Abstract
Biphenyl dioxygenase (Bph Dox) is responsible for the initial dioxygenation step during the metabolism of biphenyl. The large subunit (BphA1) of Bph Dox plays a crucial role in the determination of the substrate specificity of biphenyl-related compounds, including polychlorinated biphenyls (PCBs). Based on crystallographic analyses of naphthalene dioxygenase (B. Kauppi, K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy, Structure 6:571-586, 1998), we developed a three-dimensional model of KF707 BphA1 of Pseudomonas pseudoalcaligenes KF707. Based on structural information about the amino acids which coordinate the catalytic nonheme iron center, we constructed 12 site-directed BphA1 mutants with changes at positions 227, 332, 335, 376, 377, and 383 and expressed these enzymes in Escherichia coli. The Ile335Phe, Thr376Asn, and Phe377Leu Bph Dox mutants exhibited altered regiospecificities for various PCBs compared with wild-type Bph Dox. In particular, the Ile335Phe mutant acquired the ability to degrade 2,5,2′,5′-tetrachlorobiphenyl by 3,4-dioxygenation and showed bifunctional 2,3-dioxygenase and 3,4-dioxygenase activities for 2,5,2′-trichlorobiphenyl and 2,5,4′-trichlorobiphenyl. Furthermore, two mutants, the Phe227Val and Phe377Ala mutants, introduced molecular oxygen at the 2,3 position, forming 3-chloro-2′,3′-dihydroxy biphenyl with concomitant dechlorination.
Biphenyl-utilizing bacteria are ubiquitously distributed and isolated from various environmental samples (3, 10, 27, 38). These bacteria have been extensively studied with respect to the degradation of polychlorinated biphenyls (PCBs), a family of xenobiotic compounds that are major environmental pollutants. These studies revealed considerable differences in the congener selectivity patterns and ranges of activity of various PCB-degrading bacteria (1, 3, 12, 38). It was also demonstrated that both the relative rates of the primary degradation of PCB and the choice of the ring attacked were dependent on the bacterial strain (2, 3, 9).
It was noted that Pseudomonas pseudoalcaligenes KF707 and Burkholderia cepacia LB400 exhibit distinct differences in substrate ranges for various PCB congeners (8, 11, 13) despite the fact that the biphenyl catabolic bph operons of these two strains are nearly identical (27, 38). LB400 metabolizes PCB via both 2,3-dioxygenation and 3,4-dioxygenation, depending on the chlorine substitution of PCB. Therefore, LB400 is able to degrade many more PCB congeners than KF707 (8, 13). However, KF707 has a greater degradation activity for several double para-replaced congeners such as 4,4′-dichlorobiphenyl and 2,4,4′-trichlorobiphenyl, but the same strain is unable to degrade ortho-meta-replaced congeners such as 2,5,2′,5′-tetrachlorobiphenyl. On the other hand, LB400 degrades 2,5,2′,5′-tetrachlorobiphenyl via 3,4-dioxygenation but poorly degrades 4,4′-dichlorobiphenyl.
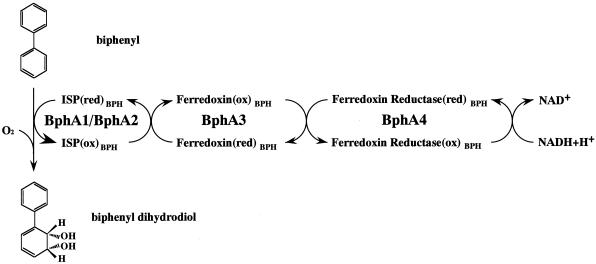
Biphenyl and PCB are oxidized to the dihydrodiol compound by biphenyl dioxygenase (Bph Dox) during the initial step in the degradative pathway (Fig. 1). Bph Dox is a multicomponent enzyme consisting of four subunits: a large (α) and a small (β) subunit of terminal dioxygenase (encoded by bphA1 and bphA2, respectively), ferredoxin (encoded by bphA3), and ferredoxin reductase (encoded by bphA4) (39). BphA1 and BphA2 are associated as an α3β3 heterohexamer and catalyze the introduction of both atoms of molecular oxygen into the aromatic nucleus of biphenyl (25). BphA1 contains the motif Cys-Xaa-His-Xaa-17-Cys-Xaa-2-His, which forms a Rieske-type [2Fe-2S] cluster involved in electron transfer. Bph Dox requires Fe(II) for activity, and oxygen activation is thought to occur at the mononuclear iron center. Ferredoxin and ferredoxin reductase act as an electron transfer system from NADH to reduce the terminal dioxygenase (26).
FIG. 1.
Initial dioxygenation of biphenyl by KF707 Bph Dox. Biphenyl is hydroxylated at the 2,3 position, yielding a 2,3-dihydrodiol compound. ox, oxidized; red, reduced.
Among these four subunits, it was found that BphA1 (α subunit) is crucially responsible for recognition and binding of the substrate and hence for the substrate specificity of Bph Dox (17). It was also reported that the β subunit influences the substrate specificity of Bph Dox for PCB (18). Previously we constructed a variety of chimeric bphA1 genes between KF707 and LB400 by using three common restriction sites (23). Then, we randomly recombined these two bphA1 genes by using DNA shuffling (24, 37). Some chimeric dioxygenases possessed enhanced PCB-degrading abilities. These results demonstrated that a relatively small number of amino acids in the carboxyl-terminal half of BphA1 are involved in the recognition of the chlorinated ring and the site of dioxygenation and are therefore responsible for the transformation of PCB.
