Abstract
Bacteria possess amino acid export systems, and Corynebacterium glutamicum excretes l-isoleucine in a process dependent on the proton motive force. In order to identify the system responsible for l-isoleucine export, we have used transposon mutagenesis to isolate mutants of C. glutamicum sensitive to the peptide isoleucyl-isoleucine. In one such mutant, strong peptide sensitivity resulted from insertion into a gene designated brnF encoding a hydrophobic protein predicted to possess seven transmembrane spanning helices. brnE is located downstream of brnF and encodes a second hydrophobic protein with four putative membrane-spanning helices. A mutant deleted of both genes no longer exports l-isoleucine, whereas an overexpressing strain exports this amino acid at an increased rate. BrnF and BrnE together are also required for the export of l-leucine and l-valine. BrnFE is thus a two-component export permease specific for aliphatic hydrophobic amino acids. Upstream of brnFE and transcribed divergently is an Lrp-like regulatory gene required for active export. Searches for homologues of BrnFE show that this type of exporter is widespread in prokaryotes but lacking in eukaryotes and that both gene products which together comprise the members of a novel family, the LIV-E family, generally map together within a single operon. Comparisons of the BrnF and BrnE phylogenetic trees show that gene duplication events in the early bacterial lineage gave rise to multiple paralogues that have been retained in α-proteobacteria but not in other prokaryotes analyzed.
Traditionally, bacterial export carriers were thought to confer resistance to antibiotics or remove waste products. However, it has become clear more recently that carriers also export primary metabolites such as sugars and amino acids. Escherichia coli, for instance, has three similar exporters encoded by the setA, setB, and setC genes, two of which (setA and setB) export lactose and glucose (21). A distant E. coli paralogue, YdeE, catalyzes arabinose export (4). Another homologue, the Erwinia chrysanthemi sotB gene product, probably exports lactose (6). The physiologically relevant functions of these exporters are poorly understood. For example, a triple setABC mutant has no observable phenotype under the conditions examined (21, 22). Possibly, sugar exporters are involved in the detoxification of nonmetabolizable sugars (22).
Two transporters in E. coli are known to catalyze amino acid export. One is encoded by the rhtB gene, whose overexpression results in increased extracellular accumulation of l-homoserine (42). The other is encoded by ydeD. Upon its overexpression, the concentration of extracellular l-cysteine metabolites is augmented (8). Whereas the primary functions of these E. coli exporters are not obvious, the basic amino acid exporter LysE of Corynebacterium glutamicum can clearly protect the cell against toxic levels of cytoplasmic cationic amino acids (3, 39). The absence of LysE results in growth arrest at elevated intracellular l-lysine or l-arginine concentrations which occur during growth on complex medium or in the presence of basic amino acid-containing peptides. Expression of this exporter is strictly controlled by the intracellular concentration of the basic amino acids (3). Since LysE homologues are present in a large number of bacteria (40), one can anticipate that many bacteria will prove to exhibit this type of intracellular amino acid control. Indeed, amino acid export during growth of Lactococcus lactis on milk has been observed (16), although the exporters responsible for this activity have not yet been identified.
We have studied amino acid export in C. glutamicum and have identified the aforementioned basic amino acid exporter LysE (39) and the l-threonine exporter ThrE (34). Surprisingly, in both cases, new large families of translocator proteins of previously unknown function were identified (40, 41). Another amino acid of long-standing interest is l-isoleucine. This amino acid is excreted by C. glutamicum after deregulation of its biosynthetic pathway (10, 26). l-Isoleucine fluxes across the bacterial membrane are of particular interest from a mechanistic point of view since this amino acid is highly hydrophobic. In fact, net flux may result from diffusion and carrier-mediated export counteracting carrier-mediated import (44). The l-isoleucine importer, BrnQ, mediates Na+-dependent import of the three branched-chain amino acids (9, 35). The exporter, known to be driven by the proton motive force (14), has not been identified. We describe here the molecular identification of a translocator exporting l-isoleucine from the cell and show that this exporter represents the prototype of a novel two-gene-encoded translocator family widely distributed in bacteria and archaea.