Abstract
Four distinct Escherichia coli immunoglobulin-binding (eib) genes, each of which encodes a surface-exposed protein that binds immunoglobulins in a nonimmune manner, are carried by separate prophages in E. coli reference (ECOR) strain ECOR-9. Each eib gene was transferred to test E. coli strains, both in the form of multicopy recombinant plasmids and as lysogenized prophage. The derived lysogens express little or no Eib protein, in sharp contrast to the parental lysogen, suggesting that ECOR-9 has an expression-enhancing activity that the derived lysogens lack. Supporting this hypothesis, we cloned from ECOR-9 overlapping genes, ibrA and ibrB (designation is derived from “immunoglobulin-binding regulator”), which together activated eib expression in the derived lysogens. The proteins encoded by ibrA and ibrB are very similar to uncharacterized proteins encoded by genes of Salmonella enterica serovar Typhi and E. coli O157:H7 (in a prophage-like element of the Sakai strain and in two O islands of strain EDL933). The genomic segment containing ibrA and ibrB has been designated the IbrAB island. It contains regions of homology to the Shiga toxin-converting prophage, Stx2, as well as genes homologous to phage antirepressor genes. The left boundary between the IbrAB island and the chromosomal framework is located near min 35.8 of the E. coli K-12 genome. Homology to IbrAB was found in certain other ECOR strains, including the other five eib-positive strains and most strains of the phylogenetic group B2. Sequencing of a 1.1-kb portion of ibrAB revealed that the other eib-positive strains diverge by ≤0.1% from ECOR-9, whereas eib-negative ECOR-47 diverges by 16%.
The Escherichia coli immunoglobulin (Ig)-binding (Eib) proteins are members of a larger family of surface-exposed bacterial proteins that includes YadA of Yersinia (39, 44, 45), UspAII of Moraxella (3, 4), and DsrA of Haemophilus ducreyi (15). The Eib proteins have several phenotypic features in common with these proteins, such as the ability to impart resistance to human serum complement and a tendency to exist as highly stable multimers. In addition to the properties shared with other members of this protein family, the Eib proteins can bind Igs such as IgA and/or the Fc fragment of human IgG (IgG Fc) in a nonimmune manner (40, 41). The eib genes are strongly expressed as the cells enter stationary phase at 37°C (42). The Eib proteins were originally identified in six of the 72 strains of the E. coli reference (ECOR) strain collection (33). One of these six strains, ECOR-9, was found to produce several distinct Ig-binding proteins, each encoded by a different member of a set of related prophages. Four genes, eibA, eibC, eibD, and eibE, were cloned from ECOR-9. In multicopy each imparted Ig-binding activity and enhanced serum resistance to a test recipient. In addition a series of lysogenic derivatives of E. coli C was established using eib-bearing phages derived from ECOR-9 (P-EibB, P-EibC, P-EibD, and P-EibE) (40). The derived lysogens, bearing from one to three of the prophages, expressed little or no Eib, in sharp contrast to the parental ECOR-9 lysogen, suggesting that ECOR-9 has an expression-enhancing activity that the derived lysogens lack. The goal of the present study was to clone from ECOR-9 a gene or genes that would activate expression of Ig-binding activity in the lysogens.
MATERIALS AND METHODS
Strains and culture conditions.
The E. coli K-12 strain DH5α was used for cloning of all constructs. All lysogens were derivatives of E. coli strain C, a nonrestricting strain (6), and are listed (see Table 2). Cultures used for protein extraction were grown in Luria-Bertani (LB) broth for 18 to 24 h at 37° with agitation and harvested by centrifugation at 4°C. LB broth containing ampicillin, 50 μg per ml, was used for maintenance of pUC21 derivatives. LB broth containing kanamycin, 50 μg per ml, was used to maintain pOK12 derivatives.
TABLE 2.
E. coli C derivative strains
| Strain | Prophage(s) or other source of eib genes | ibrAB single-copy insertionb |
|---|---|---|
| CH6249a | P-EibE from ECOR-9 | |
| CH6252a | P-EibD from ECOR-9 | |
| CH6254a | P-EibB, P-EibD, and P-EibE from ECOR-9 | |
| CH6256a | P-EibC, P-EibD, and P-EibE from ECOR-9 | |
| CH6304a | P-EibB from ECOR-9 | |
| CH6306a | P-EibC from ECOR-9 | |
| CH7124 | P-EibG from ECOR-2 | |
| CH7138 | None | + |
| CH7140 | P-EibD from ECOR-9 | + |
| CH7142 | P-EibB from ECOR-9 | + |
| CH7144 | P-EibC, P-EibD, and P-EibE from ECOR-9 | + |
| CH7146 | P-EibB from ECOR-9 | + |
| CH7248 | P-EibC from ECOR-9 | + |
| CH7259 | Inserted single copy of eibF |
See reference 40.
See Materials and Methods.
Protein extraction and Ig binding.
Preparation of cell extracts, determination of protein concentration, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were as described previously (42). Our standard immunoblotting procedure entails a one-step incubation with nonimmune antibody, such as the Fc fragment of human IgG (IgG Fc) conjugated with horseradish peroxidase (HRP). It is important to note that there is no incubation with primary antibody specifically directed against an antigen. Purified Fc fragment of human IgG conjugated with HRP (IgG Fc-HRP) (Rockland) was used at a concentration of 50 ng of antibody per ml.
DNA cloning and analysis.