It has also been reported that even small amino acid changes near the active site lead to altered substrate specificity or regiospecificity in some oxygenases. In the toluene 4-monooxygenase of Pseudomonas mendocina KR1, a multicomponent diiron enzyme, the mutation of Gln-141 and Phe-205, which lie in a hydrophobic region closer to the FeA iron site, caused the changes in regiospecificity for aromatic hydroxylation (33). The TecA chlorobenzene dioxygenase of Burkholderia sp. strain PS12 exhibits dechlorination activity for tetrachlorobenzene. In this reaction, amino acid residue 220, which is located in the region comprising the ligands of the mononuclear ferrous iron, has been reported to be important (4).
The structure of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 has been solved by X-ray crystallography (22). The enzyme is an α3β3 heterohexamer, and each α subunit contains a Rieske [2Fe-2S] center and mononuclear nonheme iron which is believed to be the site of molecular oxygen activation (6). His-208, His-213, and Asp-362 coordinate the active-site iron, forming a 2-His-1-carboxylate facial triad that is the common structural motif (16). Several amino acids were identified near the active site. Although KF707 BphA1 and NCIB 9816-4 NahAc (the large subunit of naphthalene dioxygenase) exhibit rather low amino acid sequence identity (32%), certain regions, such as the mononuclear nonheme iron center and Rieske center, are highly conserved (21), implying that the entire structure would be conserved.
Based on these findings, we developed a three-dimensional model of the α subunit of KF707 Bph Dox and mutated six amino acids near a 2-His-1-carboxylate facial triad in BphA1. We now report that several mutant Bph Doxs exhibit altered regiospecificity for certain PCB congeners.
MATERIALS AND METHODS
Bacterial strains, plasmid, and growth conditions.
The Escherichia coli strains were grown in Luria-Bertani (LB) medium or on LB agar medium (1.5% agar). Antibiotics (50 μg of ampicillin per ml) were added when needed to select for the presence of a plasmid in the E. coli transformants. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM to induce the expression of the bph genes. pJHF18ΔMluI containing disrupted bphA1 (ΔbphA1)-bphA2A3A4BC was constructed as previously described (24). pJHF18ΔMluI was used for the replacement of ΔbphA1 with the bphA1 gene variants.
Molecular modeling of BphA1.
The three-dimensional structure of KF707 BphA1 was built based on the crystal structure of the naphthalene dioxygenase of Pseudomonas sp. strain NCIB 9816-4 by using Insight II/Homology modules (Molecular Simulations, Inc.). The crystallographic coordinates of the naphthalene dioxygenase were obtained from the Protein Data Bank. Energy minimization and molecular dynamics calculations were carried out with the Insight II/Discover module (Molecular Simulations, Inc.) with the consistent valence force field. The structures were displayed on a Silicon Graphics workstation.
Site-directed mutagenesis.
Site-directed mutagenesis was performed with a Quickchange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's instructions. KF707 bphA1 DNA was amplified from plasmid pKTF18 (39) by using the following oligonucleotide primers: 5′-CCGAATTCAAGGAGACGTTGAATCATG-3′ (#18) for the forward sequence, where the EcoRI site is underlined and the start codon is boldfaced, and 3′-TCTAGACAGTTGGCCTTCTTAAGTT-5′ (#20) for the reverse primer, which includes the intervening region between bphA1 and bphA2, where the EcoRI site is underlined and the BglII site is italicized (24). PCR was performed with a total volume of 50 μl, which contained 50 ng of a plasmid as the DNA template, PCR buffer, a 100 μM concentration of each of the four deoxynucleoside triphosphates, a 1 μM concentration of each oligoprimer, and 0.5 U of Taq DNA polymerase (Promega Corp). Amplification of the DNA was carried out for 25 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min.
The PCR product was digested by EcoRI and inserted at the EcoRI site of pUC19 (Takara Shuzo) to obtain pSSF7071. This plasmid was amplified by PCR by using the various mutagenesis oligonucleotides for the following amino acid changes: Phe227Leu, Phe227Val, Leu332Ala, Ile335Phe, Ile335Ala, Thr376Asn, Phe377Ala, Phe377Leu, Phe383Ala, and Phe383Leu. The primer sequences will be provided upon request. After mutations were confirmed with a DNA sequencer (model 4000L; Li-cor), the bphA1 mutants were double digested with SacI and BglII and ligated at the same site of pJHF18ΔMluI, replacing ΔbphA1 with the mutant bphA1. To examine the mode of oxygenation of the mutant Bph Dox for PCB, the bphB and bphC genes were deleted by digestion with PpuMI and subsequently self-ligated. These plasmids were transformed into E. coli JM109.
Assay for quantitative degradation of PCB.
The recombinant E. coli JM109 cells expressing wild-type KF707 and LB400 and mutant Bph Doxs were grown to logarithmic phase (turbidity of 0.8 to 1.2 at 600 nm), washed twice in 50 mM phosphate buffer (pH 7.5), and resuspended in 1 ml of the same buffer to adjust the turbidity to 1.0. Biphenyl and PCB (AccuStandard Inc.) dissolved in dimethyl sulfoxide were added at a final concentration of 20 μg/ml. After being shaken at 200 rpm for 20 h at 30°C, the entire incubation mixture was extracted with the same volume (1 ml) of ethylacetate. One microliter of sample was injected into a gas chromatograph equipped with an electron capture detector (ECD-GC; Shimadzu GC-14B with 63Ni, 370 MBq) as previously described (35). The column used was a coiled glass column (2.1 m by 3.2 mm internal diameter) packed with silicon OV1. The column temperature was increased from 140 to 300°C at a rate of 10°C/min. The amounts of substrate depletion were calculated by normalization to the recovery of 2,4,6,2′,4′-pentachlorobiphenyl, a nondegradable internal standard, extracted from the heat-treated control cells and quantified by using a standard curve relating the peak area (1, 24).