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The strains and plasmids used are listed in Table 1. Strains were grown as described previously (34). The minimal medium for C. glutamicum was usually CGXII (26), but to quantify amino acid export, the minimal medium MMI was used (14), with (NH4)2SO4 reduced to 0.5 g liter−1. When appropriate, carbenicillin (50 μg ml−1), kanamycin (KAN; 15, 25, or 50 μg ml−1), or chloramphenicol (20 μg ml−1) was added to the medium.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Reference or sourceb |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | endA1 supE44 recA1 gyrA96 relA1 deoR U169 φ80dlacZΔM15 mcrA Δ(mrr-hsdRMS-mcrBC) | 12 |
| GM2929 | dam-13::Tn9 dcm-6 hsdR2 recF143 mcrA mcrB | 27 |
| C. glutamicum | ||
| ATCC 13032 | Wild type | ATCC |
| ATCC 14752 | Wild type | ATCC |
| 13032ΔbrnFE | Wild type deleted of a 942-nt fragment of brnFE | This work |
| 13032Δlrp | Wild type deleted of a 409-nt fragment of lrp | This work |
| 13032::lrp | Wild type with lrp disrupted by pK18moblrpint | This work |
| Plasmids | ||
| pCGL0040 | Donor of Tn5531 (IS1207, Kmr), AproriVEc | U53587 |
| pET2 | Promoter-probe vector, Cmr | 38 |
| pZ1 | E. coli-C. glutamicum shuttle vector, KmroriVEc oriVCg | 23 |
| pJC1 | E. coli-C. glutamicum shuttle vector, KmroriVEc oriVCg | 7 |
| pK18mob | Integration vector, KmroriVEc oriT | 33 |
| pK19mobsacB | Integration vector, KmroriVEc oriT sacB | 33 |
| pK19mobsacBΔbrnFE | pK19mobsacB with a 942-nt internal fragment deleted in brnFE | This work |
| pK19mobsacBΔbrnF | pK19mobsacB with a 642-nt internal fragment deleted in brnF | This work |
| pK19mobsacBΔlrp | pK19mobsacB with a 409-nt internal fragment deleted in lrp | This work |
| pK18moblrpint | pK18mob with a 203-nt internal fragment of lrp | This work |
| pJC1brnFE | pJC1 with a 1.3-kb PCR fragment containing brnFE | This work |
| pJC1brnF | pJC1 with a 0.96-kb PCR fragment containing brnF | This work |
| pZ1brnE | pZ1 with a 1.2-kb EcoRI/XbaI fragment of pUC18brnE containing brnE | This work |
| pZ1lrp | pZ1 with a 0.7-kb EcoRI/XbaI fragment of pUC18lrp containing lrp | This work |
| pET2brnF-lrp | pET2 with a 219-bp BamHI PCR fragment containing the promoter of brnF and lrp | This work |
| pET2brnE | pET2 with a 239-bp BamHI-KpnI PCR fragment containing the promoter of brnE | This work |
| pUC18brnFE-lrp | pUC18 with a 1.8-kb PCR fragment containing brnFE-lrp | This work |
| pUC18brnE | pUC18 with a 1.2-kb PCR fragment containing the brnE gene | This work |
| pUC18lrp | pUC18 with a 0.65-kb PCR fragment containing the lrp gene | This work |
Kmr, kanamycin resistant; Apr, ampicillin resistant; Cmr, chloramphenicol resistant. Subscripts: Ec, E. coli; Cg, C. glutamicum.
ATCC, American Type Culture Collection.
Isolation of peptide-sensitive mutants and localization of transposon insertion sites.
The transposon (Tn) delivery vector pCGL0040 was isolated from E. coli GM2929 grown in the presence of 50 μg of KAN and 20 μg of chloramphenicol ml−1 (1). The plasmid was used to transform C. glutamicum ATCC 14752 to KAN resistance by using LBHIS plates with 15 μg of KAN ml−1 (19). The resulting Tn mutants were transferred to CGXII plates containing 25 μg of KAN ml−1 and either 3 mM of the dipeptide, Ile-Ile, or no peptide. The clones that exhibited retarded growth in the presence of Ile-Ile but grew normally in the absence of peptide were retrieved from the LBHIS master plate and retested in liquid CGXII medium. For that purpose, mutants were precultivated on brain heart infusion medium (Difco) containing 25 μg of KAN ml−1. Minimal medium cultures (CGXII ± 3 mM Ile-Ile, 25 μg of KAN ml−1) were inoculated at an initial optical density at 600 nm (OD600) of 0.1 or 0.5. Mutants that confirmed the growth delay in the presence of Ile-Ile were saved for further studies. The cloning and sequencing of the Tn insertion site was performed as described previously (1, 34).
Construction of plasmids.