Techniques used for DNA isolation, cloning, and sequence analysis involve minor modifications of those indicated elsewhere (22, 40, 51). The plasmid vectors for cloning were pOK12 and pUC21 (48). Cloning of the Ig-binding regulator (ibrAB) genes utilized a partial Sau3AI digest of ECOR-9 genomic DNA. Fragments in the desired size range were purified by agarose gel electrophoresis and use of a Qiaex II Gel Extraction Kit (Qiagen). The fragments were ligated into the BamHI site of pOK12 and were electroporated into strain CH6256, a triple lysogen that harbors P-EibC, P-EibD, and P-EibE (see Table 2) and does not express detectable amounts of Ig-binding proteins (40). Colony blots of transformants were screened for IgG binding by procedures based on published protocols (17, 40). Briefly, colonies were blotted to nitrocellulose and lysed in situ with 1% SDS at 65° for 30 min. The membranes were blocked with 10% (wt/vol) nonfat dry milk in phosphate-buffered saline for 1 h, washed, and incubated for 1 h with affinity-purified, HRP-conjugated anti-rabbit Ig developed in donkey (50 ng per ml). The blots were washed and used to expose film. Deletions within the original cloned fragment were made using convenient restriction sites; key plasmids are listed in Table 1. Oligonucleotide synthesis and automated DNA sequencing were done by the Macromolecular Core Facility of the Pennsylvania State College of Medicine. For all new sequences, both strands were sequenced; for repetitive sequencing of essentially identical segments, sequencing was sometimes limited to a single strand. To disrupt the open reading frame (ORF) of ibrA, pCS7067 was digested with BsaHI; pCS7067 was digested with MfeI to disrupt ibrB. The positions of the restriction sites are shown in Fig. 1A. The digestions were followed by treatment with the Klenow fragment of DNA polymerase and religation. This introduced a 4-bp addition to ibrA (pCS7184) and an 8-bp addition to ibrB (pCS7206) (Fig. 1C). For Southern analysis an IbrAB probe was generated by PCR using left primer pr236 (CCTCTGTATG ATTTGATGTA CC) and right primer pr237 (CTTTAACCTC AGAACTTCAT CC). See Fig. 1A for its location. The enhanced chemiluminescence random-prime labeling and detection systems (Amersham) were used to label the IbrAB probe and detect hybrids. This pair of primers was used for PCR with the following thermal cycling protocol: 30 cycles (94°C for 2 min, 58°C for 1 min, and 72°C for 2 min). These primers and protocol were also used to amplify an ibrAB-related sequence from the strains ECOR-2, -5, -12, -43, -72, and -47. For ECOR-47 two rounds of PCR were performed using the same pair of primers for both rounds. Nucleic acid and amino acid sequence similarity searches were done with the Blast programs (5) without filters. Relationships to protein families were analyzed with the Conserved Domain Database and Search Service (27).
TABLE 1.
Plasmidsa
| Plasmid | Insert source | Insert identification |
|---|---|---|
| pCS6489 | ECOR-9 | 6-kb partial Sau3AI (ibrAB primary clone) |
| pCS6490 | ECOR-9 | 6.2-kb partial Sau3AI (ibrAB primary clone) |
| pCS7004 | pCS6490 | 2.7-kb Sau3AI-XmnI (lacks the last 34 codons of ibrA; lacks all of ibrB) |
| pCS7029 | pCS6489 | 1.4-kb SnaBI-Sau3AI (contains all of ibrB + 482-bp leader DNA) |
| pCS7038 | pCS6490 | 3.3-kb Sau3AI-BglII (contains all of ibrA; lacks the last 48 codons of ibrB) |
| pCS7052 | pCS6490 | 1.4-kb SacI-ScaI (complete ibrA ORF + 116-bp leader DNA; lacks all of ibrB) |
| pCS7067 | pCS6489 | 3-kb PvuII-BamHI (complete ibrA and ibrB ORFs + 363-bp leader DNA) |
| pCS7088 | pCS6489 | 3-kb PvuII-BamHI (complete ibrA and ibrB ORFs + 363-bp leader DNA) |
| pCS7184 | pCS7067 | 3-kb PvuII-BamHI with reading frame of ibrA ORF shifted by +4 bp at BsaHI (nt 592), causing termination after aa 235 |
| pCS7206 | pCS7067 | 3-kb PvuII-BamHI with reading frame of ibrB ORF shifted by +8 bp at MfeI (nt 279), causing altered sequence after aa 93 and termination after aa 110 |
All plasmids are based on the vector pOK12, except pCS7088, which is based on pUC21.
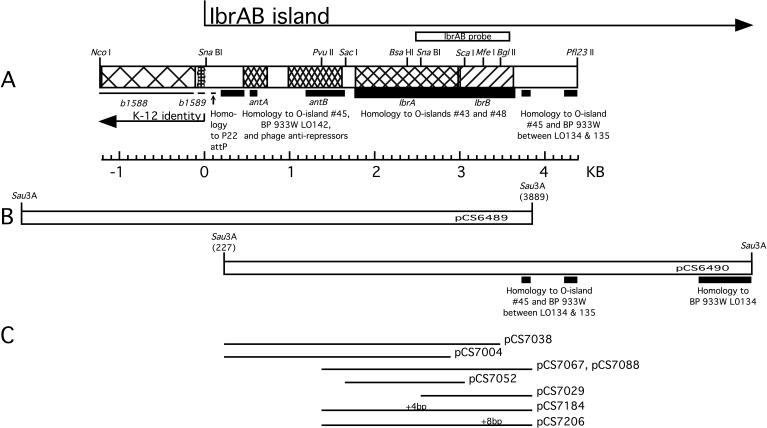
FIG. 1.