Identification of metabolites.
The production of the dihydrodiol compounds was analyzed for various PCBs after extraction with the same volume of ethylacetate. One milliliter of extract was evaporated to dryness by a centrifugal concentrator (model VC-36S; Taitec) and derivatized with 100 μg of n-butylboronic acid in 10 μl of acetone-dimethyl formamide (15). The samples were analyzed by using a gas chromatograph-mass spectrometer (GC-MS, model QP5000; Shimadzu) with a coiled-capillary glass column (0.22 mm inner diameter, 50 m long) packed with HT8 (SGE Japan Inc.). The column temperature was increased from 150 to 300°C at a rate of 10°C/min.
RESULTS
Construction of various Bph Dox variants.
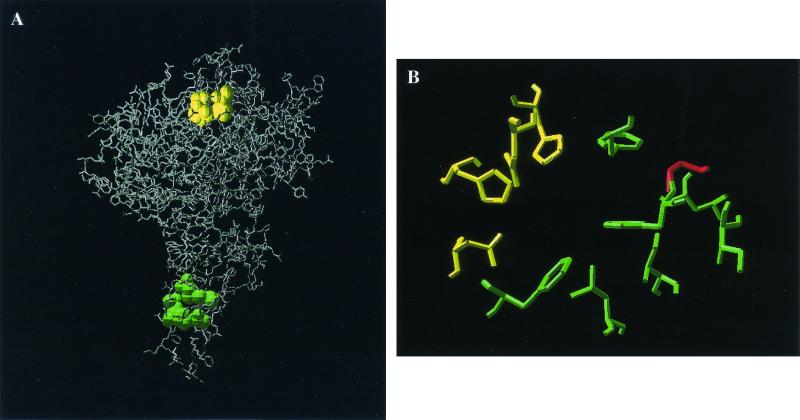
Based on the crystallographic coordinates of the naphthalene dioxygenase of Pseudomonas sp. strain NCIB 9816-4, the three-dimensional structure of KF707 BphA1 was modeled (Fig. 2A). His-233, His-239, and Asp-387 in the Bph Dox of KF707 (colored yellow in Fig. 2B), which correspond to His-208, His-213, and Asp-362, respectively, in the naphthalene dioxygenase, are considered to coordinate the active-site iron (22). Asp-230 in KF707 Bph Dox, corresponding to Asp-205 in the naphthalene dioxygenase, is considered necessary for the major pathway of electron transfer to the mononuclear iron at the active site (30). Seven positions near both the active site (colored green in Fig. 2B) and Thr-376 (colored red in Fig. 2B), which is known to play a very important role in substrate specificity (23, 35, 36), were chosen, and 12 variants of Bph Dox were constructed by site-directed mutagenesis. It has been reported that changing amino acids to smaller and more hydrophobic ones resulted in an enhanced PCB degradation activity (29). Therefore, in most cases, we selected hydrophobic amino acids and replaced them with smaller hydrophobic ones.
FIG. 2.
Proposed structure of BphA1 based on crystallographic analyses of the naphthalene dioxygenase (A) and the proposed structure near the active site in BphA1 (B). (A) The amino acids surrounding the active iron site are shown in yellow, and those coordinated by the Rieske cluster are shown in green. (B) The yellow amino acids are considered to coordinate the active iron site and be involved in electron transfer. The amino acids in green and red are targets for site-directed mutagenesis.
The Bph Doxs of P. pseudoalcaligenes KF707 and B. cepacia LB400 exhibit distinct differences in their substrate specificities for the PCB congeners (8, 13), although these two large subunits of Bph Dox (KF707, BphA1; LB400, BphA) show 95.6% identity in their amino acid sequences (23). KF707 BphA1 differs from LB400 BphA at 20 positions, including 19 amino acid substitutions and one glycine deletion in KF707 BphA1. The steric information revealed that two amino acids, Ile-335 and Thr-376, which are different between KF707 BphA1 and LB400 BphA also exist near the catalytic center. Thus, the amino acids at positions 335 and 376 were replaced with the corresponding amino acids of LB400 BphA. All the E. coli transformants tested produced almost the same amounts of Bph Dox, as judged by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (data not shown).
Oxidation of biphenyl and 4,4′-dichlorobiphenyl.
All E. coli strains expressing the modified Bph Dox formed 2,3-dihydroxy-4-chloro-1[4-chlorophenyl]-cyclohexa-4,6-diene (2,3-dihydrodiol compound) from biphenyl. However, substitution of any one of the Phe residues at positions 227, 377, and 383 reduced the biphenyl-transforming activity of the enzyme. It was previously noted that KF707 has a greater degradation activity for several double para-replaced congeners such as 4,4′-dichlorobiphenyl than does LB400 (13). E. coli expressing the wild-type KF707 Bph Dox produced a 2,3-dihydrodiol compound (m/z 322 as the butylboronate derivative) from 4,4′-dichlorobiphenyl. With the exception of the Leu332Ala mutant, all the other mutant strains lost their degradation ability for 4,4′-dichlorobiphenyl. Among these mutants, the Phe227Leu, Phe383Ala, and Phe383Leu mutants did not detectably degrade the other four PCB congeners tested (Table 1).
TABLE 1.
