Abstract
Xanthomonadins are membrane-bound, brominated, aryl-polyene pigments specific to the genus Xanthomonas. We have characterized a genetic locus (pig) from Xanthomonas oryzae pv. oryzae which contains four open reading frames (ORFs) that are essential for xanthomonadin production. Three of these ORFs are homologous to acyl carrier proteins, dehydratases, and acyl transferases, suggesting a type II polyketide synthase pathway for xanthomonadin biosynthesis. The fourth ORF has no homologue in the database. For the first time, we report that a putative cytoplasmic membrane protein encoded in the pig locus is required for outer membrane localization of xanthomonadin in X. oryzae pv. oryzae. We also report the identification of a novel 145-bp palindromic Xanthomonas repetitive intergenic consensus element that is present in two places in the pig locus. We estimate that more than 100 copies of this element might be present in the genome of X. oryzae pv. oryzae and other xanthomonads.
Members of the genus Xanthomonas cause diseases in a wide range of host plants (25). Many of them can also survive and multiply on plant leaf surfaces as epiphytes without affecting the host (15, 44). A characteristic feature of most xanthomonads is the production of yellow, membrane-bound (46), halogenated, aryl-polyene pigments (2, 3) called xanthomonadins, which serve as useful chemotaxonomic (45) and diagnostic markers (41). Xanthomonadin is not essential for in planta growth of bacterial cells (13, 35, 48). However, it is being established progressively that xanthomonadin protects bacteria against photooxidative damage. Pigment-deficient mutants of Xanthomonas juglandis (20), Xanthomonas oryzae pv. oryzae (37), and Xanthomonas campestris pv. campestris (33) are more susceptible to photooxidative damage than are wild-type strains. In vitro, xanthomonadin protects membrane lipids in egg-phosphatidylcholine liposomes against peroxidation (37). A transposon-induced xanthomonadin-deficient mutant of X. campestris pv. campestris has been recently shown to be 1,000-fold reduced for epiphytic survival in conditions of high light intensity (33). These observations suggest that xanthomonadin might aid the survival of xanthomonads on plant leaf surfaces by providing protection against photooxidative damage. In spite of their apparent importance, very little is known about the biosynthesis and mechanism of action of xanthomonadins.
An 18.6-kb genomic region from X. campestris pv. campestris that contains seven transcriptional units (pig genes) required for xanthomonadin production has been isolated (34, 35). One of the transcriptional units, pigB, is involved in biosynthesis of a diffusible factor that affects the production of xanthomonadin as well as extracellular polysaccharide (34). The unavailability of nucleotide sequences for these transcriptional units, however, limits further insight into the mechanism of xanthomonadin biosynthesis. X. oryzae pv. oryzae mutants that are defective in shikimate dehydrogenase activity have been shown to be deficient for pigment production (14). It was suggested that the aryl ring in the pigment might be derived from the aromatic amino acid biosynthetic pathway.
We have sequenced and characterized by insertional inactivation a 21.0-kb genomic region from X. oryzae pv. oryzae that is homologous to the previously identified X. campestris pv. campestris pig gene cluster. Our data suggest that the X. oryzae pv. oryzae pig cluster contains three open reading frames (ORFs) that might be part of a novel type II polyketide synthase (PKS) pathway involved in the biosynthesis of xanthomonadin. We demonstrate that the pigment localizes to the outer membrane in X. oryzae pv. oryzae and that a putative cytoplasmic membrane protein encoded in the pig cluster is required for this localization.
MATERIALS AND METHODS
Bacterial strains and media.
All bacterial strains and plasmids used are listed in Table 1. X. oryzae pv. oryzae strains were grown at 28°C in either peptone-sucrose medium (48) or modified Miller's minimal medium M4 (21). Escherichia coli strains were grown in Luria-Bertani medium (28) at 37°C. The concentrations of the antibiotics used were as follows: rifampin, 50 μg ml−1; spectinomycin, 50 μg ml−1; ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; chloramphenicol, 20 μg ml−1; streptomycin, 50 μg ml−1; tetracycline, 20 μg ml−1; and cycloheximide, 75 μg ml−1.
TABLE 1.
List of plasmids and strains used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Plasmids | ||
| pBluescript KS | Apr | Stratagene, La Jolla, Calif. |
| pMOSBlue | Apr | Amersham Pharmacia Biotech |
| pBR329 | Apr Cmr Tcr | 8 |
| pUFR034 | IncW Nmr Tra− Mob+mob(P) lacZα+ Par+cos | 9 |
| pIG102 | pLAFR3 + 25.4-kb X. campestris pv. campestris DNA that encodes pig genes | 35 |
| pLR9 | pUFR034 + 16.5-kb X. oryzae pv. oryzae insert including the pig locus | This study |
| pLR10 | pUFR034 + 21.5-kb X. oryzae pv. oryzae insert including the pig locus | This study |
| pAG13 | pBluescript KS + 5.0-kb EcoRI fragment from X. oryzae pv. oryzae pig locus | This study |
| pAG14 | pBluescript KS + 4.5-kb EcoRI fragment from X. oryzae pv. oryzae pig locus | This study |
| pAG15 | pBR329 + 12.0-kb EcoRI fragment from the pig locus of X. oryzae pv. oryzae | This study |
| pAG16 | pBluescript KS + 500-bp XhoI-SmaI fragment from orf11 | This study |
| pAG17 | pMOSBlue + 200-bp PCR-amplified fragment from orf5 | This study |
| X. oryzae pv. oryzae strains | ||
| BXO1 | Laboratory wild type, an Indian isolate | Lab collection |
| BXO43 | rif-2, derivative of BXO1 | Lab collection |
| BXO717 | pig-15::mTn5 rif-2 (a mutation in orf10) | This study |
| BXO718 | pig-16::mTn5 rif-2 (a mutation in orf6) | This study |
| BXO719 | pig-17::mTn5 rif-2 (a mutation in orf3) | This study |
| BXO720 | pig-18::mTn5 rif-2 (a mutation in orf3) | This study |
| BXO1708 | pig-55::tetA rif-2 (a mutation in orf7) | This study |
| BXO1709 | pig-56::mTn7 rif-2 (a mutation in orf4) | This study |
| BXO722 | zxx-38::mTn5 rif-2 (a mutation in orf1) | This study |
| BXO731 | zxx-10::mTn5 rif-2 (a mutation in orf1) | This study |
| BXO730 | zxx-9::mTn5 rif-2 (an insertion in the orf1-orf2 intergenic region) | This study |
| BXO727 | zxx-6::mTn5 rif-2 (a mutation in orf2) | This study |
| BXO728 | zxx-7::mTn5 rif-2 (a mutation in orf2) | This study |
| BXO729 | zxx-8::mTn5 rif-2 (a mutation in orf2) | This study |
| BXO721 | zxx-39::mTn5 rif-2 (a mutation in orf2) | This study |
| BXO1710 | zxx-40::bla rif-2 (a mutation in orf5) | This study |
| BXO724 | zxx-3::mTn5 rif-2 (a mutation in orf8) | This study |
| BXO725 | zxx-4::mTn5 rif-2 (a mutation in orf9) | This study |
| BXO1711 | zxx-35::bla rif-2 (a mutation in orf11) | This study |
| BXO1712 | zxx-36::mTn7 rif-2 (a mutation in orf12) | This study |
| BXO1713 | zxx-37::mTn7 rif-2 (a mutation in orf12) | This study |
| BXO1714 | BXO1708/pLR9-pig16::mTn5 | This study |
| BXO1715 | BXO1709/pLR9-zxx7::mTn5 | This study |
rif-2 indicates a mutation that confers resistance to rifampin. pig indicates mutations that cause deficiencies in either pigment production or localization. zxx indicates mutations that do not have a visible effect on pigmentation.