Plasmids were constructed in E. coli DH5αMCR from PCR-generated fragments (Expand High Fidelity PCR kit; Roche Diagnostics) by using C. glutamicum ATCC 13032 DNA as a template. In order to construct pJC1brnFE, the upstream primer 5′-GCTCTAGAACCTTGTCAGCCAGTGCGAGAT-3′ and the downstream primer 5′-GCTCTAGAAAAAATCCGCATCCCCTTCA-3′ were used. The resulting fragment was XbaI digested and cloned into the XbaI site of pJC1. Using the same upstream primer as before and primer 5′-GCTCTAGAGCCCGGAGCGCAAAAGTAAT-3′, brnF was amplified. The fragment was cloned into the XbaI site of pJC1. brnE was amplified by using the primers 5′-ATTCATTCAAGCCTGGAGGTGTCG-3′ and 5′-AGCGCTGTCTGCTTAAGCCTTTTC-3′. The resulting fragment was blunted, cloned into the SmaI site of pUC18, reisolated as an EcoRI-XbaI fragment and, after blunting, ligated with ScaI-treated pZ1. Cloning of lrp was achieved by use of a 0.65-kb fragment obtained by PCR. The fragment was first ligated with pUC18, excised as an EcoRI-XbaI fragment, and then ligated with pZ1. For the cloning of the promoter regions of brnF-lrp and brnE, respectively, PCR fragments carrying the putative promoter regions were ligated with the promoter probe vector pET2.
To generate a chromosomal brnFE deletion, crossover PCR was applied (20). An 869-bp fragment carrying part of the 5′ end of brnF and the 3′ end of brnE with an internal 942-bp fragment deletion was generated (Fig. 2). The fragment was cloned into pK19mobsacB via its attached EcoRI and XbaI site. Similarly, pK19mobsacB derivatives were made with 642 bp of brnF deleted to allow an in-frame deletion of this gene, as was a derivative carrying a fragment with 409 bp of the lrp sequence deleted.
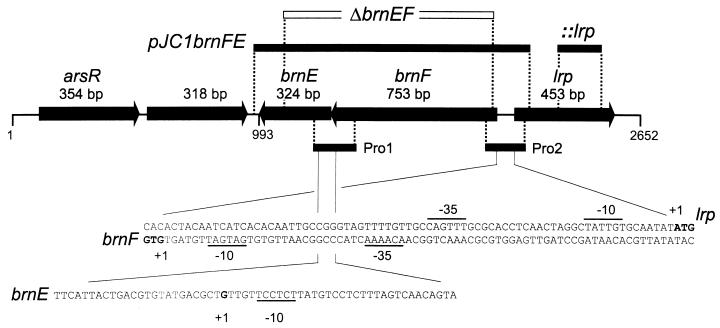
FIG. 2.
Overview on the brnE-brnF-lrp locus of C. glutamicum and regions with identified promoter activity. In the upper part of the figure, the thick arrows represent the genes with their sizes in base pairs. Above them, two fragments used for plasmid construction and lrp inactivation, respectively, are given, as well as the part of the genome deleted in the brnFE deletion mutant. In the lower part of the figure, the two fragments, Pro1 and Pro2, used for primer extension are depicted, together with the sequence of the promoters identified. The transcriptional start sites are marked “+1.” These are underlined, as are the −10 and −35 promoter regions. The initiation codons for lrp and brnF are indicated in boldface. The separate brnE-transcript initiation might not contribute for polypeptide formation (see the text).
Construction of strains.
C. glutamicum was transformed by electroporation (37), and the presence of replicative plasmids was verified by plasmid reisolation and restriction analysis. To obtain the lrp inactivation mutant, the nonreplicative plasmid pK18moblrpint was transferred to C. glutamicum. The correct integration of the vector into the chromosome was verified by PCR analysis. The brnFE deletion mutant and the lrp deletion mutant were made by using pK19mobsacBΔbrnFE and pK19mobsacBΔlrp, respectively. Clones were selected for KAN resistance to establish integration of the plasmid in the chromosome. In a second round of positive selection by using sucrose resistance, clones were selected for deletion of the vector (33). The deletions in the chromosome were verified by PCR analysis or Southern blotting.
Primer extension.
Total RNA was isolated from C. glutamicum by using extraction with hot acidic phenol (11). The transcriptional start sites were determined by primer extension by using SuperScript II reverse transcriptase (Gibco-BRL) with the primers labeled with [32P]ATP. In parallel, the respective DNAs (pET2pbrnFlrp and pET2brnE) were sequenced by using 32P-labeled primers and the Thermosequenase kit. The sequencing reactions and primer extension products were heated at 95°C for 4 min, and 2-μl samples were loaded onto a polyacrylamide gel.
Assay of amino acid export.