Map of the IbrAB island. (A) Gene names and the junction of the IbrAB island with the E. coli K-12 chromosome are indicated below the map; ORFs ibrA and ibrB are translated from left to right. Coordinate 0 marks the end of the K-12 identity. Regions of E. coli O157:H7 homology to O islands no. 43 and 48 (strain EDL933) and SpLE1 (Sakai strain) are indicated by thick lines; homology to bacteriophage 933W and O island no. 45 of E. coli O157:H7 EDL933 is indicated by lines of intermediate thickness, and regions identical to K-12 are indicated by thin lines below the map; the position of the IbrAB probe is shown above. BP, bacteriophage. (B) Primary plasmid clones. Nucleotide coordinates within the IbrAB island of the sequenced end of each primary clone are shown in parentheses. (C) Plasmids containing deletions and insertions affecting ibrA or ibrB. The positions of frameshifting insertions are indicated as +4 bp (at BsaHI) and +8 bp (at MfeI), respectively, above plasmids pCS7184 and pCS7207.
Integration of single-copy ibrAB into the genome.
The lambda InCh plasmid-chromosome shuttle system (8) was used to insert the ibrAB genes into the chromosomes of E. coli C and derived lysogens using the procedures described in the Lambda InCh Manual (http://beck1.med.harvard.edu/Lambda_InCh.html). For this purpose the minimum ibrAB insert of pCS7067 was transferred to pUC21 to produce pCS7088 (Table 1).
Nucleotide sequence accession numbers.
The GenBank accession number for the region of the IbrAB island described in this report is AF460182.
RESULTS
Cloning an element which activates eib expression in lysogens.
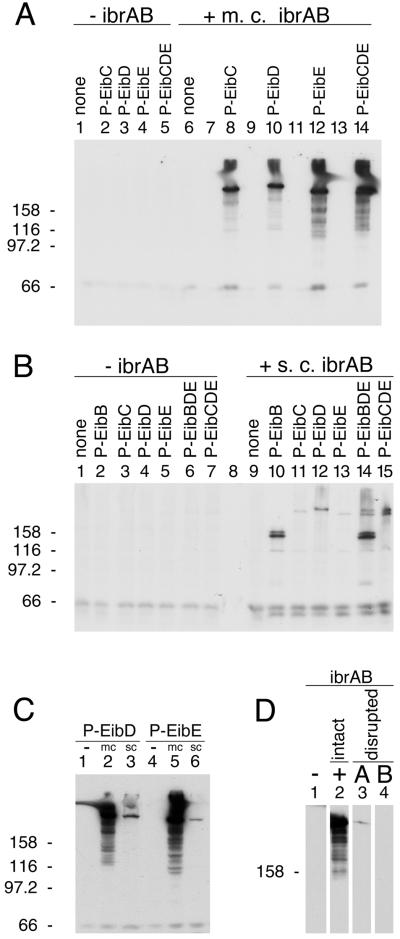
Lysogens established with phage from lysates of ECOR-9 have been shown to acquire eib genes introduced by the infecting phage, but these lysogens fail to express Ig-binding activity under conditions in which ECOR-9 is positive (40). We hypothesized that the parental lysogen, ECOR-9, harbors an activator of eib gene expression that is not present in the derived lysogens but might activate expression when introduced into them. To test this hypothesis, CH6256, a triple lysogen based on E. coli C and harboring phages P-EibC, P-EibD, and P-EibE from ECOR-9 (Table 2) was transformed with an ECOR-9 partial Sau3AI library, and the resulting transformant colonies were screened for Ig binding (Materials and Methods). This strategy yielded two overlapping recombinant plasmids (pCS6489 and pCS6490) that shared a 3.9-kb fragment (Fig. 1B). Each activated Eib expression in the test lysogen. Subcloning identified a minimal plasmid, pCS7067 (Fig. 1C and Table 1), which was sufficient to activate IgG Fc-binding activity. This recombinant plasmid activated Eib expression in single lysogens harboring P-EibC, P-EibD, or P-EibE, as well as in the triple lysogen (Fig. 2A, compare lanes 2 and 8, lanes 3 and 10, lanes 4 and 12, and lanes 5 and 14). The cloned DNA did not cause Ig-binding activity by itself when introduced into nonlysogenic E. coli C, indicating that the expression of Ig-binding activity was dependent on the presence of a P-Eib prophage (Fig. 2A, compare lanes 1 and 6). This recombinant plasmid contained an overlapping pair of ORFs, ibrA and ibrB (Fig. 1A), which we designated ibrAB.
FIG. 2.
Effect of ibrAB on eib expression in E. coli C lysogens. Lysogen strain numbers are listed in Table 2. (A) Effect of multicopy (m.c.) ibrAB on eib expression in lysogens: lanes 1 and 6, nonlysogenic E. coli C; lanes 2 and 8, P-EibC lysogen; lanes 3 and 10, P-EibD lysogen; lanes 4 and 12, P-EibE lysogen; lanes 5 and 14, P-EibC/D/E triple lysogen. The multicopy ibrAB plasmid pCS7088 was present in the strains shown in lanes 6, 8, 10, 12, and 14 and was absent from those shown in lanes 1 through 5. Lanes 7, 9, 11, and 13 were left empty for clarity. (B) Effect of single-copy (s.c.) (chromosomally inserted) ibrA/B on eib expression in lysogens: lanes 1 and 9, E. coli C; lanes 2 and 10, P-EibB; lanes 3 and 11, P-EibC; lanes 4 and 12, P-EibD; lanes 5 and 13, P-EibE; lanes 6 and 14, P-EibB/D/E triple lysogen; and lanes 7 and 15, P-EibC/D/E triple lysogen. Strains shown in lanes 9 through 15 all had a single copy of ibrAB inserted into the chromosome (Materials and Methods). (C) Comparison of multicopy (mc) and single-copy (sc) ibrAB effects on eib expression. P-EibD lysogens: lane 1, no ibrAB; lane 2, multicopy ibrAB plasmid pCS7088; and lane 3, ibrAB single-copy insertion (CH7140). P-EibE lysogens: lane 4, no ibrAB; lane 5, multicopy ibrAB plasmid pCS7088; and lane 6, ibrAB single-copy insertion (CH7138). (D) Effect of disrupting the ORFs of ibrA and ibrB on eib expression in a P-EibC/D/E triple lysogen (CH6256). Lane 1, no ibrAB; lane 2, intact ibrAB plasmid (pCS7067); lane 3, ibrA disruption (pCS7184); and lane 4, ibrB disruption (pCS7206). For all experiments, whole-cell extracts were fractionated by SDS-PAGE, blotted to polyvinylidene difluoride, and incubated with human IgG Fc-HRP (Materials and Methods). Acrylamide concentrations: 8% (A, B, and C) and 7% (D). Each lane contained approximately 30 μg of protein. Molecular mass units are kilodaltons.