Transformation activities of modified Bph Dox for biphenyl and selected PCBsa
| BphA1 | Degradation with substrate:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biphenyl
|
2,2′-Dichlorobiphenyl
|
3,3′-Dichlorobiphenyl
|
4,4′-Dichlorobiphenyl
|
2,5,2′-Trichlorobiphenyl
|
2,5,4′-Trichlorobiphenyl
|
2,5,2′,5′-Tetrachlorobiphenyl
|
||||||||
| Mode | Transformation (μg/h) | Mode | Transformation (μg/20 h) | Mode | Transformation (μg/20 h) | Mode | Transformation (μg/20 h) | Mode | Transformation (μg/20 h) | Mode | Transformation (μg/20 h) | Mode | Transformation (μg/20 h) | |
| Wild type | 2,3- | 9.9 ± 1.0 | 5,6- | 3.0 ± 0.3 | 5,6- | 18.3 ± 1.5 | 2,3- | 12.3 ± 1.8 | 5.6- | 8.8 ± 1.1 | 2,3- | 7.6 ± 1.2 | — | N.D. |
| Phe227Leu | 2,3- | 3.6 ± 0.6 | — | N.D. | — | N.D. | — | N.D. | — | N.D. | — | N.D. | — | N.D. |
| Phe227Val | 2,3- | 2.5 ± 0.3 | — | N.D. | 2,3- | 11.8 ± 0.5 | — | N.D. | — | N.D. | — | N.D. | — | N.D. |
| Leu332Ala | 2,3- | 5.8 ± 0.5 | 5,6- | 8.1 ± 0.7 | 5,6- | 14.5 ± 0.8 | 2,3- | 13.9 ± 1.1 | — | N.D. | 2,3- | 5.0 ± 0.2 | — | N.D. |
| Ile335Phe | 2,3- | 9.8 ± 0.6 | 2,3- | 6.6 ± 0.8 | 5,6- | 8.3 ± 0.3 | — | N.D. | 2,3- (24%) 3,4- (76%) | 17.1 ± 0.7 | 2,3- (18%) 3,4- (82%) | 10.9 ± 1.2 | 3,4- | 10.6 ±1.0 |
| Ile335Ala | 2,3- | 7.6 ± 1.0 | — | N.D. | 5,6- | 6.5 ± 1.6 | — | N.D. | — | N.D. | 2,3- | 5.5 ± 0.5 | — | N.D. |
| Thr376Asn | 2,3- | 11.0 ± 1.5 | 2,3- | 9.1 ± 0.2 | 5,6- | 15.9 ± 0.9 | — | N.D. | 3,4- | 7.9 ± 1.1 | 3,4- | 12.0 ± 1.3 | 3,4- | 8.1 ± 0.1 |
| Phe377Ala | 2,3- | 0.5 ± 0.1 | — | N.D. | 2,3- | 3.8 ± 1.2 | — | N.D. | — | N.D. | — | N.D. | — | N.D. |
| Phe377Leu | 2,3- | 3.2 ± 0.1 | 2,3- | 8.0 ± 1.4 | 5,6- | 6.9 ± 1.3 | — | N.D. | 3,4- | 18.1 ± 0.2 | 3,4- | 13.4 ± 1.1 | 3,4- | 7.3 ± 0.7 |
| Phe383Ala | 2,3- | 1.0 ± 0.1 | — | N.D. | — | N.D. | — | N.D. | — | N.D. | — | N.D. | — | N.D. |
| Phe383Leu | 2,3- | 9.0 ± 1.2 | — | N.D. | — | N.D. | — | N.D. | — | N.D. | — | N.D. | — | N.D. |
| LB400 | 2,3- | 9.1 ± 0.8 | 2,3- | 24.1 ± 1.1 | 4,5- | 12.9 ± 0.2 | — | N.D. | 2,3-, 3,4-, 5,6- | 24.0 ± 0.6 | 3,4- | 6.0 ± 0.9 | 3,4- | 7.8 ± 1.2 |
| 5,6- | ||||||||||||||
The substrates were used at 20 μg/ml. Mode, mode of dioxygenation (see Fig. 3 legend). LB400 data are taken from references 15 and 34. Values are from three independent experiments. N.D., metabolites not detected. —, metabolites not deleted. The numbers in parentheses are the percentages of the dioxygenation mode.
Oxidation of 2,2′-dichlorobiphenyl and 3,3′-dichlorobiphenyl.
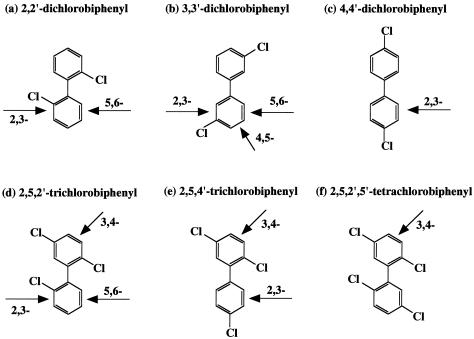
E. coli cells expressing LB400 Bph Dox converted 2,2′-dichlorobiphenyl to 2-chloro-2′,3′-dihydroxybiphenyl (m/z 286 as the butylboronate derivative) with the loss of one chlorine (34). The same substrate was converted differently by E. coli cells expressing KF707 Bph Dox. The molecular ion peak was found at m/z 322 as the butylboronate derivative, which could be a dichlorodihydrodiol compound. This result indicates that KF707 Bph Dox introduced molecular oxygen at the 5,6 position of 2,2′-dichlorobiphenyl to form 5,6-dihydroxy-2-chloro-1-[2-chlorophenyl]-cyclohexa-1,3-diene (2,2′-dichloro-5,6-dihydrodiol compound). The Leu332Ala mutant also attacked the 5,6-position in the same fashion as the KF707 Bph Dox (Table 1). The Ile335Phe, Thr376Asn, and Phe377Leu mutants formed 2-chloro-2′,3′-dihydroxybiphenyl with the loss of one chlorine, as did the LB400 enzyme. The transformation activities of these mutants for 2,2′-dichlorobiphenyl were much higher than that of the wild-type KF707 enzyme but much lower than that of the wild-type LB400 enzyme (Table 1).