Southern and colony hybridizations.
Genomic DNA was isolated from the X. oryzae pv. oryzae strains as described earlier (23). Plasmids were isolated from E. coli cultures by using the alkaline lysis method (5). Southern hybridizations were performed as described by Yashitola et al. (50). Colony blots of X. oryzae pv. oryzae clones were prepared (6) and hybridized with pIG102 (35) as described previously (50). Restriction digestion, ligation, transformation, and [α-32P]dATP labeling of DNA fragments were performed as per Sambrook et al. (40). Restriction enzymes were purchased from New England Biolabs, Inc. (NEB), Beverly, Mass., and the Hybond N membranes were purchased from Amersham Life Science, Little Chalfont, Buckinghamshire, England.
Mutagenesis, marker exchange, and complementation.
Thirteen independent mini-Tn5 (mTn5) (Tn5gusA40) (49) insertions were obtained on the cloned DNA in pLR9 and marker exchanged into the BXO43 background (38) to obtain the mutant strains listed in Table 1. The 5.0- and 4.5-kb EcoRI fragments from pLR10 (Table 1) were cloned into pBluescript KS to obtain pAG13 and pAG14, respectively. The 12.0-kb fragment was cloned into pBR329 to obtain pAG15 (Table 1). The plasmids pAG13 and pAG15 were mutagenized with mTn7 (genome priming system; NEB) in order to sequence the cloned DNA as well as to isolate insertions within individual ORFs. The mTn7 insertions zxx-36::mTn7 and zxx-37::mTn7 obtained in orf12 and pig-56::mTn7 obtained in orf4 were electroporated into BXO43 cells by selecting Kmr Aps colonies (mTn7 encodes Kmr). The plasmid vectors pBluescript KS and pBR329 (Apr vectors) do not replicate in X. oryzae pv. oryzae, therefore, the Kmr Aps colonies are expected to have undergone a marker exchange event. The mutant strains BXO1709 (pig-56::mTn7), BXO1712 (zxx-36::mTn7), and BXO1713 (zxx-37::mTn7) were obtained in this manner.
After the mTn5 and mTn7 mutagenesis, no insertions could be obtained in ORFs 5, 7, and 11. In order to mutagenize orf7 (Fig. 1), a BamHI-cut tetA cassette was cloned into a BclI restriction site in this ORF to obtain the plasmid pAG14-pig55::tetA (BclI and BamHI produce compatible ends that can be ligated together). This plasmid was electroporated into BXO43 to obtain BXO1708 (Aps Tcr Pig−). Mutagenesis of orf11 and orf5 was done by cloning DNA fragments, truncated at the 5′ and 3′ termini of these ORFs, into plasmid vectors that do not replicate in X. oryzae pv. oryzae. A 500-bp XhoI-SmaI fragment from orf11 was cloned into pBluescript KS to obtain the plasmid pAG16. Similarly, a 200-bp PCR-amplified fragment from orf5 was cloned into pMOSBlue (Table 1) to obtain pAG17. The plasmids pAG16 and pAG17 were then electroporated into BXO43 electrocompetent cells to obtain the Apr strains BXO1711 and BXO1710, respectively. The Apr strains were expected to have undergone a single recombination event that would create a gene disruption. All of the mutants obtained were analyzed by Southern hybridization to confirm that the gene disruption events had occurred as expected (data not shown).
FIG. 1.
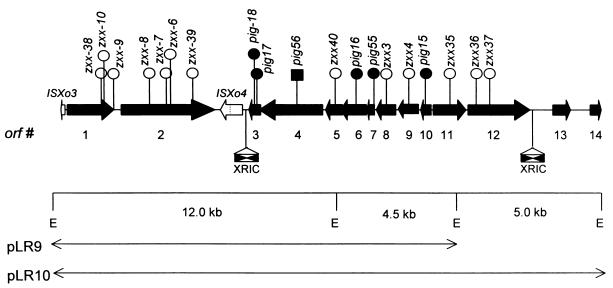
Genetic organization of the pig locus of X. oryzae pv. oryzae. An EcoRI restriction map of two cosmid clones, pLR9 and pLR10, is shown (E, EcoRI site). The solid black arrows represent the 14 ORFs identified in the 20,310-bp region. The direction of the arrow indicates the orientation of the ORF. The white arrows represent two ISs, ISXo3 and ISXo4. A frameshift mutation in the ISXo4 transposase is indicated by a dotted line. The filled and empty circles above the ORFs indicate the insertions that cause the Pig− and Pig+ phenotypes, respectively. An insertion in Orf4 that affects the transport of the pigment to the outer membrane but does not affect the visible levels of pigmentation is represented by a solid rectangle. Two copies of a 145-bp XRIC sequence are shown in the orf3-ISXo4 and orf12-orf13 intergenic regions. The oppositely oriented solid triangles within these elements represent the imperfect inverted repeats.
For complementation analysis, clones pLR9 and pLR10 were mobilized by using biparental matings (14) into the pigment-deficient mutants BXO717, BXO718, BXO719, BXO720, and BXO1708. These clones could not be directly introduced into the transport-defective mutant BXO1709 (pig-56::mTn7) (see Results), as mTn7, pLR9, and pLR10 (Table 1) encode kanamycin resistance. Therefore, a pLR9 derivative, pLR9-zxx-7::mTn5 (Kmr Spr), was mobilized into BXO1709 to obtain BXO1715 (mTn5 encodes Spr).
DNA sequence analysis.
The sequence of the 20,310-bp X. oryzae pv. oryzae region was obtained by using primers directed outward from mTn7 (provided in the genome priming system kit from NEB) and mTn5 (10). Primer walking was performed to fill gaps. The sequencing reactions, electrophoresis, and sequence data analyses were performed with the ABI Prism 3700 automated DNA sequencer (Perkin Elmer, Foster City, Calif.). ORF analysis and homology searches were performed by using ORF finder and the BLAST algorithm (1), respectively, which are available at www.ncbi.nlm.nih.gov. Analysis of the transmembrane domains (TMDs) for Orf4 was performed by using TMpred software (16), which is available at www.expasy.ch. The palindromic structure (ΔG = −44.9 kcal mol−1) of the orf3 (X. oryzae pv. oryzae) Xanthomonas repetitive intergenic consensus (XRIC) element was predicted by using SeqWeb software. Sequence alignment of the XRIC sequences was performed by using the MULTALIN software, which is available at http://prodes.toulouse.inra.fr/multalin/multalin.html (7).