Although the presence of ibrAB was not expected in any of the P-Eib prophages, we nevertheless tested for it by attempting to amplify by PCR a 1.1-kb region spanning portions of ibrA and ibrB (i.e., the region comprising the IbrAB probe in Fig. 1A; see Materials and Methods). This procedure failed to produce a PCR product when DNA from any of the lysogens was used as the template, whereas a parallel PCR using ECOR-9 DNA as a positive control template yielded a product of the expected size. These results indicated that ibrAB was absent from the P-Eib prophage (not shown).
Role of ibrAB in eib expression in lysogens.
Genes ibrA and ibrB overlap by 16 bp, suggesting possible functional interdependence as well as cotranscription. To determine whether both ORFs are required for activation of eib expression, we made a series of deletions affecting each (Fig. 1C) and introduced the modified constructs into the triple lysogen CH6256. The transformants were tested for Ig-binding activity by SDS-PAGE and blotting. The following modifications disrupted the ibrAB activity: deletion of the C-terminal 34 codons of ibrA and all of ibrB (pCS7004); deletion of the C-terminal 48 codons of ibrB retaining intact ibrA (pCS7038); retention of ibrA alone (pCS7052); and retention of ibrB alone (pCS7029) (not shown). Of the conditions examined, eib expression was activated only when both ORFs were present and intact (pCS7067 or pCS7088). Due to its orientation, the construct retaining only ibrB (pCS7029) might not retain transcription signals necessary for ibrB expression. In summary, our results show that ibrB is required but that the requirement for ibrA is not established. To test whether the requirement might be for RNA rather than protein, a small addition was made to each ibr ORF to disrupt each reading frame but to introduce only a small (4- or 8-nucleotide [nt]) change in the nucleotide sequence (Materials and Methods and Fig. 1C). The resulting plasmids were pCS7184, which contains a 4-bp addition after codon 235 (BsaHI site) in ibrA, and pCS7206, which contains an 8-bp addition after codon 93 (MfeI site) in ibrB (Fig. 1A and C). The disruption of ibrB completely eliminated the activation effect imparted by intact ibrAB (Fig. 2D, compare lanes 2 and 4). Disruption of ibrA greatly reduced but did not completely eliminate the activation effect (Fig. 2D, compare lanes 2 and 3). These results are consistent with a requirement for protein rather than for RNA. The low level of Eib activity observed upon ibrA disruption is open to two interpretations. It may be that the relatively large (235-amino-acid [aa]) N-terminal portion of IbrA maintains a low level of activity. Alternatively, the effect may be due to a polar effect that reduces the expression of downstream ibrB. Consequently, the question of whether ibrA is absolutely required remains open.
Activation by single-copy ibrAB.
The 2.5-kb ibrAB-containing insert of pCS7088 (Table 1) was introduced in single copy into the chromosome of each single and the triple P-Eib lysogen by use of lambda InCh methodology (Materials and Methods). The lysogens, with and without single-copy ibrAB, were then assessed for eib expression (Fig. 2B). Single-copy ibrAB clearly activated eib expression (Fig. 2B, compare lanes 2 and 10, lanes 3 and 11, lanes 4 and 12, lanes 5 and 13, lanes 6 and 14, and lanes 7 and 15). Notably, the positive effect of ibrAB on eib expression in single lysogens harboring P-EibD and P-EibE, respectively, was much greater in multicopy than in single copy (Fig. 2C, compare lane 2 to lane 3 and lane 5 to lane 6). Comparable differences between multicopy and single-copy ibrAB effects were also noted in the single lysogen harboring P-EibC (not shown). As can be seen in Fig. 2A and C, multicopy ibrAB-induced eib expression led to the appearance of material with apparent sizes greater than those for the forms induced by single-copy ibrAB. In all cases bands comparable in apparent size to those resulting from single-copy ibrAB expression were also present. This was a consistent observation that appeared to result from an overabundance of Eib protein whenever multicopy ibrAB was used. The smear-like zone between this material and the smaller forms was another consistent observation.
Effect of ibrAB on expression of eib genes derived from ECOR-2.