The GC-MS analysis found that LB400 Bph Dox attacked 3,3′-dichlorobiphenyl, forming two cis-dihydrodiols with two chlorines as previously reported (15, 34). The major productwas 5,6-dihydroxy-3-chloro-1-[3-chlorophenyl]-cyclohexa-1,3-diene (5,6-dihydrodiol), and the minor one was 4,5-dihydroxy-3-chloro-1-[3′-chlorophenyl]-cyclohexa-2,6-diene (4,5-dihydrodiol). Thus, in this oxidative reaction, no dechlorination was observed. The KF707 Bph Dox converted 3,3′-dichlorobiphenyl to a 5,6-dihydrodiol compound (m/z 322) without the elimination of chlorine (Table 1). Likewise, five mutants, the Leu332Ala, Ile335Phe, Ile335Ala, Thr376Asn, and Phe377Leu mutants, attacked the same position as did the KF707 enzyme. On the other hand, two mutants, the Phe227Val and Phe377Ala mutants, converted a 3,3′-dichlorobiphenyl to 3-chloro-2,3-dihydroxybiphenyl accompanied by the loss of one chlorine (m/z 286). Furthermore, it was revealed that E. coli strains expressing these two Bph Dox mutants produced a yellow ring meta-cleavage compound from 3,3′-dichlorobiphenyl when BphB and BphC were provided. These results suggested that Phe227Val and Phe377Ala led to the different modes of dioxygenation for 3,3′-dichlorobiphenyl. The transformation activities of these two mutants were lower than those of the KF707 and LB400 enzymes (Table 1).
Oxidation of 2,5,2′-trichlorobiphenyl and 2,5,4′-trichlorobiphenyl.
Previously, it was reported that the LB400 Bph Dox oxidizes 2,5,2′-trichlorobiphenyl to three products (15). One major product has a molecular mass of m/z 320 as a butylboronate derivative with two chlorines, which could be 2,5-dichloro-2′,3′-dihydroxybiphenyl, losing one chlorine at the 2′ position. The second major product exhibited a molecular mass of m/z 356 with three chlorines. This was identified as 3,4-dihydroxy-2,5-dichloro-1-[2′-chlorophenyl]-cyclohexa-1,5-diene (2,5,2′-trichloro-3,4-dihydro-3,4-diol compound). The third minor product was estimated to be 5,5′-dihydroxy-2′-chloro-1-[2,5-dochlorophenyl]-cyclohexa-1,3-diene (2,5,2′-trichloro-5′,6′-dihydro-5′,6′-diol). The GC-MS analysis revealed that KF707 Bph Dox attacked the 5′,6′ position for 2,5,2′-trichlorobiphenyl, forming 2,5,2′-trichloro-5′,6′-dihydro-5′,6′-diol (Table 1). On the other hand, 2,5,2′-trichloro-3,4-dihydro-3,4-diol was formed by the Ile335Phe, Thr376Asn, and Phe377Leu mutants via 3,4-dioxygenation (Fig. 3). Interestingly, the product with the molecular ion peak at m/z 320 was also detected from 2,5,2′-trichlorobiphenyl by the Ile335Phe mutant. The product was estimated to be 2,5-dichloro-2′,3′-dihydroxybiphenyl, which was formed via 2,3-dioxygenation, accompanying the spontaneous dechlorination at the 2′ position.
FIG. 3.
Mode of dioxygenation of various PCB congeners by the wild-type and the mutant Bph Doxs. The thick arrow indicates the site of dioxygenation. 2,3-, 2,3-dioxygenation; 3,4-, 3,4-dioxygenation; 5,6-, 5,6-dioxygenation.
It has been shown previously that the KF707 wild-type Bph Dox primarily recognizes the 4′-chlorinated ring and binds in order to introduce molecular oxygen at the 2′,3′ position of 2,5,4′-trichlorobiphenyl, while LB400 Bph Dox recognizes the 2,5-dichlorinated ring and introduces molecular oxygen at the 3,4-position (23). Based on the GC-MS analysis results, the Leu332Ala, Ile335Phe, and Ile335Ala mutants formed 3,4-di-hydroxy-2,5-dichloro-1-[4′-chlorophenyl]-cyclohexa-1,5-diene(2,5,4′-trichloro-2′,3′-dihydro-2′,3′-diol), as in the case of the wild-type Bph Dox. On the other hand, the Phe377Leu and Thr376Asn mutants produced 2′,3′-dihydroxy-4′-chloro-1′-[2,5-dichlorophenyl]-cyclohexa-1′,3′-diene (2,5,4′-trichloro-3,4-dihydro-3,4-diol) via 3,4-dioxygenation. Interestingly, the Ile335Phe mutant exhibited both 2,3- and 3,4-dioxygenation activities for 2,5,4′-trichlorobiphenyl, producing 2,5,4′-trichloro-2′,3′-dihydro-2′,3′-diol and 2,5,4′-trichloro-3,4-dihydro-3,4-diol.