Separation of cytoplasmic and outer membranes.
The cytoplasmic and outer membranes of X. oryzae pv. oryzae were separated as described previously for X. campestris pv. campestris (18). In brief, the X. oryzae pv. oryzae cultures were grown to the late exponential phase in peptone-sucrose medium. Cells were harvested by centrifugation, washed with ice-cold distilled water, and resuspended in 10 mM HEPES, pH 7.5 (approximately 1 ml for 100 to 150 mg [wet weight] of cells), containing 1 mM MgCl2, DNase (50 μg ml−1), and RNase (50 μg ml−1). The cells were disrupted by being passed twice through a French pressure cell at 18,000 to 20,000 lb/in2. Unbroken cells were removed by centrifugation at 3,500 × g for 10 min. The total-membrane preparation was pelleted by centrifugation at 342,000 × g for 45 min, washed once with 10 mM HEPES, pH 7.5, and resuspended in the same buffer. Concentrated membranes were then loaded on a step sucrose gradient (25 to 65%) and centrifuged at 130,000 × g for 20 h at 4°C. Densities of the fractions were calculated by using the refractive index values determined with a refractometer. Samples from each fraction were loaded on Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (24) and stained with silver nitrate (47). The samples were also treated with protease, and the protease-resistant bands on Tricine-SDS-PAGE gels were compared with lipopolysaccharide (LPS) profiles. LPS was prepared from X. oryzae pv. oryzae cells by using the protocol of Yi and Hackett (51). Succinate dehydrogenase activity was assayed by using the protocol described previously for Salmonella enterica serovar Typhimurium (31). One unit of enzyme activity was defined as the amount of enzyme used to produce 1 nmol of thiazolyl formazan (λmax = 550) per minute. Membranes were precipitated with 10% trichloroacetic acid and extracted with methanol. The pigment was quantified as the optical density at 445 nm (OD445) of the methanolic extracts (14).
Nucleotide sequence accession number.
The 20,310-bp X. oryzae pv. oryzae sequence described in this report has been submitted to the GenBank database and assigned the accession number AY010120.
RESULTS
Cloning of a xanthomonadin biosynthetic locus from X. oryzae pv. oryzae.
A 25.4-kb genomic clone, pIG102, containing seven pig transcriptional units has been shown to be involved in xanthomonadin production in X. campestris pv. campestris (35). Southern hybridization indicated that three EcoRI fragments of 12.0, 4.5, and 5.0 kb in the genomic DNA isolated from a wild-type X. oryzae pv. oryzae strain BXO1 are homologous to the pIG102 clone (data not shown). Colony hybridizations were performed to identify cosmid clones from a BXO1 genomic library (36) that contain the three fragments. Six out of 1,728 clones tested were found to hybridize with pIG102. An EcoRI restriction map of two cosmid clones, pLR9 and pLR10, that were identified in this screen and used for further studies is shown in Fig. 1.
Mutagenesis of the X. oryzae pv. oryzae pig cluster was performed as described in Materials and Methods. Thirteen independent mTn5, three mTn7, two Apr cassettes, and one Tcr cassette insertion in the pig cluster were marker exchanged into BXO43, an Rfr derivative of BXO1. The mutant strains BXO717, BXO718, BXO719, BXO720, and BXO1708 obtained with the insertions pig-15::mTn5, pig-16::mTn5, pig-17::mTn5, pig-18::mTn5, and pig-55::tetA, respectively (Table 1), did not produce visible amounts of pigment. All of the other insertions resulted in strains that are visibly indistinguishable from the wild-type strain, BXO43. Also, the absorption spectra of methanolic extracts from these strains are identical to that of BXO43 (data not shown). The clones pLR9 and pLR10 complement all five of the Pig− mutants for pigment production (data not shown).
Sequence analysis of the 20,310-bp pig region was performed as described in Materials and Methods. Fourteen putative ORFs have been predicted in the sequenced region. The insertions pig-15::mTn5, pig-55::tetA, and pig-16::mTn5, which caused the Pig− phenotype, were found to disrupt orf10, orf7, and orf6, respectively (Fig. 1). Two insertions, pig-17::mTn5 and pig-18::mTn5, that affect pigmentation were found to be at different sites in orf3 (Fig. 1). As shown in Fig. 1, insertions in orf9 and orf8 which are downstream of orf10 do not cause the Pig− phenotype. This suggests that the phenotype of the insertion in orf10 is not due to a polar effect on these genes and that orf10, by itself, is required for pigment production. Similarly orf6 is essential for pigment production as insertions in the downstream ORFs (orf5 and orf4) do not affect visible levels of pigmentation (Fig. 1). The Pig− phenotype of the strain BXO1708 (pig-55::tetA) (Table 1) in orf7 could be due to the polar effect of this insertion on orf6 (Fig. 1). Therefore, a pLR9 derivative, pLR9-pig-16::mTn5, containing an insertion in orf6 was mobilized into BXO1708. The resultant strain, BXO1714 (Table 1), produces wild-type levels of the pigment (data not shown). This suggests that orf7, per se, is required for pigment production. The intergenic region between orf3 and orf2 shows homology to a transposase-like protein (Table 2), which is very unlikely to affect pigment production. Moreover, the homology extends in two different reading frames (amino acids [aa] 1 to 178 and 188 to 317), which suggests that the transposase homologue is a pseudogene with a frameshift mutation in the coding region. Therefore, we conclude that orf3 is also required for pigment production. It is evident from these results that four putative ORFs, orf10, orf7, orf6, and orf3, in the X. oryzae pv. oryzae pig cluster are required for xanthomonadin biosynthesis.
TABLE 2.
Homologous proteins for ORFs encoded in the pig locus of X. oryzae pv. oryzaea
| ORF (size [aa]) | Predicted function | Homologous protein (size [aa]) | Organism | I/S (E value)b | Accession no. |
|---|---|---|---|---|---|
| Orf1 (593) | Outer membrane protein | Conserved hypothetical protein (617) | Xylella fastidiosa | 70/81 (0.0) | AE003957 |
| Orf2 (1,184) | Periplasmic protein | Hypothetical protein (1,179) | Xylella fastidiosa | 59/71 (0.0) | AE003957 |
| ISXo4c | Pseudogene | Putative transposase (345) | Burkholderia cepacia | 33/49 (e−31) | U67938 |
| Orf3 (145) | Unknown | Hypothetical protein (169) | Ralstonia solanacearum | 60/61 (e−22) | AL646059 |
| Orf4 (807) | Membrane transport | Membrane protein (788) | Xylella fastidiosa | 59/67 (0.0) | AE003918 |
| Orf5 (215) | Unknown | Putative transmembrane protein (211) | Ralstonia solanacearum | 28/42 (e−09) | AL646059 |
| Orf6 (324) | Acyltranferase | Hypothetical protein (326) | Xylella fastidiosa | 70/80 (0.0) | AE003918 |
| Orf7 (95) | Dehydratase | Hypothetical protein (98) | Xylella fastidiosa | 57/68 (e−23) | AE003918 |
| Orf8 (249) | Acyltransferase | Lysophosphatidic acid acyltransferase (250) | Borrelia burgdorferi | 32/70 (e−16) | AE001117 |
| Orf9 (284) | Unknown | Hypothetical protein (246) | Xylella fastidiosa | 61/71 (e−73) | AE003918 |
| Orf10 (132) | ACP | ACP (106) | Xylella fastidiosa | 82/92 (e−32) | AE003918 |
| Orf11 (414) | Halogenase | Halogenase (449) | Pseudomonas fluorescens | 29/46 (e−35) | AF081920 |
| Orf12 (789) | Dipeptidyl peptidase | Dipeptidyl peptidase (795) | Xylella fastidiosa | 69/79 (0.0) | AE003856 |
| Orf13 (231) | Unknown | Hypothetical protein (180) | Xylella fastidiosa | 36/54 (e−12) | AE003856 |
| Orf14 (145)d | Unknown | Hypothetical protein (155) | Vibrio cholerae | 37/55 (e−12) | AE004199 |
ORFs 3, 6, 7, and 10 are essential for xanthomonadin production, and ORF4 is involved in the outer membrane localization of xanthomonadin.