To determine the amino acid export rates, cells pregrown in brain heart infusion medium (Difco) were washed once with ice-cold 0.9% NaCl and transferred into MMI (14) to give an initial OD600 of 10, which corresponds to 3.0 mg (dry weight) ml−1. After preincubation for 20 min at 30°C with rapid stirring (ca. 700 rpm on a magnetic stirrer), the assay of amino acid excretion was initiated by addition of dipeptide in final concentrations as indicated in Results. Processing of samples for separation of extra- and intracellular fluid was performed by using silicone oil centrifugation (17). In the resulting fractions, amino acids were quantified as their o-phthaldialdehyde derivatives via high-pressure liquid chromatography. The intracellular volume used to calculate the internal amino acid concentration was 1.6 μl mg (dry weight)−1.
Calculations.
Observed efflux rates of branched-chain amino acids are due to diffusion, active export, and active import (44). Therefore, the carrier-mediated export rate is calculated as: Vex = Veff − Kd · [aa]in + Vin, where Veff is the observed efflux rate as provided in the graphs. Kd · [aa]in is the efflux due to diffusion, which is dependent on the determined internal amino acid concentration ([aa]in) in nanomoles per microliter. Due to the low external concentrations of the branched-chain amino acids accumulating in the experiments, these were neglected in the calculations. The known diffusion constants (Kd values) are 0.13 for l-Ile and l-Leu and 0.09 for l-Val in μl mg (dry weight)−1 min−1 (18, 24). Vin is the rate of uptake of the branched-chain amino acids. Uptake was invariably observed under all conditions assayed (9, 14, 35, 44). The uptake rates measured were 1.1, 0.94, and 1.3 nmol min−1mg (dry weight)−1 for l-Ile, l-Leu, and l-Val, respectively. The active export rates resulting from this approximation are expressed in nanomoles per minute per milligram (dry weight).
Computer methods.
The programs and computer methods used here were as described previously (41).
Nucleotide sequence accession number.
The sequence data have been submitted to the GenBank database under accession numbers AF454053 (brnFE-lrp), AF454055 (chl1), and AF454054 (htaA).
RESULTS
Isolation of peptide-sensitive mutants and characterization of Tn insertion loci.
After the observation that the growth of a lysE-null mutant of C. glutamicum unable to export l-lysine is sensitive to the presence of lysine-containing peptides (39) and, similarly, that the growth of a thrE mutant is sensitive to the presence of threonine-containing peptides (34), we screened 1,800 Tn mutants of C. glutamicum ATCC 14752 for sensitivity to 3 mM of the isoleucine dipeptide, Ile-Ile, on plates. This strategy was based on the knowledge that C. glutamicum can hydrolyze cytoplasmic Ile-Ile and that internally accumulated l-isoleucine is toxic (44). The screening procedure yielded three clones that were studied in liquid culture (Fig. 1A). Clone 1-75 showed weak growth retardation upon addition of Ile-Ile that was comparable to that of clone 1-25 (not shown), whereas clone 1-8 exhibited strong growth retardation. Cloning and analysis of the Tn adjacent sequences revealed that in clone 1-25 a polypeptide encoding a putative heme transporter, htaA, was disrupted, and that in clone 1-75 the Tn was inserted into the promoter region of a putative magnesium chelatase subunit, chl1. Although the only currently known function of this latter gene is to incorporate Mg2+ into the protoporphyrin skeleton of photosynthetic organisms, attempted inactivation in C. glutamicum was not possible. From this observation we suggest that Mg2+ incorporation into an unidentified cellular component is essential for the growth of C. glutamicum.
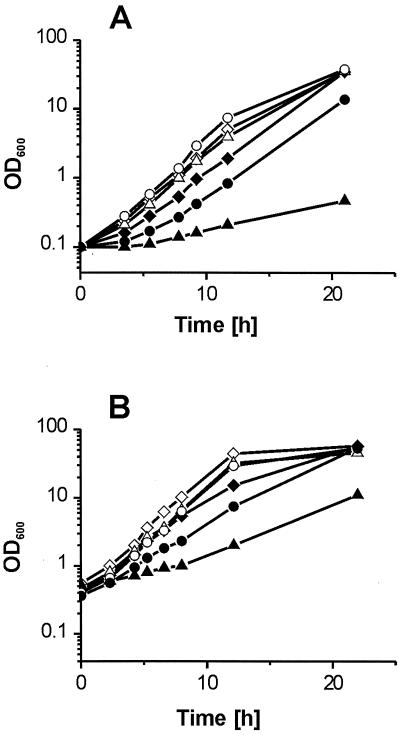
FIG. 1.
