein in the soluble fraction is normal when these PBPs are highly overexpressed. For example, when overproduced, wild-type PBP 5 fractionated between the soluble and membrane fractions (Fig. 2, lanes 2), whereas when produced at wild-type levels, PBP 5 was entirely membrane associated in the host strain (Fig. 2, lanes 1).
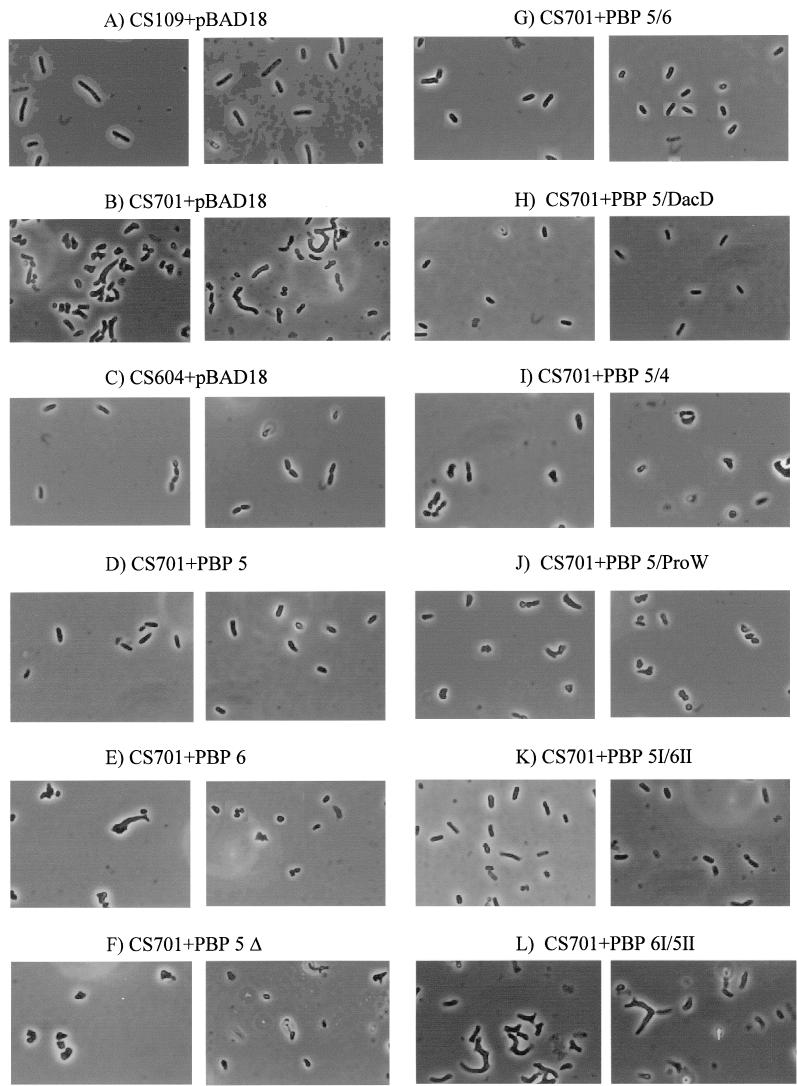
As reported previously (18), E. coli CS701-1(pBAD-Cam), from which seven PBPs are deleted (PBPs 4, 5, 6, and 7, DacD, AmpC, and AmpH), exhibited severe alterations in morphology (Fig. 3B), while the isogenic dacA+ (PBP 5+) strain, CS604-2 (Fig. 3C), was virtually indistinguishable from the original parent, CS109 (Fig. 3A). The gross morphological defects of CS701-1 were reversed by expression of PBP 5 in trans (Fig. 3D) but not by expression of PBP 6 (Fig. 3E) or anchorless PBP 5 (Fig. 3F) (18). In contrast to the latter two enzymes, expression of the PBP 5/6 and PBP 5/DacD fusion proteins complemented the defective morphology of CS701-1 (Fig. 3G and H, respectively). In all cases, when grown in the presence of 0.2% glucose to inhibit expression of cloned PBPs, each strain was morphologically aberrant, indicating that complementation was the direct result of expressing the cloned genes (data not shown). Although PBP 5 (Fig. 3D) and PBP 5/6 (Fig. 3G) did return the mutants to normal shape, diameter, and overall contour, close inspection revealed that many cells still had very slight abnormalities at their poles (partially squared or sometimes tapered). We do not know if this phenomenon is real or if it is the result of different expression levels of the complementing proteins among the cell population.