I and S indicate identity and similarity, respectively. E values were obtained by using the BLAST algorithm.
ISXo4 has a −1 frameshift mutation after codon 178.
orf14 has been sequenced partially.
ORFs 10, 7, and 6 in the X. oryzae pv. oryzae pig locus appear to be components of a novel PKS-fatty acid (FAS) biosynthetic pathway.
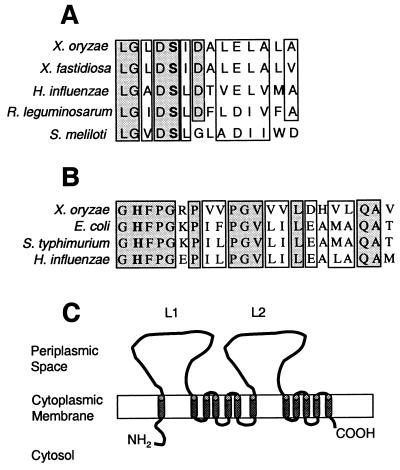
Sequence similarity searches were performed by using the BLAST algorithm for the ORFs that are encoded in the sequenced 20,310-bp genomic region. The results of the homology searches are summarized in Table 2. orf10 is homologous to acyl carrier proteins (ACPs) from various prokaryotic and eukaryotic organisms. The ACPs are important constituents of PKS-FAS biosynthetic pathways. A conserved phosphopantethein attachment domain (pfam00550) has been identified in this ORF (Fig. 2A). Phosphopantethein is the prosthetic group that attaches to a conserved serine residue (Fig. 2A) in ACPs and serves as a swinging arm for the attachment of activated fatty acyl groups. The homology of the orf10 product to ACPs is significant because, to our knowledge, it is the first genetic evidence to indicate that PKS-FAS biosynthetic pathways may be involved in xanthomonadin production.
FIG. 2.
Sequence features of putative ACP, dehydratase, and membrane transporter involved in xanthomonadin biosynthesis and localization in X. oryzae pv. oryzae. (A) Sequence alignment of a conserved domain (pfam00550) in the ACPs from X. oryzae pv. oryzae, X. fastidiosa, Haemophilus influenzae, R. leguminosarum, and Sinorhizobium meliloti. The GenBank accession numbers for these sequences are AY010120, AE003918, U32701, AF083916, and AE007237, respectively. (B) Sequence alignment of a conserved domain (pfam01377) in FabZ proteins [3-hydroxy-myristoyl-(ACP)-dehydratase] from X. oryzae pv. oryzae, E. coli, S. enterica serovar Typhimurium, and H. influenzae. The GenBank accession numbers for these sequences are AY010120, AE000127, U32786, and C37083, respectively. The conserved residues, serine (which is required for phosphopantethein attachment in ACPs) in panel A and histidine (which is known to act as the catalytic site in FabZ proteins) in panel B are shown in boldface type. The shaded and empty boxes indicate the identical and similar amino acids, respectively, in panels A and B. (C) Predicted structure of the orf4-encoded putative membrane transporter. The 12 TMDs are shown by solid cylinders spanning the cytoplasmic membrane. L1 and L2 represent the two periplasmic loops. The N- and C-terminal ends are indicated as NH2 and COOH, respectively.
Orf7 is homologous to a hypothetical protein in Xylella fastidiosa, a bacterial pathogen that causes citrus variegated chlorosis (43). It also exhibits moderate homology to the bacterial gene fabZ, which encodes 3-hydroxy-myristoyl-(ACP)-dehydratase, which is required during the elongation step in FASs and lipid A biosynthesis. The sequence alignment of a conserved domain (pfam01377) present in FabZ proteins from various bacterial species and the X. oryzae pv. oryzae homologue, Orf7, is shown in Fig. 2B. A conserved histidine residue present in this domain (Fig. 2B) is known to act as the catalytic site (29). orf6 exhibits strong homology to another hypothetical protein-encoding gene from X. fastidiosa and moderate homology (E value = 9e−7) to a conserved domain of late-stage acyl transferase in the conserved domain database (www.ncbi.nlm.nih.gov). The late-stage acyl transferase domain, identified by the Clusters of Orthologous Groups staff (www.ncbi.nlm.nih.gov), contains 278 aa and is present in bacterial proteins that are putatively involved in the biosynthesis of the lipid A region of LPSs. The homologies to dehydratases and acyl transferases may be significant as these are important constituents of PKS-FAS biosynthetic pathways. orf3 exhibits significant homology with a conserved protein from the plant pathogenic bacterium Ralstonia solanacearum (39). No functional domains could be identified in orf3, and its role in xanthomonadin biosynthesis in X. oryzae pv. oryzae is not clear.
orf4 encodes a putative membrane transporter.
orf4 is homologous to a putative membrane protein from X. fastidiosa (Table 2). Hydropathy plot and secondary structure analysis predicted 12 α-helical TMDs and two periplasmic loops (between TMDs 1 and 2 and 7 and 8) in Orf4 (Fig. 2C). The presence of hydrophobic α-helical TMDs suggests that the protein is localized to the cytoplasmic membrane of X. oryzae pv. oryzae. It is known that the membrane transport proteins of the resistance, nodulation, and division (RND) family (32) are localized to the cytoplasmic membrane and contain 12 TMDs and two periplasmic loops, between the first and second and seventh and eighth TMDs. However, we find that the two periplasmic loops in RND proteins (average size, 330 aa each) are usually much longer than those of Orf4 (225 and 250 aa). Based on the predicted localization and structural features of Orf4, we hypothesize its involvement in cellular transport and demonstrate (see below) its role in the localization of xanthomonadin to the outer membrane of X. oryzae pv. oryzae.
orf11 is homologous to bacterial halogenases but is not essential for xanthomonadin production and bromination
orf11 is homologous to prnC, a halogenase that is required for production of the antifungal antibiotic pyrrolnitrin, a dichlorinated tryptophan derivative in Pseudomonas fluorescens (22). PrnC belongs to a novel class of bacterial halogenases that utilize either NADH or reduced flavin adenine dinucleotide as cofactors (22, 30). An NADH binding site, GXGX2(G/A)X3(G/A)X6G (42), characteristic of these enzymes has been identified in the N-terminal region of the putative protein product of orf11. The insertion zxx-35::bla in this ORF, however, does not affect either visible levels of pigmentation or the absorption profile and molecular weights of xanthomonadin as determined by mass spectroscopy (data not shown). The majority of the isolated compound in purified xanthomonadin from the wild-type and the orf11 mutant X. oryzae pv. oryzae strains was found to be monobrominated (molecular weight of 551), and a minor dibrominated fraction had a molecular weight of 631 (data not shown). The molecular weights obtained are in a range similar to that reported for the dibrominated form of xanthomonadin (molecular weight, 576) from X. juglandis (2). Therefore, although orf11 is homologous to halogenases, it does not appear to be essential for halogenation of xanthomonadin.