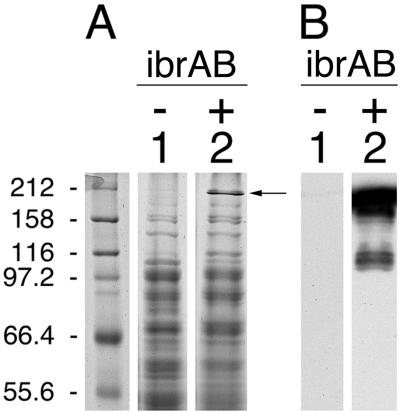
The observation that ibrAB activated expression of all of the eib genes that have been identified in ECOR-9 led us to test whether that ability extended to the activation of an eib gene from another strain of E. coli. We have cloned from ECOR-2 a gene that also encodes an Eib protein, EibF, but one with a strong preference for binding human serum IgA (41). We used lambda InCh methodology to integrate this gene, eibF, into the chromosome in single copy, producing CH7259 (Materials and Methods). CH7259 did not express IgA-binding activity, as evidenced by blotting (Fig. 3B, lane 1), nor did it produce proteins of a size expected of EibF (Fig. 3A, lane 1). Provision of multicopy ibrAB (pCS7088) to CH7259 cells resulted in expression of large proteins that bound IgA (Fig. 3B, lane 2). We also established a lysogen of E. coli C, CH7124, using a phage from a UV-induced lysate of ECOR-2, and tested it for Ig-binding activity. As had been observed for all of the lysogens derived from ECOR-9 prophage, CH7124 failed to express Ig-binding activity (Fig. 4, lane 2). Introduction of ibrAB in multicopy activated the lysogen to express IgG Fc-binding activity with an apparent molecular mass that is characteristic of Eib proteins (Fig. 4, lane 3). With the caveat that the gene encoding Ig-binding activity of CH7124 has not yet been cloned or sequenced, we conclude that ibrAB from ECOR-9 can activate at least one and probably two eib genes from ECOR-2 in addition to those from ECOR-9. Expression of Ig-binding activity in the lysogen containing multicopy ibrAB was much greater than in source strain ECOR-2 (Fig. 4, compare lanes 1 and 3).
FIG. 3.
Effect of multicopy ibrAB on expression of chromosomally inserted single-copy eibF. Whole-cell extracts were fractionated on duplicate gels by SDS-10% PAGE. One gel was stained with Coomassie brilliant blue (A); the other was blotted to polyvinylidene difluoride and incubated with IgG Fc-HRP (B). Lane 1, single-copy eibF (CH7259); and lane 2, single-copy eibF with multicopy ibrAB (CH7259/pCS7067). Molecular mass standards are shown in the leftmost lanes in kilodaltons. The arrow indicates the most prominent band evident in lane 2 that is not present in lane 1.
FIG. 4.
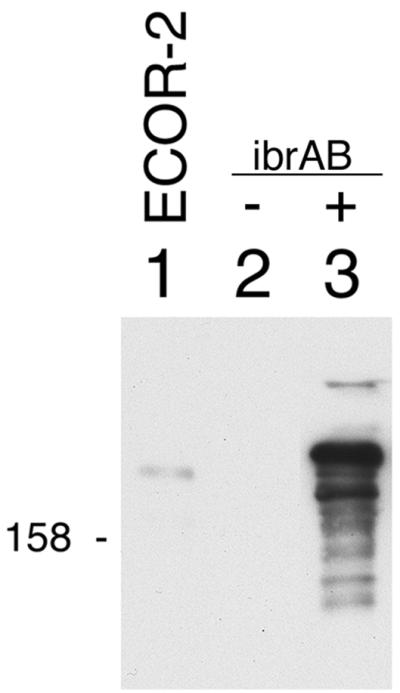
Effect of multicopy ibrAB on expression of Ig-binding activity in a lysogen (CH7124) established using phage from a lysate of ECOR-2. Blots were prepared from whole-cell extracts and were incubated with IgG Fc-HRP as described for Fig. 1. Lanes: 1, ECOR-2; 2, CH7124; and 3, CH7124/pCS7067. 158, 158 kDa.
IbrAB homology in ECOR strains.
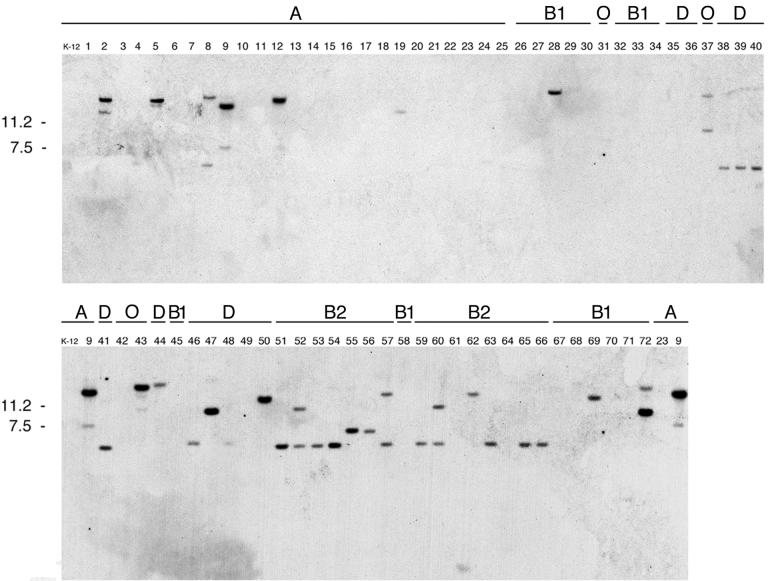
The ability of ibrAB cloned from ECOR-9 to activate expression of eib genes from both ECOR-2 and ECOR-9 raised the question of whether ibrAB was generally present in all eib-positive E. coli strains and/or whether it was restricted to them. We tested all 72 ECOR strains for homology to ibrAB by Southern hybridization using a DNA probe, IbrAB, spanning 537 bp of ibrA and 579 bp of ibrB (Fig. 1A). We found that each of the six eib-positive ECOR strains (strains 2, 5, 9, 12, 43, and 72) showed a strong IbrAB hybridization signal (Fig. 5). Somewhat unexpectedly, each displayed an additional, weaker hybridization signal. A number of additional strains that lack eib gene homology and Ig-binding activity also showed hybridization signals with the IbrAB probe. Notably, a particularly high proportion of hybridizing strains was found in phylogenetic group B2. Thirteen of the 15 group B2 strains showed strong or moderately strong hybridization signals. Five of the 12 group D strains also gave positive signals. In contrast, only six of the 25 group A strains showed hybridization, and of the six, four were the eib-positive strains. E. coli K-12 was negative. Only three of the 16 group B1 strains (including eib-positive strain 72) showed evidence of ibrAB homology. Of the four outlying group E strains, only two showed ibrAB homology. These were ECOR-37, which is phylogenetically similar to E. coli strain O157:H7 (38), and the eib-positive strain ECOR-43.