Among the mutant strains tested, the Ile335Phe and Phe377Leu mutants exhibited higher transformation activities for 2,5,2′-trichlorobiphenyl than the KF707 enzyme but lower activities than the LB400 enzyme. For 2,5,4′-trichlorobiphenyl, the Phe383Leu, Thr376Asn, and Ile335Phe mutants exhibited greater transformation activities than the KF707 and LB400 enzymes (Table 1).
Oxidation of 2,5,2′,5′-tetrachlorobiphenyl.
LB400 Bph Dox transformed 2,5,2′,5′-tetrachlorobiphenyl, producing large amounts of a 3,4-dihydrodiol compound (15). No metabolite was obtained from 2,5,2′,5′-tetrachlorobiphenyl with E. coli cells expressing KF707 Bph Dox. E. coli cells expressing the Ile335Phe, Thr376Asn, and Phe377Leu mutant enzymes produced a dihydrodiol compound with m/z 390 as the molecular ion peak, indicating that these three mutants had acquired 3,4-dioxygenation activity for 2,5,2′,5′-tetrachlorobiphenyl. The transformation activities of two mutants, the Ile335Phe and Thr376Asn mutants, were higher than that of the LB400 enzyme (Table1).
DISCUSSION
Modeling the enzyme structure based on the crystallographic coordinates of a known structure has been considered useful when combined with experimental data (14, 19, 20). In this study, we built a model of KF707 BphA1 based on the structure of the naphthalene dioxygenase of Pseudomonas sp. strain NCIB 9816-4 (Fig. 2). According to information about the amino acids near the mononuclear iron center, which is supposed to be the catalytic site, we engineered BphA1, a large subunit of terminal dioxygenase, which is significantly involved in substrate specificity. Some of the Bph Dox variants obtained exhibited altered regiospecificity for PCB congeners.
Based on the pattern of degradation abilities, the PCB-degradative strains are categorized into two groups (29). P. pseudoalcaligenes KF707 is included with the strains having a relatively narrow range of PCB substrates. This group of PCB degraders is superior in degradation of double para-replaced congeners such as 4,4′-dichlorobiphenyl but are unable to degrade 2,5,2′,5′-tetrachlorobiphenyl. On the other hand, B. cepacia LB400, categorized with the strains having a broad substrate specificity, tends to attack 2,5,2′,5′-tetrachlorobiphenyl via 3,4-dioxygenation, but these strains hardly transform 4,4′-dichlorobiphenyl. Between these two categories of PCB degraders, all strains with broad specificity contain Asn at the 376 position in BphA1, whereas the strains with narrow specificity contain Thr at this site (Table 2). In fact, the replacement of Thr-376 with Asn in KF707 BphA1 led to the acquisition of 3,4-dioxygenase activity for 2,5,4′-trichlorobiphenyl and 2,5,2′,5′-tetrachlorobiphenyl (23, 35). This amino acid corresponds to Thr-351 in the large subunit of naphthalene dioxygenase. However, the change of Thr-351 to Asn in naphthalene dioxygenase had a minor effect on product formation (32).
TABLE 2.
Amino acids important for enzymatic activities in the large subunits of various dioxygenasesa
| Origin | Amino acids (positions) | Reference |
|---|---|---|
| KF707 | Phe (227), Leu (332), Ile (335), Thr (376), Phe (377), Phe (383) | 42 |
| LB400 | Phe (227), Leu (333), Phe (336), Asn (377), Phe (378), Phe (384) | 28 |
| 9816-4 | Phe (202), Leu (307), Ser (310), Thr (351), Phe (352), Trp (358) | 32 |
KF707, P. pseudoalcaligenes KF707; LB400, B. cepacia LB400; 9816-4, Pseudomonas sp. strain NCIB 9816-4. Numbers in parentheses show the positions of the corresponding amino acids in the large subunit of each dioxygenase.
The steric information revealed that the structure of the active-site cavity can accommodate substrates like naphthalene and indole (7). The side chains of 17 residues contribute to the topology of the substrate cavity, in which the walls are hydrophobic residues suitable for interactions with an aromatic substrate. Table 2 shows the corresponding amino acids important to the activity of the related dioxygenases. With the exception of Thr-351, all other corresponding amino acids of the naphthalene dioxygenase listed in Table 2 contribute to form part of the binding pocket. It has been reported that Thr-351 plays a minor role in the shape of the binding pocket and substrate specificity in naphthalene dioxygenase (32). In the KF707 Bph Dox, on the other hand, the Thr376Asn mutation had a critical effect on the regiospecificity for various PCB congeners. The same mutant acquired 3,4-dioxygenation activity (38). These results are different between KF707 Bph Dox and naphthalene dioxygenase of NCIB 9816-4 with respect to the role of Thr at this site.
Bph Dox mutants with changes at positions 335 (Ile335Phe and Ile335Ala) and 377 (Phe377Ala and Phe377Leu) showed different patterns of dioxygenation for 2,5,4′-trichlorobiphenyl and 3,3′-dichlorobiphenyl, respectively (Table 1). This is distinct evidence that a specific amino acid is important for the determination of the regioselectivity of Bph Dox. In contrast to the Ile335Ala mutant, the Ile335Phe variant transformed 2,5,2′-trichlorobiphenyl and 2,5,4′-trichlorobiphenyl by both 2,3- and 3,4-dioxygenations. Furthermore, the same variants catalyzed the 3,4-dioxygenation of 2,5,2′,5′-tetrachlorobiphenyl and did not detectably transform 4,4′-dichlorobiphenyl. A replacement of Phe-336 by Ile in LB400 Bph Dox significantly improved the ability to degrade 4,4′-dichlorobiphenyl (29). These results clearly indicate that Ile-335 in KF707 Bph Dox plays a crucial role in the degradation of 4,4′-dichlorobiphenyl. This is in agreement with the report by Brühlmann and Chen that substitutions Thr335Ala and Phe336Ile in LB400 BphA1 are important in substrate specificity (5).