ORFs 1, 2, 5, 8, 9, 12, and 13 are not essential for xanthomonadin production, and their respective homologies are indicated in Table 2. The transposase gene homologue located between ORFs 3 and 2 is not related at the nucleotide level to previously identified insertion sequence (IS) elements, suggesting that this may be a part of a novel IS element. A partial sequence of ISXo3 (130 bp), obtained within the sequenced region in this study, is identical to the corresponding region of the fully sequenced element (GenBank accession no. AF339839).
Orf4 is involved in localization of xanthomonadin to the outer membrane of X. oryzae pv. oryzae.
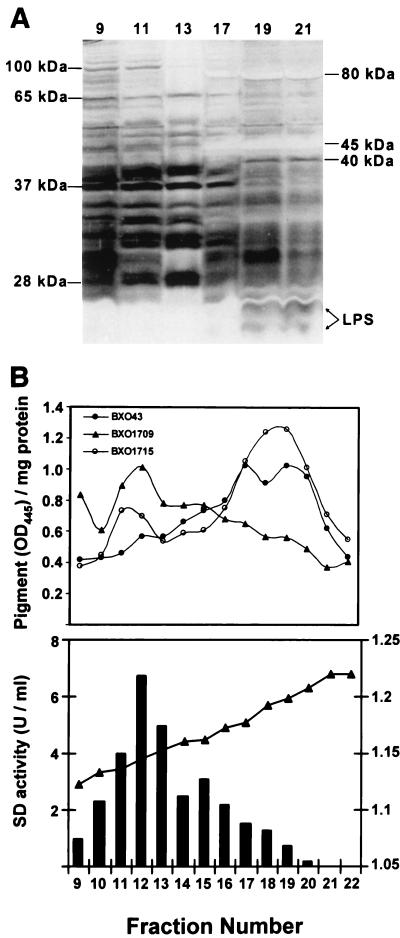
We determined the localization of xanthomonadin in X. oryzae pv. oryzae by separating the cytoplasmic and outer membranes of strain BXO43 on sucrose density gradient columns. Twenty-two fractions of 0.5 ml each were collected from the columns and analyzed for LPS and succinate dehydrogenase activity, the outer and cytoplasmic membrane markers, respectively. Protein profiles of fractions 9 to 13 and 17 to 21 are distinct from each other as seen on silver nitrate-stained SDS-PAGE gels (Fig. 3A). Four bands of approximately 25, 37, 65, and 100 kDa are specific to fractions 9 to 13 (Fig. 3A). Similarly, at least three bands of 40, 45, and 80 kDa are specific to fractions 17 to 21 (Fig. 3A). Interestingly, some of the bands present in fractions 9 and 11 do not appear in fraction 13 (Fig. 3A). The fractions between fractions 13 and 17 contained bands characteristic of both classes (data not shown). Two bands of <25 kDa that are protease resistant and comigrate with purified low-molecular-mass X. oryzae pv. oryzae LPS bands (data not shown) are present in fractions 17 to 21 and absent from fractions 9 to 13 (Fig. 3A). Succinate dehydrogenase activity was found to be maximum in fractions 9 to 13, whereas little or no activity was found in fractions 17 to 21 (Fig. 3B). The fractions 9 to 13 and 17 to 21 correspond to sucrose densities of 1.12 to 1.16 and 1.18 to 1.22 g/ml, respectively (Fig. 3B). The distribution of LPS (31) and succinate dehydrogenase activity (18) and the corresponding sucrose densities (18) suggest that cytoplasmic membranes are present in fractions 9 to 13, whereas fractions 17 to 21 contain the outer membranes. Methanolic extracts were prepared for each of the fractions, and the pigment was quantified as the OD445 per milligram of protein. The absorption spectra of the methanolic extracts exhibit peaks and shoulders characteristic of xanthomonadin (data not shown). As shown in Fig. 3B, most of the xanthomonadin in BXO43 is present in fractions 17 to 21, indicating that the pigment in X. oryzae pv. oryzae is mainly localized in the outer membrane.
FIG. 3.
The product of the orf4 gene is involved in the localization of xanthomonadin in the outer membrane of X. oryzae pv. oryzae. The cytoplasmic and outer membranes of X. oryzae pv. oryzae strains were separated on sucrose density gradients as discussed in Materials and Methods. BXO43, wild type; BXO1709, orf4::mTn7; BXO1715, BXO1709 plus a complementing plasmid (pLR9-zxx7::mTn5). The experiments were repeated at least three times, and similar results were obtained. (A) Protein profiles of the representative membrane fractions of BXO43. Twenty-five micrograms of protein from each fraction was subjected to Tricine-SDS-PAGE and stained with silver nitrate reagent. The fraction numbers are indicated above the lanes. The molecular masses and locations of proteins and LPS bands that are characteristic of different membrane fractions of X. oryzae pv. oryzae are indicated. Similar profiles were obtained with strains BXO1709 and BXO1715. (B) The upper panel indicates the amount of the pigment (OD445) per milligram of protein for the membrane fractions of different X. oryzae pv. oryzae strains. Methanol extraction and quantitation of the pigment are described in Materials and Methods. The lower panel shows succinate dehydrogenase activity (in units/milliliter) and sucrose density (in grams/milliliter) for the corresponding membrane fractions.
Membrane localization of xanthomonadin in the orf4 insertional mutant (BXO1709) (pig-56::mTn7) was studied as discussed for BXO43. The distribution of LPS and succinate dehydrogenase activity and the protein profiles obtained with the cytoplasmic and outer membrane preparations of BXO1709 were similar to those of BXO43 (data not shown). As shown in Fig. 3B, most of the pigment in strain BXO1709 is present in the cytoplasmic membrane. A complementing clone, pLR9-zxx-7::mTn5, was mobilized into BXO1709 (to obtain the strain BXO1715), and this restores outer membrane localization of xanthomonadin (Fig. 3B). BXO1715 produces greater amounts of the pigment, possibly due to the presence of the pig genes on a multicopy plasmid (Fig. 3B). The significantly higher amounts of xanthomonadin retained in the cytoplasmic fractions of BXO1715 than in those of BXO43 (Fig. 3B) could be due to the inability of the transport system to cope with the extra amount of the pigment produced, as the transport system might require several other proteins not encoded by pLR9.
Identification of a novel repeat element in the X. oryzae pv. oryzae pig locus.