FIG. 5.
IbrAB homology in the ECOR strains. BamHI digests were fractionated by agarose gel electrophoresis and were transferred for Southern analysis as indicated in Materials and Methods. The probe used was a 1.1-kb segment from the ibrAB locus (Fig. 1A). ECOR strain numbers are shown above each lane. The phylogenetic group (20) of each strain is shown above its number. Kilodaltons are given on the left.
The segment defined by the 1,100-bp IbrAB probe (Fig. 1A) was amplified by PCR from eib-positive ECOR strains (strains 2, 5, 12, 43, and 72) and from eib-negative ECOR-47 (a member of phylogenetic group D) (Materials and Methods). The resulting amplicons were sequenced and compared to the sequence of this region from ECOR-9. The following results were obtained: no nucleotide changes in ECOR-2, -5, and -12; one synonymous change each in ECOR-43 and ECOR-72; and 162 changes resulting in 36-aa differences in ECOR-47. In spite of the divergence observed within ECOR-47, the ORFs of ibrA and ibrB were maintained within the region analyzed (not shown).
Definition and location of the IbrAB island.
A 5.6-kb segment spanning ibrAB was sequenced (see Fig. 1A). The first 1,194 bp were identical to sequences of E. coli K-12 near min 35.8 between speG and dgsA (7), while the remainder was divergent. The junction between the unique sequence and that common to E. coli K-12 occurred within gene b1589, which is predicted to encode the Fe-S subunit of an oxidoreductase. Based on this relationship, we define the left end of the IbrAB island as that sequence divergent from the E. coli K-12 sequence beginning near min 35.8, extending rightward through ibrAB, and including the homology with prophage 933W (between ORFs LO134 and LO135 [Fig. 1B]). The right boundary of the island has not been defined.
To the left of the ibrAB genes within the IbrAB island were two ORFs designated antA (bp 445 to 729) and antB (bp 979 to 1608) (Fig. 1A). The products of these two loci have homology to putative antirepressors found in numerous bacteriophages. Comparison of proteins AntA (94 aa) and AntB (209 aa) with each other revealed 62% amino acid identities near their N termini. AntA appears to be encoded by a truncated and divergent copy of antB. Both AntA and AntB are homologous to the antirepressor protein encoded by gene LO142 of bacteriophage 933W (37) (amino acid identities of 73 and 84%, respectively [Table 3 ]). Other homologies within the IbrAB island to sequences in the databases are addressed in the Discussion.
TABLE 3.
Protein homology
| Gene | Coordinates in IbrAB island | Accession no. (reference) of related protein | Source and function of related protein | % Identity |
|---|---|---|---|---|
| antA | 445-729 | NP_050579 (30) | Bacteriophage VT2-Sa, ORF 80 | 74 |
| NP_309278 (18) | E. coli O157:H7 Sakai strain gene Ecs1251: putative antirepressor protein | 74 | ||
| NP_049540 (37) | Bacteriophage 933W, ORF LO142 | 73 | ||
| NP_287009 (36) | E. coli O157:H7 EDL933 gene Z1503 (O island no. 45) | 73 | ||
| antB | 978-1608 | NP_050579 (30) | Bacteriophage VT2-Sa, ORF 80 | 84 |
| NP_309278 (18) | E. coli O157:H7 Sakai strain gene Ecs1251; putative antirepressor protein | 84 | ||
| NP_049540 (37) | Bacteriophage 933W, ORF LO142 | 84 | ||
| NP_287009 (36) | E. coli O157:H7 EDL933 gene Z1503 (O island no. 45) | 84 | ||
| ibrA | 1805-3031 | NP_286738 (36) | E. coli O157:H7 EDL933 gene Z1203 (O island no. 43) | 90 |
| NP_287146 (36) | E. coli O157:H7 EDL933 gene Z1643 (O island no. 48) | 90 | ||
| NP_309413 (18) | E. coli O157:H7 Sakai gene Ecs1386 | 90 | ||
| CAD09357 (35) | S. enterica serovar Typhi gene STY4583 | 85 | ||
| AAL19556 (28) | S. enterica serovar Typhimurium LT2 gene STM0605 (ybdN); putative 3′ phosphoadenosine 5′-phosphosulfate sulfotransferase (PAPS reductase)/FAD synthetase | 58 | ||
| CAD05081 (35) | S. enterica serovar Typhi gene STY0649 | 57 | ||
| NP_308668 (18) | E. coli O157:H7 Sakai gene Ecs0641 | 56 | ||
| NP_415135 (7) | E. coli K-12/MG1655 gene b0602 (ybdN) | 56 | ||
| NP_286329 (36) | E. coli O157:H7 EDL933 gene Z0746 (ybdN) | 56 | ||
| ibrB | 3016-3660 | NP_286739 (36) | E. coli O157:H7 EDL933 gene Z1204 (O island no. 43) | 86 |
| NP_287147 (36) | E. coli O157:H7 EDL933 gene Z1644 (O island no. 48) | 86 | ||
| NP_309414 (18) | E. coli O157:H7 Sakai gene Ecs1387 | 86 | ||
| NP_085259 (47) | Shigella flexneri large virulence plasmid pWR501 gene S0105a | 84 | ||
| CAD09358 (35) | S. enterica serovar Typhi gene STY4584 | 75 | ||
| AAL19555 (28) | S. enterica serovar Typhimurium gene STM0604; putative transcriptional regulator | 60 | ||
| CAD05080 (35) | S. enterica serovar Typhi gene STY0648 | 60 | ||
| NP_286328 (36) | E. coli O157:H7 EDL933 gene Z0744 (ybdM) | |||
| NP_308667 (18) | E. coli O157:H7 Sakai gene Ecs0640 | 61 | ||
| NP_415134 (7) | E. coli K-12/MG1655 gene b0601 (ybdM) | 60 |
Because of the homology of portions of the IbrAB island to bacteriophage 933W as well as fragmentary homologies to other phages, we attempted to determine whether the IbrAB island could be moved from ECOR-9 to the test triple lysogen, CH6256, via infectious transfer. Repeated attempts to accomplish this were negative in spite of success in obtaining PFU from the ECOR-9 lysates (not shown). We surmise that the IbrAB island, although clearly containing phage-like genes, is defective for replication. Interestingly, the homologs of ibrA and ibrB of E. coli O157:H7 strains EDL933 and Sakai are also linked to phage genes but are not located within the genomes of replication-proficient phages (18, 36).