FIG.3.
Complementation of morphological defects by wild-type and hybrid dd-carboxypeptidase PBPs. Overnight cultures of E. coli strains with or without cloned PBP genes were diluted 1:250 into fresh LB broth and grown at 37°C until it reached an A600 of 0.2. Cells were collected, prepared for microscopy, and photographed at a magnification of ×1,000. Two representative fields are shown for each strain, and all photographs are at equal magnification. E. coli CS109 is the parental strain; CS604-2 is missing six PBPs but expresses wild-type levels of PBP 5; CS701-1 is missing the same six PBPs as CS604-2 but also carries a deletion of dacA so that it makes no PBP 5. All plasmids were derived from pBAD18-CAM (A, B, and C). Hybrid proteins were produced from the following plasmids (described in Table 1): pPJ5C (D), pAG6 (E), pPJ5D (anchorless PBP 5 [PBP 5 Δ]) (F), pPJ5/6 (G), pPJ5/DacD (H), pPJ5/4 (I), pPJ5/W (J), pPJ5I/6II (K), and pPJ6I/5II (L).
Because we recently discovered that strain CS701-1 was missing two non-PBP genes in addition to its documented PBP mutations (17), we introduced the plasmids into strain CS703-1, which is missing the same PBPs as CS701-1 but has no other mutations (17). CS703-1 exhibited the same morphological abnormalities as CS701-1, and the PBP 5/6 and PBP 5/DacD hybrid proteins reversed these effects (data not shown). Thus, the morphological phenotype depended only on the presence or absence of the PBPs and not on any other mutation in the original CS701-1 strain. Overall, the data indicate that the membrane anchors of PBP 5, PBP 6, and DacD are functionally interchangeable.
The putative membrane anchor of PBP 4 does not restore function to anchorless PBP 5.
To determine if the membrane anchor is functionally conserved in a more distantly related dd-carboxypeptidase, we constructed the plasmid pPJ5/4, in which the carboxy terminus of PBP 5 was replaced with the putative membrane anchor of PBP 4. To do this without altering protein domain II, the first 385 amino acids (aa) of PBP 5 (up to and including β barrel 22) were fused to the final 17 aa of PBP 4 (Table 1). The plasmid pPJ5/4 expressed a PBP of the expected size (Fig. 2, lanes 5), and about half of the hybrid PBP was membrane bound (Fig. 2, lanes 5) and lethal when overexpressed (data not shown), indicating that the protein retained dd-carboxypeptidase activity. However, even though some of the protein was membrane associated, the PBP 5/4 hybrid did not complement the morphological defects of CS701-1 (Fig. 3I).
Anchoring PBP 5 with a heterologous transmembrane domain does not restore function.
Since the amphipathic carboxy-terminal residues of PBP 6 and DacD restored activity to truncated PBP 5, it was possible that anchoring was simply a mechanical means of localizing the protein to the outer face of the inner membrane. We hypothesized that if this were true, then anchoring PBP 5 with an unrelated membrane-spanning sequence would also restore activity to the truncated protein. To test this, we created plasmid pPJ5/W, which encodes a protein in which the carboxy-terminal 18 aa of PBP 5 were replaced by the final transmembrane segment of the osmosensor protein, ProW (Table 1). The membrane orientation of this ProW peptide was characterized by Haardt and Bremer, who created an active PhoA-ProW fusion protein at this point (8). Thus, the signal sequence of PBP 5 exports the PBP5/ProW hybrid to the periplasm, but the protein remains tethered to the outer face of the inner membrane by the transmembrane segment of ProW (as observed for the PhoA-ProW fusion) (8).