Peptide-sensitive growth of Tn mutants. (A) Growth of Tn mutants 1-75 (circles) and 1-8 (triangles) compared to the control strain, C. glutamicum ATCC 14752/pZ1(diamonds), in the presence (solid symbols) or absence (open symbols) of 3 mM Ile-Ile. (B) Growth of the deletion mutant 13032ΔbrnFE with control vector pJC1 (triangles) or pJC1brnFE (circles) compared to the wild-type 13032/pZ1 (diamonds) in the presence (solid symbols) or absence (open symbols) of 5 mM Ile-Ile.
In clone 1-8, with the strongest peptide sensitivity, the Tn was inserted into a gene whose homologue in Bacillus subtilis is azlC and which when deleted together with azlD renders B. subtilis more susceptible to azaleucine (2). The Tn-inactivated gene in C. glutamicum ATCC 13032 is 756 nucleotides (nt) in size and encodes a polypeptide with a molecular weight of 27,300 (Fig. 2). It overlaps by 4 bp the downstream gene of 327 nt that is predicted to encode a polypeptide of 11,500 Da. In accordance with the subsequent functional analyses, the two corresponding genes in C. glutamicum were termed brnF and brnE for branched-chain amino acid export. Both polypeptides exhibit pronounced local hydrophobicities (see below). Divergently transcribed from brnFE is a putative regulatory gene that encodes a protein exhibiting 37% identity to Lrp of E. coli (13).
Deletion of brnFE and growth characteristics.
With the objective of deleting the chromosomal brnFE locus, the mobilizable replacement vector pK19mobsacBΔbrnFE was constructed carrying sequences flanking brnFE (Fig. 2). It was transferred into C. glutamicum ATCC 13032, and two rounds of positive selection were applied to integrate and subsequently remove the vector (33). PCR analysis of 10 clones revealed that in 3 of the clones brnFE was in fact deleted, whereas in 7 clones the wild-type locus was reconstituted. These numbers indicate that these genes may not play a major housekeeping function for cellular metabolism or growth of C. glutamicum. Growth of the deletion mutant in liquid culture was indistinguishable from that of the wild type (Fig. 1B), thus confirming this conclusion. Only when the Ile-Ile dipeptide was present in the medium was growth reduced. When the deletion mutant was transformed with pJCbrnFE, the strain regained its peptide-resistant growth phenotype (Fig. 1B). This effect was not observed when brnF or brnE was individually overexpressed, providing evidence that both genes together promote resistance to the peptide (not shown). In view of the fact that deletion of the brnFE homologues, azlCD, abolishes azaleucine resistance in B. subtilis (2), we also assayed growth of the recombinant C. glutamicum strains in response to azaleucine. Although the growth rate of the wild-type strain was reduced from 0.34 to 0.15 h−1 in the presence of 0.5 mM azaleucine, deletion or overexpression of brnFE did not influence growth either with or without azaleucine (not shown).
Determination of the transcriptional start sites of brnF, brnE, and lrp.
To define the genes, transcript initiation sites were measured. For this purpose a 219-bp BamHI fragment carrying the divergent promoter regions of lrp and brnF (Pro2 in Fig. 2) was cloned in the two possible orientations into the promoter probe vector pET2 (38). Similarly, a 239-bp BamHI-KpnI fragment (Pro1) carrying a possible start site of brnE was cloned. The resulting plasmids rendered C. glutamicum resistant to chloramphenicol. The MICs caused by the brnF and lrp promoters were 60 and 50 μg ml−1, respectively; that of the brnE promoter was 25 μg ml−1.
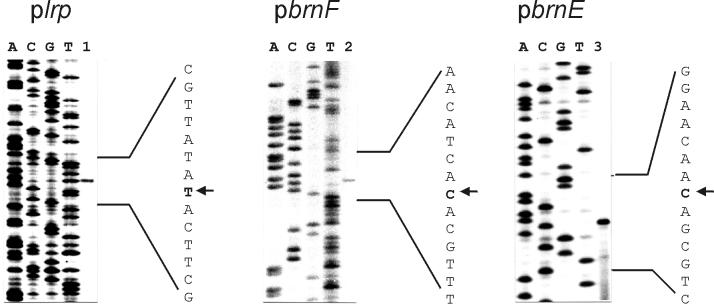
The results of the primer extension experiments with the sequencing reactions carried out in parallel are shown in Fig. 3. In the case of brnF and lrp, the transcript initiation sites are identical to those of their deduced polypeptides (Fig. 2). Such leaderless genes are a frequent feature of C. glutamicum and related organisms (25). The close proximity of the genes separated by just 74 nt, as well as their overlapping promoters, indicates that brnF expression might be a target of the lrp-encoded regulator (see below). By using fragment Pro1, an additional transcript initiation site was observed within brnE (Fig. 2). However, the corresponding “−10” consensus sequence is located 2 nt closer to the determined transcript initiation site than in most other C. glutamicum promoters (28). This fact and the low MIC make it unlikely that this site contributes to transcript formation. Instead, we assume that there is a single message for both brnF and brnE.