DISCUSSION
This report documents the cloning of overlapping genes, ibrA and ibrB, which together activate expression of otherwise silent eib genes that have been introduced into E. coli C either by lysogenization or, in the case of eibF, lambda InCh-mediated insertion. Both ibrA and ibrB were shown to be required for activation of eib expression, and additions of 4 or 8 nt that disrupted the reading frame of either gene near its midpoint abrogated activation. This result favors a requirement for protein rather than for RNA, since the frameshifting insertions would be expected to have a relatively small effect on RNA structure while having a profound effect on protein structure. Our data demonstrate a requirement for IbrB, but the requirement for IbrA remains open.
The copy number of ibrAB had a major effect on the amount of Eib produced. When ibrAB was present in multicopy in a given lysogen, the amount of Ig-binding activity observed was much greater than was observed when it was present in single copy (Fig. 3C and 4). The ibrAB element activated expression of all four eib genes, eibA, eibC, eibD, and eibE, from source strain ECOR-9 and of one, eibF, from a second strain, ECOR-2. In an effort to identify features that would provide insight into a potential common control mechanism, we have examined the sequence upstream from each eib coding sequence (40, 41). Comparison of the nucleotide sequence located between the translational start codon of each eib gene and the ORF immediately upstream revealed two distinct patterns. The leader sequences for genes eibA and eibF comprise 545 nt and have 98% identities (the A-F pattern). Genes eibC and eibE have identical 358-nt leaders, while the leader of eibD is identical to those of eibC and eibE except for having an insertion of 5 nt and 2-nt differences (the C-D-E pattern). Comparison of the A-F and the C-D-E patterns revealed only 63% identities over the 141 nt nearest the translational start. One invariant sequence was identified in all five leader regions: a 19-nt sequence (ATTTGTATACAGATAATTT) extending from −34 to −16 nt with respect to the translation initiation codon. In addition, each leader has a stem-loop structure centered between −110 and −120 nt from the translational start. The mechanism by which eib gene expression is silenced in the lysogens and the way that ibrAB abrogates silencing remain to be discovered. It should be emphasized that it has not been established whether ibrAB directly affects eib transcription or whether the effect is at some other level.
A search for similarities to families of protein domains showed that IbrA is a member of the phosphoadenosine phosphosulfate reductase family of proteins and that IbrB contains a ParB-like nuclease domain. ParB-like nuclease domains occur in the following DNA-binding bacterial proteins: transcriptional activator VirB of Shigella (2, 49), transcriptional repressor protein KORB of plasmid RK2 (25, 46), and numerous chromosome-partitioning proteins and replication proteins. The fact that IbrB belongs to a family of DNA-binding proteins is consistent with a role in regulating eib gene expression, possibly at the level of transcription.
A database search identified several proteins with amino acid sequence similarity to IbrA and IbrB (Table 3). The closest matches were proteins of unknown function encoded by genes of E. coli O157:H7 and Salmonella enterica serovar Typhi. The homologous proteins of the Sakai strain of E. coli O157:H7 are encoded by a prophage-like element, SpLE1 (18). The homologous proteins of the EDL933 strain of E. coli O157:H7 are encoded by genes in O islands no. 43 and 48 (36).
A search of the databases for similar nucleotide sequences was conducted using the 4.4-kb sequence of the IbrAB island as the query. This search produced hits within the attP site of bacteriophage P22 and several O islands of E. coli O157:H7 strains Sakai (18) and EDL933 (36). The homology is mosaic (occurring in six regions totaling 2,977 nt and ranging from 81 to 96% identities), as would be expected if multiple recombination events had occurred in the region.
There is much evidence from DNA sequence analysis for phage-mediated horizontal transfer of genes encoding virulence factors (reviewed in references 9, 12, 14, 26, 29, 31, 32, 34, and 43) and for recombination among phages (11, 19, 21, 23). Transduction of the prophage gene encoding Shiga toxin 1 from one strain of E. coli to another has been experimentally demonstrated in vivo in the mouse intestinal tract (1). Gene transfer among pathogens can contribute to the emergence of new pathogenic variants as new combinations of virulence factors are assembled in bacteria, and phages are potential key players in this process (16). ECOR-9 is considered to be a commensal strain, since it was originally derived from the feces of a healthy schoolchild (33). Yet this strain harbors considerable homology to virulence-associated genetic islands found in the genomes of pathogens such as E. coli O157:H7. To our knowledge the virulence of ECOR-9 has not been studied in an animal model. Although the Eib proteins from ECOR-9 are known to bind IgG from humans and from large animals, such as donkey and sheep, they do not bind detectable amounts of IgG from mouse. It is tempting to speculate that the genetic makeup of ECOR-9 reflects extensive reshuffling of phage-encoded genes, leading to increased fitness for a commensal lifestyle in the intestinal tract.