A penicillin-binding PBP5/ProW hybrid protein was successfully expressed in E. coli CS701-1 (Fig. 2A, lane 9) and, like the other fusion constructs, was lethal when overexpressed (data not shown). This hybrid was even more strongly membrane bound than the previous fusion proteins, with very little appearing in the soluble fraction even when overexpressed (Fig. 2, lanes 9). The hybrid protein did not restore normal morphology to CS701-1 at any level of expression (Fig. 3J), suggesting that the proper physiological function of PBP 5 requires something more than simply being anchored to the membrane face.
It should be noted that, unlike the previously described fusion proteins, approximately half of the PBP5/ProW hybrid was degraded to a pair of shorter molecules at these expression levels (Fig. 2A, lane 9). The upper band in the figure is a molecule of approximately 49 kDa, the predicted size of the PBP5/ProW hybrid protein (48.66 kDa). The lower of the doublet bands is ∼45 kDa. If the ProW segment of the hybrid was degraded by proteolysis so that the carboxy-terminal cytoplasmic portion was removed up to the inner face of the inner membrane, the protein would be shortened by 34 aa (residues 321 to 354) (8) and would be reduced in size by 3.94 kDa while retaining the transmembrane domain. This corresponds exactly to what is observed: the smaller proteins are membrane associated and bind penicillin. The opposite possibility, removal of 34 aa at the amino terminus, would decrease the activity of or completely destroy the active site of PBP 5. Therefore, degradation at the carboxy terminus as described is the most likely explanation for the appearance of these bands. Even so, at least half of the PBP5/ProW hybrid protein was full length, and even the smaller hybrid proteins retained PBP 5 activity and were tethered to the correct face of the inner membrane.
Site-directed mutagenesis of the membrane anchor of PBP 5.
The results implied that in order to restore normal cellular morphology, active PBP 5 should be attached to the inner membrane by a specific mechanism. Comparison of several closely related PBP 5 homologues suggested that three residues in the carboxy-terminal membrane anchor might be important for proper orientation of this domain or for mediating interactions with the membrane or other proteins (see Discussion). To determine if these conserved residues were essential to the function of PBP 5, we altered seven different residues to create 11 mutations of the dacA gene in plasmid pPJ5 (Table 3). Each plasmid was transformed by electroporation into CS701-1 or CS703-1, and the ability of mutant proteins to restore normal morphology was scored visually by microscopy (Table 3 and morphological data not shown).
Mutations that interrupted the hydrophobic face of the amphipathic helix by inserting a charged amino acid (Ile9-Asp and Phe2-Asp) destroyed the ability of PBP 5 to complement aberrant morphology (Table 3). A mutant protein in which the conserved phenylalanine residue was converted to alanine (Phe2-Ala) retained wild-type activity, but complementation was destroyed by altering the same residue to leucine (Phe2-Leu) (Table 3). Among the homologous membrane anchors, the two most strongly conserved residues were Lys4 and Asp7 (Fig. 4 and 5). Conversion of these residues to similarly charged amino acids resulted in active proteins (Lys4-His and Asp7-Glu), as might be expected if the preservation of charge at that position were important. However, mutant proteins in which each of these residues was converted to the uncharged amino acid alanine (Lys4-Ala and Asp7-Ala) complemented the morphological phenotype as well as the wild-type protein (Table 3), suggesting that specific charges were not necessary for protein function. Likewise, conversion of the charged residue His14 to alanine did not damage the activity of PBP 5, but replacing His15 with alanine did prevent complementation (Table 3).
FIG. 4.