The Phe377Leu mutant demonstrated altered regiospecificity for 2,2′-dichlorobiphenyl, 2,5,2′-trichlorobiphenyl, and 2,5,4′-trichlorobiphenyl and also gained a novel ability to transform 2,5,2′,5′-tetrachlorobiphenyl. On the other hand, the Phe377Ala mutant hardly attacked any of the PCB substrates tested. Because the side chain of Ala is much smaller than that of Phe, it may become difficult to properly place the substrate due to the change in the binding pocket. In the naphthalene dioxygenase of NCIB 9816-4, site-directed mutagenesis also revealed that Phe-352, corresponding to Phe-377 in KF707 Bph Dox, plays an important role in determining the regioselectivity of biphenyl and phenanthrene oxidation (31, 32). The same amino acids were also involved in the stereochemistry of naphthalene cis-dihydrodiol formed from naphthalene and the enantioselectivity with naphthalene and biphenyl.
3,3′-Dichlorobiphenyl was a poor substrate for LB400 Bph Dox, but a 5,6-dihydrodiol and 4,5-dihydrodiol were formed without the elimination of chlorine (15, 34). The KF707 Bph Dox converted 3,3′-dichlorobiphenyl to a 5,6-dihydrodiol. On the other hand, the Phe227Val and Phe377Ala mutants exhibited different modes of dioxygenation. These two mutants introduced molecular oxygen at the 2,3 position, forming 3-chloro-2′,3′-dihydroxybiphenyl with dechlorination. This novel reaction for 3,3′-dichlorobiphenyl is unexpected, because such a dechlorination was not shown in either the KF707 or LB400 Bph Dox. These results raise the possibility that site-directed mutagenesis near the active site creates a novel catalytic activity. Although the two variants of Phe227Val and Phe377Ala exhibited weak activity for biphenyl, the same variants were able to attack only 3,3′-dichlorobiphenyl among the PCB congeners tested. It is likely that the shape of the binding pocket for the 2,3-dioxygenation for 3,3′-dichlorobiphenyl might be significantly different from those of the biphenyl and other PCB substrates.
Changing the amino acids to smaller and more hydrophobic ones resulted in enhanced PCB degradation activity (29). The transformation activity of the Leu332Ala mutant for 2,2′-dichlorobiphenyl and 4,4′-dichlorobiphenyl was higher than that of KF707 Bph Dox. A Phe377Leu mutant acquired novel activity for 2,5,2′,5′-tetrachlorobiphenyl. Substitutions with smaller amino acids may facilitate access of PCB substrates to the active site. However, the change to a larger amino acid, as in the Ile335Phe mutant, caused enhanced and extended degradation activity for certain PCB substrates. The change to a smaller amino acid in Ile335Ala decreased the degradation ability compared to that of the wild-type enzyme. It seems that the proper size of the amino acid in the binding pocket is important for enzymatic activity. A suitable amino acid substitution may reshape the binding pocket and result in relaxation of the binding capacity.
We have previously reported that the four-amino-acid substitution in KF707 BphA1 permits great expansion of the ability to degrade single aromatic compounds such as benzene, toluene, and alkylbenzenes (37). From the structural information of KF707 BphA1, these four amino acids (His-255, Val-258, Gly-268, and Phe-227) are situated surrounding an active site (data not shown). Thus, site-directed mutagenesis based on steric information obtained by structure modeling provides valuable insights relevant to the rational design of enzymes with altered specificity.
Acknowledgments
We thank Takatugu Hirokawa, Ryoka System, Inc., Urayasu, Japan, for helpful discussion about modeling BphA1.
REFERENCES
- 1.Bedard, D. L., R. Unterman, L. H. Bopp, M. J. Brennan, M. L. Haberl, and C. Johnson. 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol. 51:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedard, D. L., R. E. Wagner, M. J. Brennan, M. L. Haberl, and J. F. Brown, Jr. 1987. Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl. Environ. Microbiol. 53:1094-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard, D. L., and M. L. Haberl. 1990. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb. Ecol. 20:87-102. [DOI] [PubMed] [Google Scholar]
- 4.Beil, S., J. R. Mason, K. N. Timmis, and D. H. Pieper. 1998. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J. Bacteriol. 180:5520-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brühlmann, F., and W. Chen. 1999. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol. Bioeng. 63:544-551. [DOI] [PubMed] [Google Scholar]
- 6.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 7.Carredano, E., A. Karlsson, B. Kauppi, D. Choudhury, R. E. Parales, J. V. Parales, K. Lee, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2000. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J. Mol. Biol. 296:701-712. [DOI] [PubMed] [Google Scholar]
- 8.Erickson, B. D., and F. J. Mondello. 1993. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl. Environ. Microbiol. 59:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa, K., N. Tomizuka, and A. Kamibayashi. 1979. Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl. Environ. Microbiol. 38:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa, K. 1982. Microbial degradation of polychlorinated biphenyls, p. 33-57. In A. M. Chakrabarty (ed.), Biodegradation and detoxification of environmental pollutants. CRC Press, Boca Raton, Fla.