FIG. 3.
Mapping of the transcriptional start sites of lrp (plrp), brnF (pbrnF), and brnE (pbrnE) by primer extension analysis. In each experiment, the primer extension product was run in the numbered lane. The sequencing ladder (labeled A, C, G, and T) of the coding strand was generated by using the same primer that was used for primer extension. The respective transcriptional start site is indicated by the arrow.
Increased l-isoleucine export is due to brnFE overexpression.
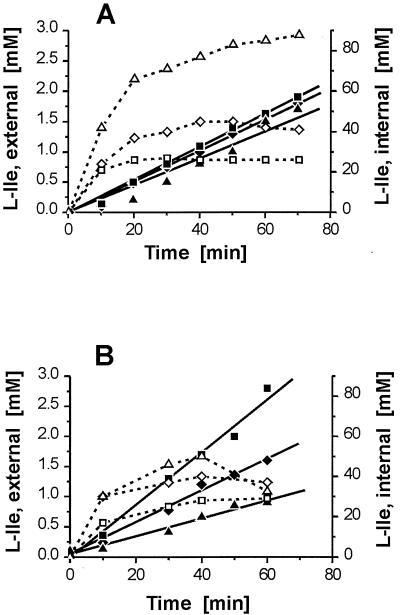
In order to perform functional analyses of brnFE, we measured export rates in the presence of Ile-Ile. Addition of this peptide leads to increased intracellular l-Ile steady-state levels (44). The actual cytoplasmic concentrations are determined by the rates of import and hydrolysis of the peptide, as well as the l-Ile efflux rate. Upon addition of 3 mM Ile-Ile, the internal l-Ile concentration increased up to 23 mM in strain 13032/pJCbrnFE, up to 45 mM in strain 13032/pZ1, and up to 87 mM in strain 13032ΔbrnFE/pJC1 (Fig. 4A).
FIG. 4.
Internal l-isoleucine concentrations and isoleucine efflux after brnFE expression. (A) Efflux (solid symbols) and internal l-isoleucine concentrations (open symbols) upon addition of 3 mM Ile-Ile to 13032/pJC1brnFE (squares) and 13032ΔbrnFE/pJC1 (triangles) compared to the wild-type strain 13032/pZ1 (diamonds). (B) Same as panel A, except that 10 mM Ile-Ile was used with strain 13032/pJC1brnFE and 0.5 mM Ile-Ile was used with strain 13032ΔbrnFE/pJC1.
The rates of l-isoleucine efflux from these strains are fairly comparable, which may at first seem surprising. However, this is due to the significant l-Ile efflux component caused by diffusion (44). This carrier-independent flux component is determined by the concentration gradient across the plasma membrane. Based on the known diffusion constant, at the high internal concentration of 87 mM in the deletion mutant, efflux due to diffusion is 6.4 nmol min−1 mg (dry weight)−1. At the lower concentration of 23 mM in the overexpressing strain, it is only 2.6 nmol min−1 mg (dry weight)−1. The l-isoleucine uptake rate of 1.1 nmol min−1 mg (dry weight)−1 detracts from the apparent l-isoleucine efflux rate (9, 35). Accordingly, the active export rate is the difference of the total efflux minus diffusion plus uptake. When we calculated the active export rates (see Materials and Methods), values of 7.8 nmol min−1 mg (dry weight)−1 for the overexpressing strain, 0.4 nmol min−1 mg (dry weight)−1 for the deletion mutant, and 4.2 nmol min−1 mg (dry weight)−1 for the wild type were derived (Table 2). Upon overexpression of either brnF or brnE, active export was not increased (Table 2), in accordance with the failure of each of these genes singly to relieve peptide-sensitive growth. Also, with a brnF in-frame deletion, no export was obtained (Table 2).
TABLE 2.