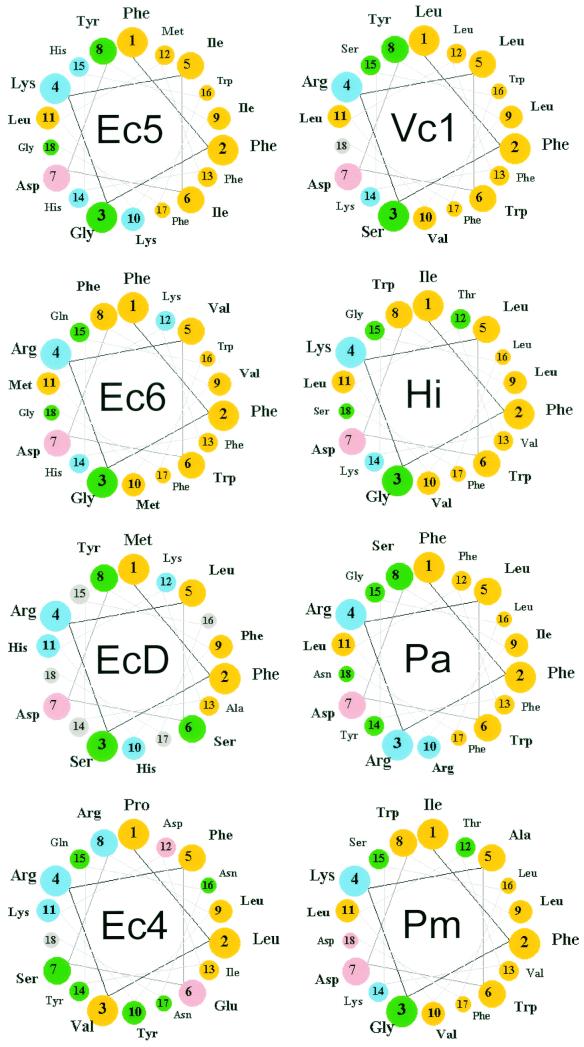
Helical wheel representations of the carboxy-terminal amino acid sequences of PBP 5 homologues. The carboxy-terminal α-helices of several dd-carboxypeptidase PBPs were determined by alignment with the sequence of PBP 5 (Fig. 5) and represented in a helical-wheel format. The first residue of the membrane anchoring sequence was defined as the amino acid immediately following the PEGN residues that mark the end of the crystal structure of PBP 5 in E. coli (4). Color key: yellow, nonpolar; green, polar uncharged; pink, acidic; blue, basic. Abbreviations (with accession numbers, when applicable): Ec5, E. coli PBP 5; Ec6, E. coli PBP 6; EcD, E. coli DacD; Ec4, E. coli PBP 4; Vc1, Vibrio cholerae group O1 strain N16961 (VC0947); Hi, Haemophilus influenzae PBP 5 (HI0029); Pa, Pseudomonas aeruginosa (PA3999); Pm, Pasteurella multocida DacA (PM1927).
FIG. 5.
Sequence alignment of carboxy termini from PBP 5 homologues. The amino acid sequences of the dd-carboxypeptidase PBPs most closely related to PBP 5 from E. coli were aligned, beginning with the PEGN sequence that marks the end of the crystal structure (4). Identical amino acids are highlighted in black, and similar amino acids are highlighted in gray. Consensus residues at the most highly conserved positions are noted below the alignments. Abbreviations (with accession numbers, when applicable): Ec5, E. coli PBP 5; Ec6, E. coli PBP 6; Vc1, V. cholerae group O1 strain N16961 (VC0937); Pm, P. multocida DacA (PM1927); Hi, H. influenzae PBP 6 (HI0029); Pa, P. aeruginosa (PA3999); Xf, Xylella fastidiosa PBP 6 (XF2230); St, Salmonella enterica serovar Typhimurium LT2 PBP 6b; Mll, Mesorhizobium loti (mll0426).
Domain I contributes morphological specificity to PBP 5.
The majority of amino acid sequence differences between PBP 5 and related dd-carboxypeptidases are located in domain II, whereas the proteins are more highly conserved in domain I (Fig. 1) (4). To determine which domain imparted to PBP 5 its specificity in creating morphologically normal bacterial cells, we constructed a hybrid protein in which domain I of PBP 5 was fused to domain II of its most nearly identical relative, PBP 6, and the analogous hybrid in which domain I of PBP 6 was fused to domain II of PBP 5 (Table 1). The boundary between these two domains spans α-helix 10 (Fig. 1), where the sequence is identical in PBPs 5 and 6 (4). Therefore, this site served as a natural junction for the fusion proteins.