- 11.Furukawa, K., and T. Miyazaki. 1986. Cloning of gene cluster encoding biphenyl degradation in Pseudomonas pseudoalcaligenes. J. Bacteriol. 166:392-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa, K. 1994. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation 5:289-300. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, D. T., D. L. Cruden, J. D. Haddock, G. J. Zlstra, and J. M. Brand. 1993. Oxidation of polychlorinated biphenyls by Pseudomonas sp. strain LB400 and Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 175:4561-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham-Lorence, S., M. W. Khalil, M. C. Lorence, C. R. Mendelson, and E. R. Simpson. 1991. Structure-function relationships of human aromatase cytochrome P-450 by using molecular modeling and site-directed mutagenesis. J. Biol. Chem. 266:11939-11946. [PubMed] [Google Scholar]
- 15.Haddock, J. D., J. R. Horton, and D. T. Gibson. 1995. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegg, E. L., and L. Que, Jr. 1997. The 2-His-1-carboxylate facial triad—an emerging structural motif in mononuclear nonheme iron(II) enzymes. Eur. J. Biochem. 250:625-629. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, J., A. Suyama, S. Hayashida, and K. Furukawa. 1994. Construction of hybrid biphenyl (bph) and toluene (tod) genes for functional analysis of aromatic ring dioxygenases. Gene 138:27-33. [DOI] [PubMed] [Google Scholar]
- 18.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1998. Involvement of the terminal oxygenase β subunit in the biphenyl dioxygenase reactivity pattern for chlorobiphenyls. J. Bacteriol. 180:5828-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki, M., T. A. Darden, L. G. Pedersen, D. G. Davis, R. O. Juvonen, T. Sueyoshi, and M. Negishi. 1993. Engineering mouse P450coh to a novel corticosterone 15 alpha-hydroxylase and modeling steroid-binding orientation in the substrate pocket. J. Biol. Chem. 268:759-762. [PubMed] [Google Scholar]
- 20.Iwasaki, M., T. A. Darden, C. E. Perker, K. B. Tomer, L. G. Pedersen, and M. Negishi. 1994. Inherent versatility of P450 oxygenase. Conferring dehydroepiandrosterone hydroxylase activity to P450 2a-4 by a single amino acid mutation at position 117. J. Biol. Chem. 269:9079-9083. [PubMed] [Google Scholar]
- 21.Jiang, H., R. E. Parales, N. A. Lynch, and D. T. Gibson. 1996. Site-directed mutagenesis of conserved amino acids in the α subunit of toluene dioxygenase: potential mononuclear nonheme iron coordination sites. J. Bacteriol. 178:3133-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, N., A. Nishi, M. Goto, and K. Furukawa. 1997. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J. Bacteriol. 179:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumamaru, T., H. Suenaga, M. Mitsuoka, T. Watanabe, and K. Furukawa. 1998. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol. 16:663-666. [DOI] [PubMed] [Google Scholar]
- 25.Maeda, T., Y. Takahashi, H. Suenaga, A. Suyama, M. Goto, and K. Furukawa. 2001. Functional analyses of Bph-Tod hybrid dioxygenase, which exhibits high degradation activity for trichloroethylene. J. Biol. Chem. 276:29833-29838. [DOI] [PubMed] [Google Scholar]
- 26.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 27.Masse, R., F. Messier, L. Peloquin, C. Ayotte, and M. Sylvestre. 1984. Microbial biodegradation of 4-chlorobiphenyl, a model compound of chlorinated biphenyls. Appl. Environ. Microbiol. 47:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequence that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parales, R. E., J. V. Parales, and D. T. Gibson. 1999. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 181:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parales, R. E., S. M. Resnick, C.-L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the α subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 34.Poulos, T. L., B. C. Finzel, I. C. Gunsalus, G. C. Wagner, and J. Kraut. 1985. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J. Biol. Chem. 260:16122-16130. [PubMed] [Google Scholar]
- 35.Poulos, T. L., B. C. Finzel, and A. Howard. 1987. High-resolution crystal structure of cytochrome P450. J. Mol. Biol. 195:687-700. [DOI] [PubMed] [Google Scholar]
- 36.Seeger, M., M. Zielinski, K. N. Timmis, and B. Hofer. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suenaga, H., A. Nishi, T. Watanabe, M. Sakai, and K. Furukawa. 1999. Engineering a hybrid Pseudomonad to acquire 3,4-dioxygenase activity for polychlorinated biphenyls. J. Biosci. Bioeng. 87:430-435. [DOI] [PubMed] [Google Scholar]
- 38.Suenaga, H., M. Goto, and K. Furukawa. 2001. Emergence of multifunctional oxygenase activities by random priming recombination. J. Biol. Chem. 276:22500-22506. [DOI] [PubMed] [Google Scholar]
- 39.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sylvestre, M., and M. Sondossi. 1994. Selection of enhanced polychlorinated biphenyl-degrading bacterial strains for bioremediation: consideration of branching pathways, p. 47-73. In G. R. Chaudhry (ed.), Biological degradation and bioremediation of toxic chemicals. Dioscorides Press, Portland, Oreg.
- 41.Szklarz, G. D., Y. A. He, and J. R. Halpert. 1995. Site-directed mutagenesis as a tool for molecular modeling of cytochrome P450 2B1. Biochemistry 34:14312-14322. [DOI] [PubMed] [Google Scholar]
- 42.Taira, K., J. Hirose, S. Hayashida, and K. Furukawa. 1992. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 267:4844-4853. [PubMed] [Google Scholar]