Export rates due to brnF, brnE, and lrp expression
| C. glutamicum strain | Export rate (nmol min−1 mg [dry wt]−1)a of:
|
||
|---|---|---|---|
| l-Isoleucine | l-Valine | l-Leucine | |
| 13032/pJCbrnFE | 7.8 ± 1.3a | 3.3, 3.2 | 6.3 |
| 13032ΔbrnFE/pJC1 | 0.4 ± 0.2 | −0.1 ± 0.9 | 0.6 ± 0.4 |
| 13032ΔbrnF/pJC1 | −0.9 ± 0.6 | ND | ND |
| 13032/pZ1 | 4.2 ± 0.4 | 2.4, 2.0 | 4.0 |
| 13032ΔbrnFE/pJCbrnFE | 6.9 | 4.5 | 8.1 |
| 13032/pJC1brnF | 002d>4.6 | ND | ND |
| 13032/pJC1brnE | 3.7 | ND | ND |
| 13032ΔbrnFE/pJC1brnE | 0.5, 0.3 | ND | ND |
| 13032ΔbrnFE/pJC1brnF | 0.4, 0.2 | ND | ND |
| 13032/pZ1lrp | 3.6 | ND | ND |
| 13032Δlrp | 0.0, −0.7 | ND | ND |
| 13032lrp::pK18moblrpint | 0.7 | ND | ND |
Values for the calculation of standard deviations (provided where applicable) were derived from three independent measurements. ND, not done.
In separate experiments, 10 mM Ile-Ile was used with the overexpressing strain and 0.5 mM Ile-Ile was used with the deletion mutant in order to achieve more comparable internal concentrations of l-Ile (Fig. 4B). Total efflux proved to be highest with pJC1brnFE and lowest with the deletion mutant. Nevertheless, the calculated export rates were almost identical to those determined in the experiment described earlier (Table 2). These experiments (i) confirm that l-isoleucine export is due to brnFE expression, (ii) suggest that both genes are required for export, and (iii) make it unlikely that an additional carrier contributes appreciably to l-isoleucine export. In C. glutamicum, under identical conditions but without peptide, the internal concentrations of the branched-chain amino acids do not exceed 2 mM (not shown).
l-Val and l-Leu export.
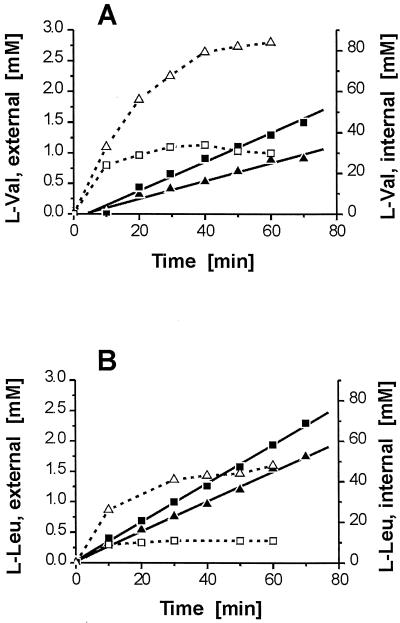
As previously reported, loading C. glutamicum from the external side with l-Leu or l-Val resulted in reduced l-Ile efflux by up to 25% (14). We used the identified exporter and peptides to assay more directly whether BrnFE accepts the other branched-chain amino acids. With 5 mM Ala-Val in the deletion mutant, up to 82 mM l-Val accumulated, and in the deletion mutant with pJC1brnFE, ca. 30 mM l-Val accumulated (Fig. 5A). The total l-Val efflux rate by the deletion mutant was substantially lower than that by the overexpressing strain. This may be due to the known low membrane diffusibility of l-Val (24), as becomes evident from comparison of the l-Ile and l-Val efflux rates in the deletion mutant under otherwise identical conditions (compare Fig. 4A with Fig. 5A). Using a diffusion rate constant of 0.09 μl mg (dry weight)−1 min−1 (18, 24), the calculated export rates for l-Val are −0.1 nmol min−1 mg (dry weight)−1 with the deletion mutant and 4.5 nmol min−1 mg (dry weight)−1 with the same strain but overexpressing brnFE (Table 2). In addition, with the wild-type strain carrying pJC1brnFE, the l-Val export rate proved to be 3.3 nmol min−1 mg (dry weight)−1. This confirms the conclusion that l-Val export is mediated by the brnFE-encoded exporter. This rate is determined to be ca. 60% of that of l-Ile export.
FIG. 5.
Efflux (solid symbols) and internal concentrations (open symbols) of l-valine and l-leucine by C. glutamicum ATCC 13032ΔbrnFE/pJC1 (triangles) and ATCC 13032ΔbrnFE/pJC1brnFE (squares). (A) Response in the presence of 5 mM Ala-Val. (B) Response in the presence of 5 mM Leu-Leu.