Plasmid pPJ5I/6II encoded a protein containing the amino-terminal 294 residues of PBP 5 fused to the final 113 carboxy-terminal residues of PBP 6, and plasmid pPJ6I/5II encoded the amino-terminal 287 aa of PBP 6 fused to the final 109 carboxy-terminal residues of PBP 5 (Table 1). Each hybrid was successfully expressed as an active PBP (Fig. 2A, lanes 10 and 11). The PBP 5I/6II hybrid protein was as lethal upon overexpression as wild-type PBP 5, but the PBP 6I/5II hybrid was less lethal (data not shown), mirroring the reduced lethality of wild-type PBP 6 (19).
The PBP 5I/6II hybrid protein complemented the morphological defects of the dacA mutant CS701-1 just as well as wild-type PBP 5 (Fig. 3K). On the other hand, the PBP 6I/5II hybrid did not restore normal morphology to CS701-1 (Fig. 3L). The results indicate that the ability of PBP 5 to create morphologically normal cells, and the inability of PBP 6 to do so, is determined primarily by differences in the enzymatically active domain I.
DISCUSSION
Because multiply mutated strains lacking PBP 5 form abnormally shaped cells (18, 19), understanding this protein is a useful approach to answering the question of how bacteria create and maintain a defined and uniform shape. However, E. coli and other bacteria express multiple dd-carboxypeptidase PBPs that share similar enzymatic capabilities. Therefore, the easiest way to explain the fact that one protein plays a dominant role in cellular morphology is to invoke differences in subcellular localization, protein interactions or timing of expression. The two structural components outside the active site that might impart such special properties to PBP 5 are the carboxy-terminal membrane anchor and an elongated domain of β-sheets that can be viewed as a stalk holding up the more globular enzymatic domain (4). As a first step toward describing this mechanism of morphological determination, we used hybrid molecules to identify the functional modules of PBP 5 responsible for influencing cell shape.
Function of the carboxy-terminal amphipathic helix.
PBP 5 is localized to the outer face of the cytoplasmic membrane by an amphipathic helix formed by its carboxy-terminal 18 aa (13, 14, 21). Although it appears that PBP 5 must be membrane bound to produce normally shaped cells (19), questions remain about the specificity of attachment and whether it regulates the biochemical activity of PBP 5. For example, the membrane anchor might promote participation in a multiprotein complex (20), or binding to penicillin or peptidoglycan might modulate membrane attachment (6, 20). So far, no in vivo data exist for either possibility. Instead, the results reported here call into question whether either of these possibilities occurs or is required for PBP 5 function.
First of all, the composition or function of the anchoring domain does not distinguish PBP 5 from the other dd-carboxypeptidases in E. coli, because the morphological defects of a dacA mutant were complemented equally well by PBP 5 variants possessing the anchors of PBP 6 and DacD. This is important because even though PBP 6 is the PBP most closely related to PBP 5, its carboxy-terminus shares only 9 of 18 aa with PBP 5 (Fig. 4 and 5). The sequence of the DacD anchor is even less similar: the carboxy terminus is shorter (13 aa instead of 18), and only 4 of 13 residues are identical to those in PBP 5 (Fig. 4, wheel EcD). That both PBP 6 and DacD sequences restored activity to anchorless PBP 5 implies that a general structure, not a specific sequence, may be more important for membrane anchoring and protein function.
This conclusion is supported by examining the helical wheel representations of the dd-carboxypeptidases most likely to act as PBP 5 homologues in related bacteria (Fig. 4). Two structural similarities stand out. First, all are strongly amphipathic, with a broad hydrophobic face opposite a charged hydrophilic face. Second, the amino acids at positions 2 (Phe), 4 (Lys or Arg), and 7 (Asp) are invariant (an Fx[K/R]xxD motif), as is position 17 (Phe) among those with termini of sufficient length (Fig. 4 and 5). The exception to the motif pattern is the terminus of PBP 4, which was ineffective in restoring activity to anchorless PBP 5. The principal finding is that the sequences comprising both helix faces are extremely divergent, including the extremes of having two to five charged residues on the hydrophilic face. Nonetheless, it remained possible that the three conserved residues in the Fx[K/R]xxD motif might play specific biochemical roles. However, when each of these three residues was replaced with alanine, the mutant proteins retained wild-type function, a strong argument that these “motif” amino acids do not mediate essential protein-protein or autoregulatory interactions. On the other hand, complementation was destroyed by interrupting the amphipathic structure with charged amino acids in the hydrophobic face of the helix. Together, these data are most consistent with the idea that the carboxy terminus contributes a general structure with a simple role in membrane binding.