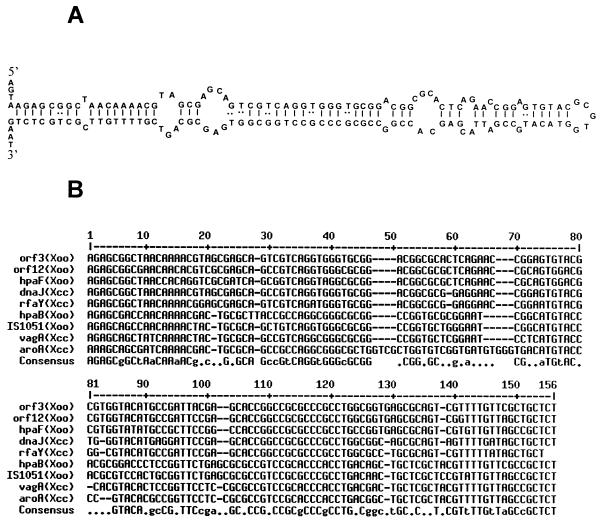
Two copies of a novel 145-bp repeat element were identified in the X. oryzae pv. oryzae pig region. These elements are located 51 and 79 bp downstream of the stop codons for orf3 and orf12, respectively (Fig. 1). The repeat is an imperfect palindrome and may adopt a stable stem-loop structure (Fig. 4A) with a free energy of −44.9 kcal mol−1. We have called this element the XRIC sequence. The G+C content (64%) of the element is characteristic of xanthomonads. A nucleotide BLAST search in the database revealed the presence of three more copies of this element in X. oryzae pv. oryzae and four copies in X. campestris pv. campestris (Fig. 4B). No other homologues of the XRIC element were found in the database. Based on the total sequence deposited to date in the database for X. oryzae pv. oryzae (approximately 165 kb) and X. campestris pv. campestris (approximately 170 kb), we estimate that there might be approximately 150 and 120 copies of the XRIC element, respectively, in these organisms, assuming the genome sizes to be 5.0 Mb.
FIG. 4.
Sequence analysis of the XRIC element. (A) Palindromic structure (ΔG = −44.9 kcal/mol) of the orf3 (X. oryzae pv. oryzae) XRIC element as predicted by using SeqWeb software (see Materials and Methods). (B) Nucleotide alignment of the XRIC elements found in the database by using MULTALIN software. The capital and lowercase letters in the consensus sequence indicate >80 and >50% consensus, respectively. A flanking gene (either 5′ or 3′) and the name of the organism are mentioned for each element. Xoo, X. oryzae pv. oryzae; Xcc, X. campestris pv. campestris. The GenBank accession numbers of the sequences, from top to bottom, are AY010120, AY010120, AB045312, AF302775, U19896, AB045311, AF329697, AF360374, and AF303107.
DISCUSSION
This report, for the first time, describes the characterization of functions specifically involved in the biosynthesis and localization of xanthomonadin. Four ORFs in a pig locus from X. oryzae pv. oryzae have been shown to be essential for xanthomonadin biosynthesis. Three of these ORFs, 10, 7, and 6, are homologous to ACP, dehydratases, and acyl transferases, respectively. These three proteins perform vital functions in PKS and FAS biosynthetic pathways. ACPs bind to the activated small fatty acyl molecules like malonyl coenzyme A (CoA) or acetyl-CoA. Each cycle in the FAS-PKS is initiated by the interaction of the acyl-ACP with a ketosynthase (condensing enzyme) that has a starter or growing fatty acyl chain attached to a cysteine residue. After condensation of the two acyl moieties, the extended fatty acyl chain is transferred back onto the ACP for further modifications, such as ketoreduction, dehydration, and enoyl reduction, etc. Dehydratases introduce the C=C bond in the growing PKS-fatty acyl chain and generate a polyene group after several cycles of chain extension in PKS biosynthesis. Acyl transferases transfer the extended PKS-fatty acyl chain from the ACP either to the ketosynthase to make the ACP available for the next round of extension or to a CoA group for chain termination (reviewed by Hopwood in reference 17). The homologies identified in this study provide a plausible role for a PKS pathway in xanthomonadin biosynthesis. However, detailed biochemical experimentation is required to confirm the roles assigned for each of the ORFs.
If indeed a PKS pathway is involved in xanthomonadin biosynthesis, it would require several other functions, such as a ketosynthase and a ketoreductase, that are not encoded in the X. oryzae pv. oryzae pig locus. It is possible that these functions are encoded elsewhere in the genome. Another possibility is that some of these functions are shared with the cellular FAS biosynthetic pathway. Rhizobium leguminosarum has been reported to synthesize a polyunsaturated fatty acid that is attached to the Nod factor by using two dedicated proteins, NodF (an ACP) and NodE (a ketosynthase), as well as a third protein, FabG (a ketoreductase), shared with the cellular FAS pathway (26).
It was suggested that xanthomonadin is localized in the cytoplasmic membrane in X. juglandis, as the kinetics of release of the pigment upon ballistic disintegration of bacterial cells was found to be the same as that of NADH oxidase, a cytoplasmic membrane marker (46). Dianese and Schaad (11) physically separated the two membranes of X. campestris pv. campestris by sucrose density centrifugation and showed that xanthomonadin resides exclusively in the outer membrane. We have also used the sucrose density gradient method for separation of the two membranes of X. oryzae pv. oryzae. The apparent discrepancy between our results and those of Stephens and Starr (46) can be explained as we find high NADH oxidase activity in the outer membrane fraction of X. oryzae pv. oryzae (data not shown). This activity, which may be due to an as yet unidentified outer membrane-associated protein, indicates that NADH oxidase is not a good cytoplasmic membrane marker for X. oryzae pv. oryzae and possibly for other xanthomonads as well.
The product encoded by orf4 is a putative membrane transporter. We demonstrate that Orf4 is essential for outer membrane localization of xanthomonadin in X. oryzae pv. oryzae. The predicted structure of this protein resembles that of the RND family of proton motive force-dependent cytoplasmic membrane proteins that transport a wide variety of substrates like antibiotics, detergents, dyes, and oligosaccharides, etc., across the outer membrane (32). In gram-negative bacteria, these proteins usually work in concert with a periplasmic protein called membrane fusion protein and an outer membrane factor. The membrane fusion protein probably brings the two membranes closer by causing fusion of the cytoplasmic and outer membranes (12), and the outer membrane factor acts as a channel for transport of the substrates across the outer membrane (27). Is Orf4 acting in concert with such proteins for outer membrane localization of xanthomonadin? Sequence analysis indicates that ORFs 1 and 2 in the X. oryzae pv. oryzae pig locus encode putative outer membrane and periplasmic proteins, respectively. However, initial results suggest that these may not be essential for transport of the pigment to the outer membrane (data not shown). The mechanism of action of the putative membrane transporter and the possible involvement of other proteins in the localization of the pigment to the outer membrane needs to be investigated.