In summary, expression of distinct eib genes on four different prophages of ECOR-9, as well as expression of eib genes on prophages of ECOR-2, is activated by unlinked genes occurring elsewhere in the ECOR-9 genome on the IbrAB island. Although no evidence has been obtained to indicate that the IbrAB island is part of a replication-proficient prophage, genes found within it are similar to genes found in diverse phages. The interdependence of activities encoded by separate phages has emerged as an important component of microbial pathogenesis (reviewed in reference 9). A salient example is the pathogenicity of Vibrio cholerae, which depends on the cholera toxin phage (CTXΦ)-encoded cholera toxin, and the Vibrio pathogenicity island phage (VPIΦ)-encoded toxin-coregulated pilus, which functions both as a colonization factor and as the CTXΦ receptor (24). VPIΦ also encodes a transcription activator, ToxT (24), which directly activates expression of the cholera toxin (ctx) genes encoded by CTXΦ as part of a regulatory cascade (13, 50). Thus, V. cholerae strains that host VPIΦ and CTXΦ provide a precedent for the sort of prophage-prophage intedependence that we have observed in the activation of eib gene expression by IbrA and IbrB. Our results indicate that the extent of eib gene activation by IbrA and IbrB can vary over a wide range, resulting in large differences in Ig-binding activity (Fig. 2C). Consequently we speculate that expression of the ibrA and ibrB genes may be subject to additional regulation by factors yet to be discovered.
Acknowledgments
We thank Du Chungen for expert technical assistance.
This work was supported by Public Health Service grant GM16329 from the National Institutes of Health and an Innovative Biotechnology Seed Grant from the Pennsylvania State University Life Sciences Consortium.
REFERENCES
- 1.Acheson, D. W. K., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 3.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertani, G., and J. J. Weigle. 1953. Host controlled variation in bacterial viruses. J. Bacteriol. 65:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, E. F., B. M. Davis, and B. Hochhut. 2001. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9:137-144. [DOI] [PubMed] [Google Scholar]
- 10.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 12.Cheetham, B. F., and M. E. Katz. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201-208. [DOI] [PubMed] [Google Scholar]
- 13.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenberg, M. S., and T. S. Whittam. 2001. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Investig. 107:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 19.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Highton, P. J., Y. Chang, and R. J. Myers. 1990. Evidence for the exchange of segments between genomes during the evolution of lambdoid bacteriophages. Mol. Microbiol. 4:1329-1340. [DOI] [PubMed] [Google Scholar]
- 22.Hill, C. W., G. Feulner, M. S. Brody, S. Zhao, A. B. Sadosky, and C. H. Sandt. 1995. Correlation of Rhs elements with Escherichia coli population structure. Genetics 141:15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestad. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 24.Karaolis, D. K. R., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 25.Kornacki, J. A., P. J. Balderes, and D. H. Figurski. 1987. Nucleotide sequence of korB, a replication control gene of broad host-range plasmid RK2. J. Mol. Biol. 198:211-222. [DOI] [PubMed] [Google Scholar]
- 26.Levin, B. R., and C. T. Bergstrom. 2000. Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc. Natl. Acad. Sci. USA 97:6981-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 29.Miao, E. A., and S. I. Miller. 1999. Bacteriophages in the evolution of pathogen-host interactions. Proc. Natl. Acad. Sci. USA 96:9452-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto, H., W. Nakai, N. Yajima, A. Fujibayashi, T. Higuchi, K. Sato, and A. Matsushiro. 1999. Sequence analysis of Stx2-converting phage VT2-Sa shows a great divergence in early regulation and replication regions. DNA Res. 6:235-240. [DOI] [PubMed] [Google Scholar]
- 31.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 32.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 33.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 35.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pupo, G. M., D. K. R. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334:522-525. [DOI] [PubMed] [Google Scholar]
- 40.Sandt, C. H., and C. W. Hill. 2000. Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Escherichia coli. Infect. Immun. 68:2205-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandt, C. H., and C. W. Hill. 2001. Nonimmune binding of human immunoglobulin A (IgA) and IgG Fc by distinct sequence segments of the EibF cell surface protein of Escherichia coli. Infect. Immun. 69:7293-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandt, C. H., Y.-D. Wang, R. A. Wilson, and C. W. Hill. 1997. Escherichia coli strains with nonimmune immunoglobulin-binding activity. Infect. Immun. 65:4572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders, J. R., H. Allison, C. E. James, A. J. McCarthy, and R. Sharp. 2001. Phage-mediated transfer of virulence genes. J. Chem. Technol. Biotechnol. 76:662-666. [Google Scholar]
- 44.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 45.Tamm, A., A.-M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995-1011. [DOI] [PubMed] [Google Scholar]
- 46.Theophilus, B. D., and C. M. Thomas. 1987. Nucleotide sequence of the transcriptional repressor gene korB which plays a key role in regulation of the copy number of broad host range plasmid RK2. Nucleic Acids Res. 15:7443-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe, H., E. Arakawa, K.-I. Ito, J.-I. Kato, and A. Nakamura. 1990. Genetic analysis of an invasion region by use of a Tn 3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of InvE with ParB of plasmid P1. J. Bacteriol. 172:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, R. R., and V. J. DiRita. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J. Bacteriol. 181:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, S., C. H. Sandt, G. Feulner, D. A. Vlazny, J. A. Gray, and C. W. Hill. 1993. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J. Bacteriol. 175:2799-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]