Even though the evidence argues that the carboxy terminus probably serves only to attach PBP 5 to the membrane, it is clear that the precise mechanism of anchoring remains in question, as does the relationship between anchoring and protein function. For example, a histidine-to-alanine mutation at position 15 of the anchor hampered complementation, even though this residue is not at all conserved among PBP 5 homologues (Fig. 4) and even though this residue is different than (PBP 6) or completely absent from (DacD) the two sequences that restored functionality to anchorless PBP 5. In addition, although the carboxy-terminal sequence of PBP 4 appears to be able to form a helix almost as amphipathic as that of DacD (Fig. 4, wheels Ec4 and EcD), this potential anchor did not restore function to truncated PBP 5. Membrane binding by this amphipathic helix is not strong, as evidenced by the fact that 80% of overexpressed PBP 4 remains soluble (15) and that an artificial PBP 4 terminal oligopeptide binds poorly to membrane vesicles (9). Still, some of the PBP 5/4 fusion protein investigated here was membrane attached, suggesting that the association was ineffectual for unknown reasons.
Additional evidence for an unusual requirement for membrane binding comes from observing that the PBP 5/ProW fusion protein failed to complement the morphological defects of a dacA mutant. In contrast to the results for PBP 5/4, the PBP 5/ProW hybrid fractionated almost entirely with membrane and was, in fact, more strongly attached than wild-type PBP 5. Thus, the failure of PBP 5/ProW to complement the morphological phenotype suggests that simple mechanical tethering of the enzyme to the appropriate membrane face is not itself sufficient for proper physiological function of PBP 5. Perhaps anchoring PBP 5 via its normal amphipathic sequence allows a flexibility, freedom of movement or localization not available to the PBP 5/ProW hybrid, which is not tethered to the membrane surface but is embedded via a transmembrane domain. Or perhaps the carboxy-terminal anchor interacts with domain II of PBP 5 to orient the enzyme in the periplasm, so that the PBP 5/ProW protein is inactive because its motion is unrestrained.
Overall, the simplest conclusion is that the sole function of the carboxy terminus is to tether PBP 5 to the membrane without embedding it there. Although the specific mechanism is still in question, two things seem certain. First, several highly divergent and mutant membrane anchors work perfectly well in place of the natural terminus of PBP 5. And second, the speculative interactions between PBP 5 and other proteins must involve very few, if any, specific residues, and such interactions as may occur must be extremely forgiving.
Functions of domains I and II.
Because the dd-carboxypeptidase PBPs perform the same enzymatic reaction, though at different rates (2), we expected that the unique morphological function of PBP 5 would reside in the structural features of domain II. When we tested this proposition by creating hybrid proteins in which the two domains were exchanged between PBPs 5 and 6, we were surprised to discover that complementation of morphological defects was linked solely to PBP 5 domain I. A composite protein carrying this domain fused to domain II of PBP 6 functioned just as effectively as wild-type PBP 5, whereas domain II of PBP 5 did not impart a similar functionality to domain I of PBP 6. The different capabilities of PBPs 5 and 6 could arise because the enzymes recognize different substrates or catalyze reactions at different rates. PBP 5 is three to four times more active toward certain artificial substrates than is PBP 6 (2), but whether this is sufficient to explain the difference between the two enzymes is not clear. Also, the enzymes may have functions beyond the in vitro enzymology we know about. Identifying the specific sequence(s) within domain I that gives PBP 5 its singular ability will hopefully help narrow the list of possible mechanisms.
Davies et al. envisioned one of two functions for domain II—as a simple mechanical device elevating the active site of the enzyme to a fixed distance from the membrane, or as a participant in protein-protein interactions, possibly in a multienzyme complex (4). Although our data do not differentiate between these alternatives, it is clear that the function of domain II does not distinguish PBP 5 from PBP 6 (or, probably, from other dd-carboxypeptidases). Thus, the two forms of domain II have equivalent mechanical functions in these proteins, or, if domain II guides PBP 5 to a multienzyme complex, then PBP 6 must be capable of participating in the same complex. If the latter is true, different carboxypeptidase PBPs may compete for a common site within these complexes, raising the possibility that such competition might have regulatory consequences.