The xanthomonadin produced by Xanthomonas populi is not halogenated (19). Our results suggest that, like most xanthomonads, the xanthomonadin isolated from X. oryzae pv. oryzae is halogenated and that it can exist in either mono- or dibrominated forms. The orf11 product is a putative halogenase. A mutation in this ORF, however, does not seem to affect bromination, as determined by mass spectrometry. This suggests that either the putative halogenase is not involved in bromination or that this function is redundant, as there may be another halogenase in the X. oryzae pv. oryzae genome. Two different halogenases, prnA and prnC have been shown to be required for the production of pyrrolnitrin in P. fluorescens (22). Three putative halogenases, PltA, PltD, and PltM, have been identified in the pyoluteorin (a dichlorinated antifungal antibiotic) biosynthetic gene cluster in P. fluorescens strain Pf-5 (30).
As per the predicted gene organization (Fig. 1), ORFs 7 and 6, 6 and 5, and 4 and 3 overlap each other by 4, 77, and 14 bp, respectively. Reverse transcription-PCR analysis (data not shown) suggests that ORFs 8, 7, 6, and 5 and ORFs 4 and 3 constitute two separate transcription units. ORFs 9, 10, 11, and 12 do not appear to be cotranscribed with either each other or with ORF 8. The pigment-proficient phenotype associated with the zxx-3::mTn5 (orf8) and pig-56::mTn7 (orf4) insertions suggests that these are nonpolar on downstream genes. Complementation analysis indicates that the pig-55::tetA insertion in orf7 is not polar on orf6. It is possible that the lack of polarity is due to multiple internal promoters in the pig locus and/or promoters reading out of the insertions.
It is interesting that ORFs 10, 9, 7, 6, and 4 in the X. oryzae pv. oryzae pig locus are extremely homologous and have similar organization to ORFs 0771, 0772, 0774, 0775, and 0777, respectively, from X. fastidiosa (43). ORFs 10, 9, 7, 6, 5, 4, and 3 of the pig locus also exhibit homology and have similar organization to the ORFs 434, 433, 432, 431, 430, 429, and 425, respectively, of the plant pathogenic bacterium R. solanacearum (39). Intriguingly, R. solanacearum ORF 432 encodes a putative protein of 580 aa, of which the C-terminal 100 aa are homologous to Orf7 (putative dehydratase), which is 98 aa long. The rest of R. solanacearum ORF 432 exhibits homology to acyl or aryl-CoA ligases, which are reported to be involved in the synthesis of PKS precursors (17). Two other genes, R. solanacearum ORFs 427 and 435, from the cognate locus in R. solanacearum are homologous to ketosynthases and ketoreductases, the functions that are required for PKS-FAS biosynthesis but are missing from the X. oryzae pv. oryzae pig locus. The function of these loci in X. fastidiosa and R. solanacearum is not clear (these bacteria are not reported to produce xanthomonadin), but they might be involved in the synthesis of a xanthomonadin-like molecule. Also, X. fastidiosa does not require an epiphytic phase because it is transmitted through an insect vector, and R. solanacearum is known to infect plants through their roots. Does this suggest another role for xanthomonadin during in planta growth? Alternatively, the homology could be related to a common PKS-FAS biosynthetic pathway that has been co-opted by xanthomonads to produce xanthomonadin.
A novel repeat element, XRIC, was discovered during sequence analysis of the X. oryzae pv. oryzae pig locus. We identified nine copies of this element in the database, including two copies in the pig locus, and they were all found in either X. oryzae pv. oryzae or X. campestris pv. campestris. This element was not found in any other organism. The XRIC element, therefore, may be specific to either a few or all Xanthomonas species. Several classes of small extragenic sequences, repeated from 6 to more than 250 times have been reported in E. coli and other bacteria (reviewed by Bachellier et al. in reference 4). In E. coli, these sequences represent almost 2% of the bacterial DNA. Most of these elements are known to be transcribed and have been postulated to be involved in varied functions: mRNA stabilization, transcription termination, organization of the bacterial nucleoid, and generation of chromosomal rearrangements, etc. (4). The XRIC elements may also have roles similar to those identified earlier in extragenic, repetitive, palindromic sequences.
Previously, it was demonstrated that shikimate dehydrogenase (AroE) is required for xanthomonadin production in X. oryzae pv. oryzae (14), and it was proposed that the aromatic ring in xanthomonadin may be derived from the shikimate pathway. The results presented here suggest the possibility that the polyene chain in xanthomonadin may be derived from a PKS pathway. We have isolated several pigment-deficient mutants of X. oryzae pv. oryzae that are not complemented by clones of either the shikimate dehydrogenase gene or the pig locus isolated in this study (L. Rajagopal and R. Sonti, unpublished results). These mutants are likely to define additional functions that are involved in xanthomonadin biosynthesis, including those that might be involved in bridging the shikimate and PKS pathways. In order to gain a better understanding of these functions, we are planning to clone and characterize X. oryzae pv. oryzae genomic regions that complement these other pigment-deficient mutants.
The X. campestris pv. campestris clone pIG102 complements all naturally isolated and induced pigment-deficient mutants of X. campestris pv. campestris (35) and X. oryzae pv. oryzae (Rajagopal and Sonti, unpublished results), including the shikimate dehydrogenase mutant, and is sufficient to produce xanthomonadin in P. fluorescens (35). In X. oryzae pv. oryzae, on the other hand, the pig genes identified in this study and the shikimate dehydrogenase gene, are present at two different genomic loci. Also, several pigment-deficient mutants of X. oryzae pv. oryzae could not be complemented by genomic clones of either of these two loci (Rajagopal and Sonti, unpublished results). These observations indicate that the X. oryzae pv. oryzae pig locus has been rearranged in comparison to the X. campestris pv. campestris pig locus. The sequence of the X. campestris pv. campestris pig locus will be available after the completion of the genome sequencing project that is under way in Brazil. Sequence comparison of the pig loci from these two organisms should reveal the nature of these rearrangements and identify the genes that have been either deleted or dispersed to other locations on the genome of X. oryzae pv. oryzae.
Acknowledgments
We thank Mehar Sultana for primer synthesis and Jan E. Leach and A. R. Poplawsky for providing bacterial strains and the pIG102 clone, respectively.