In summary, the ability of PBP 5 to create uniformly shaped bacterial cells relies on a distinct mechanism of membrane attachment and on unknown specificities of its enzymatically active domain. Further characterization of this system should lead to a better understanding of how bacteria generate different shapes and may also address the question of why shape should matter in bacterial physiology and survival.
Acknowledgments
This work was supported by grant GM61019 from the National Institutes of Health. D. E. Nelson was supported by a North Dakota EPSCoR doctoral fellowship from the National Science Foundation. A. L. Paulson was supported by grant GM61019-S1 from the National Institutes of Health.
The first two authors contributed equally to this work.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amanuma, H., and J. L. Strominger. 1980. Purification and properties of penicillin-binding proteins 5 and 6 from Escherichia coli membranes. J. Biol. Chem. 255:11173-11180. [PubMed] [Google Scholar]
- 3.Baquero, M.-R., M. Bouzon, J. C. Quintela, J. A. Ayala, and F. Moreno. 1996. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J. Bacteriol. 178:7106-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, C., S. W. White, and R. A. Nicholas. 2001. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3-Å resolution. J. Biol. Chem. 276:616-623. [DOI] [PubMed] [Google Scholar]
- 5.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gittins, J. R., D. A. Phoenix, and J. M. Pratt. 1994. Multiple mechanisms of membrane anchoring of Escherichia coli penicillin-binding proteins. FEMS Microbiol. Rev. 13:1-12. [DOI] [PubMed] [Google Scholar]
- 7.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haardt, M., and E. S. O. Bremer. 1996. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J. Bacteriol. 178:5370-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, F., R. Demel, B. de Kruijff, and D. A. Phoenix. 1998. An investigation into the lipid interactions of peptides corresponding to the C-terminal anchoring domains of Escherichia coli penicillin-binding proteins 4, 5 and 6. Biochim. Biophys. Acta 1415:10-22. [DOI] [PubMed] [Google Scholar]
- 10.Harris, F., and D. A. Phoenix. 1998. The Escherichia coli low molecular mass penicillin-binding proteins and a putative membrane bound protein complex. Membr. Cell Biol. 11:591-596. [PubMed] [Google Scholar]
- 11.Henderson, T. A., P. M. Dombrosky, and K. D. Young. 1994. Artifactual processing of penicillin-binding proteins 7 and 1b by the OmpT protease of Escherichia coli. J. Bacteriol. 176:256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson, T. A., K. D. Young, S. A. Denome, and P. K. Elf. 1997. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J. Bacteriol. 179:6112-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson, M. E., and J. M. Pratt. 1987. An 18 amino acid amphiphilic helix forms the membrane-anchoring domain of the Escherichia coli penicillin-binding protein 5. Mol. Microbiol. 1:23-28. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, M. E., and J. M. Pratt. 1988. Analysis of the membrane-binding domain of penicillin-binding protein 5 of Escherichia coli. Mol. Microbiol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 15.Korat, B., H. Mottl, and W. Keck. 1991. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol. Microbiol. 5:675-684. [DOI] [PubMed] [Google Scholar]
- 16.Massova, I., and S. Mobashery. 1998. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 42:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meberg, B. M., F. C. Sailer, D. E. Nelson, and K. D. Young. 2001. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J. Bacteriol. 183:6148-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson, D. E., and K. D. Young. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson, D. E., and K. D. Young. 2001. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phoenix, D. A., and J. M. Pratt. 1993. Membrane interaction of Escherichia coli penicillin binding protein 5 is modulated by the ectomembranous domain. FEBS Lett. 322:215-218. [DOI] [PubMed] [Google Scholar]
- 21.Pratt, J. M., M. E. Jackson, and I. B. Holland. 1986. The C terminus of penicillin-binding protein 5 is essential for localisation to the E. coli inner membrane. EMBO J. 5:2399-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]