A.K.G. and L.R. were supported by fellowships from the Council of Scientific and Industrial Research (CSIR) of the government of India.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrewes, A. G., C. L. Jenkins, M. P. Starr, J. Shepherd, and H. Hope. 1976. Structure of xanthomonadin I, a novel dibrominated aryl-polyene pigment produced by the bacterium Xanthomonas juglandis. Tetrahedron Lett. 45:4023-4024. [Google Scholar]
- 3.Andrewes, A. G., S. Hertzberg, S.-L. Jensen, and M. P. Starr. 1973. Xanthomonas pigments. 2. The Xanthomonas “carotenoids”-non-carotenoid brominated aryl-polyene esters. Acta Chem. Scand. 27:2383-2395. [DOI] [PubMed] [Google Scholar]
- 4.Bachellier, S., E. Gilson, M. Hofnung, and C. W. Hill. 1996. Repeated sequences, p. 2012-2040. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Shaechter, and H. C. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buluwela, L., A. Forster, T. Boehm, and T. H. Rabbitts. 1989. A rapid procedure for colony screening using nylon filters. Nucleic Acids Res. 17:452.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covarrubias, L., and F. Bolivar. 1982. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 17:79-89. [DOI] [PubMed] [Google Scholar]
- 9.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 10.Dharmapuri, S., and R. V. Sonti. 1999. A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol. Lett. 179:53-59. [DOI] [PubMed] [Google Scholar]
- 11.Dianese, J. C., and N. W. Schaad. 1982. Isolation and characterization of inner and outer membranes of Xanthomonas campestris pv. campestris. Phytopathology 72:1284-1289. [Google Scholar]
- 12.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 176:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durgapal, J. C. 1996. Albino mutation in rice bacterial blight pathogen. Curr. Sci. 70:15. [Google Scholar]
- 14.Goel, A. K., L. Rajagopal, and R. V. Sonti. 2001. Pigment and virulence deficiencies associated with mutations in the aroE gene of Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 67:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano, S. S., and C. D. Upper. 1983. Ecology and epidemiology of foliar bacterial plant pathogens. Annu. Rev. Phytopathol. 21:243-269. [Google Scholar]
- 16.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 17.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2497. [DOI] [PubMed] [Google Scholar]
- 18.Hu, N. T., M. N. Hung, C. T. Liao, and M. H. Lin. 1995. Subcellular location of XpsD, a protein required for extracellular protein secretion by Xanthomonas campestris pv. campestris. Microbiology 141:1395-1406. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins, C. L., and M. P. Starr. 1982. The pigment of Xanthomonas populi is a nonbrominated aryl-heptaene belonging to xanthomonadin pigment group II. Curr. Microbiol. 7:195-198. [Google Scholar]
- 20.Jenkins, C. L., and M. P. Starr. 1982. The brominated aryl polyene (xanthomonadin) pigments of Xanthomonas juglandis protect against photobiological damage. Curr. Microbiol. 7:323-326. [Google Scholar]
- 21.Kelemu, S., and J. E. Leach. 1990. Cloning and characterization of an avirulence gene from Xanthomonas campestris pv. oryzae. Mol. Plant-Microbe Interact. 3:59-65. [Google Scholar]
- 22.Kirner, S., P. E. Hammer, D. S. Hill, A. Altmann, I. Fischer, L. J. Weislo, M. Lanahan, K.-H. van Pée, and J. M. Ligon. 1998. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach, J. E., F. F. White, M. L. Rhoads, and H. Leung. 1990. A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris. Mol. Plant-Microbe Interact. 3:238-246. [Google Scholar]
- 24.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium-dodecyl sulfate polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-111. [DOI] [PubMed] [Google Scholar]
- 25.Leyns, F., M. DeCleene, J. G. Swings, and J. Deley. 1984. The host range of the genus Xanthomonas. Bot. Rev. 50:308-356. [Google Scholar]
- 26.Lopez-Lara, I. M., and O. Geiger. 2001. The nodulation protein NodG shows the enzymatic activity of an 3-oxoacyl-acyl carrier protein reductase. Mol. Plant-Microbe Interact. 14:349-357. [DOI] [PubMed] [Google Scholar]
- 27.Ma, D., D. N. Cook, J. E. Hearst, and H. Nikaido. 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Mohan, S., T. M. Kelly, S. S. Eveland, C. R. H. Raetz, and M. S. Anderson. 1994. An Escherichia coli gene (fabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. J. Biol. Chem. 269:32896-32903. [PubMed] [Google Scholar]
- 30.Nowak-Thompson, B., N. Chaney, J. S. Wing, S. J. Gould, and J. E. Loper. 1999. Characterization of a pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 181:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborn, M. J., J. E. Gander, and E. Parisi. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. J. Biol. Chem. 247:3973-3986. [PubMed] [Google Scholar]
- 32.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poplawsky, A. R., S. C. Urban, and W. Chun. 2000. Biological role of xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl. Environ. Microbiol. 66:5123-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poplawsky, A. R., and W. Chun. 1997. pigB determines a diffusible factor needed for extracellular polysaccharide slime and xanthomonadin production in Xanthomonas campestris pv. campestris. J. Bacteriol. 179:439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poplawsky, A. R., M. D. Kawalek, and N. W. Schadd. 1993. A xanthomonadin-encoding gene cluster for the identification of pathovars of Xanthomonas campestris. Mol. Plant-Microbe Interact. 6:545-552. [Google Scholar]
- 36.Rajagopal, L., S. Dharmapuri, A. T. Sayeepriyadarshini, and R. V. Sonti. 1999. A genomic library of Xanthomonas oryzae pv. oryzae in the broad host range mobilizing Escherichia coli strain S17-1. Int. Rice Res. Notes 24:20-21. [Google Scholar]
- 37.Rajagopal, L., C. S. Sundari, D. Balasubramanian, and R. V. Sonti. 1997. The bacterial pigment xanthomonadin offers protection against photo-damage. FEBS Lett. 415:125-128. [DOI] [PubMed] [Google Scholar]
- 38.Ray, S. K., R. Rajeshwari, and R. V. Sonti. 2000. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant-Microbe Interact. 13:394-401. [DOI] [PubMed] [Google Scholar]
- 39.Salanoubat, M., et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schaad, N. W., and R. E. Stall. 1988. Xanthomonas, p. 81-94. In N. W. Schaad (ed.), Laboratory guide for plant pathogenic bacteria, 2nd ed. The American Phytopathological Society, St. Paul, Minn.
- 42.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature (London) 343:38-43. [DOI] [PubMed] [Google Scholar]
- 43.Simpson, A. J., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 44.Starr, M. P. 1981. The genus Xanthomonas, p. 742-763. In M. P. Starr, H. Stolp, G. H. Truper, A. Balows, and G. H. Schlegel (ed.), The prokaryotes, vol. 1. Springer-Verlag, Berlin, Germany.
- 45.Starr, M. P., C. L. Jenkins, L. B. Bussey, and A. G. Andrewes. 1977. Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas. Arch. Microbiol. 113:1-9. [DOI] [PubMed] [Google Scholar]
- 46.Stephens, W. L., and M. P. Starr. 1963. Localization of carotenoid pigment in the cytoplasmic membrane of Xanthomonas juglandis. J. Bacteriol. 86:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai, C.-M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchiya, K., T. W. Mew, and S. Wakimoto. 1982. Bacteriological and pathological characteristics of wild type and induced mutants of Xanthomonas campestris pv. oryzae. Phytopathology 72:43-46. [Google Scholar]
- 49.Wilson, K. J., A. Sessitsch, J. C. Corbo, K. E. Giller, A. D. Akkermans, and R. A. Jefferson. 1995. Beta-glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141:1691-1705. [DOI] [PubMed] [Google Scholar]
- 50.Yashitola, J., D. Krishnaveni, A. P. K. Reddy, and R. V. Sonti. 1997. Genetic diversity within the population of Xanthomonas oryzae pv. oryzae in India. Phytopathology 87:760-765. [DOI] [PubMed] [Google Scholar]
- 51.Yi, E. C., and M. Hackett. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651-656. [DOI] [PubMed] [Google Scholar]