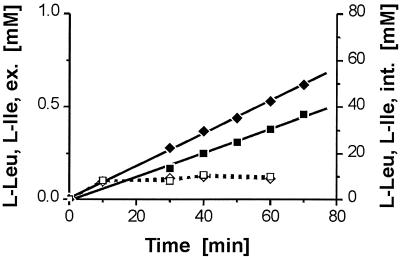
Interestingly, with Leu-containing peptides (Leu-Leu or Ala-Leu) in the brnFE-overexpressing strain, the internal l-Leu concentration never exceeded 15 mM. Nevertheless, with 5 mM Leu-Leu, a high efflux rate (8.6 nmol min−1 mg [dry weight]−1) resulted with 13032ΔbrnFE/pJC1brnFE (Fig. 5B). Thus, from the calculated export rate (Table 2), we conclude that l-Leu and l-Ile are exported at comparable rates. For further confirmation, we conducted a competition experiment in which both amino acids were simultaneously present as a substrate of the exporter. The dipeptide, Leu-Ile, was used for this purpose (Fig. 6). In spite of a comparatively low concentration of ca. 10 mM for both amino acids, which could be due to limited peptide uptake, comparable export rates were observed for l-Ile and l-Leu of 2.5 and 1.8 nmol min−1 mg (dry weight)−1, respectively. This proved not to be true for the deletion mutant (data not shown). This finding shows that the exporter does not discriminate between the two hydrophobic amino acids. In contrast to the experiment shown in Fig. 4, a low l-isoleucine export rate suggests that the carrier is not saturated at the low internal concentrations present. This is in accordance with the observed low affinity of the system (observable Km, 21 mM [44]). It suggests a comparable affinity of the system for these two amino acids.
FIG. 6.
Efflux (solid symbols) and internal concentrations (open symbols) of l-leucine (squares) and l-isoleucine (diamonds) with C. glutamicum ATCC 13032 after the addition of 10 mM Leu-Ile.
Involvement of lrp in export.
lrp maps upstream of brnFE (Fig. 2). To reveal whether lrp is involved in regulation of the exporter, we constructed an lrp disruption mutant. Strain 13032lrp::pK18moblrpint no longer proved to export l-isoleucine (Table 2). An additionally used lrp deletion mutant confirmed this result, indicating that Lrp is essential for the export of the branched-chain amino acids, probably by acting as an activator of brnFE expression.
Homologues of BrnFE in other organisms.
Table 3 lists homologues of BrnF (left) and BrnE (right) retrieved from the databases. These proteins are derived exclusively from prokaryotic organisms, with gram-negative and gram-positive bacteria as well as archaea being represented. Most organisms with fully sequenced genomes that are represented in Table 3 have just one pair of these proteins. However, the closely related α-proteobacteria Agrobacterium tumefaciens, Sinorhizobium meliloti, and Mesorhizobium loti have multiple paralogues. The two Helicobacter pylori pairs of homologues are orthologues derived from two different strains of this organism. It can be seen from the data summarized in Table 3 that most BrnF homologues are of 218 to 256 amino acids (aa) long, whereas all BrnE homologues are 98 to 118 aa. However, the Methanococcus jannaschii BrnF homologue, as well as the Neisseria meningitidis BrnF homologue, is truncated, and no BrnE homologue could be identified in the two strains of the latter organism that have been sequenced.
TABLE 3.
Sequenced proteins of the branched-chain amino acid exporter (LIV-E) familya
| Protein abbreviation | Organism | BrnF homologues
|
BrnE homologues
|
||
|---|---|---|---|---|---|
| Size (aa) | GI no. | Size (aa) | GI no. | ||
| Atu1 | Agrobacterium tumefaciens | 240 | 15157197 | 104 | 15157198 |
| Atu2 | Agrobacterium tumefaciens | 234 | 15156113 | 100 | 15156114 |
| Atu3 | Agrobacterium tumefaciens | 275 | 15156429 | 114 | 15156430 |
| Afu | Archaeoglobus fulgidus | 219 | 11499344 | 100 | 11499343 |
| Bha | Bacillus halodurans | 237 | 10175532 | 102 | 10175533 |
| Bsu | Bacillus subtilis | 254 | 3121821 | 110 | 3121820 |
| Cgl | Corynebacterium glutamicum | 251 | 14273894 | 108 | 14273895 |
| Dra | Deinococcus radiodurans | 235 | 7471172 | 98 | 7473007 |
| Dgi | Desulfovibrio gigasb | 235 | 12802727 | 103 | |
| Eco | Escherichia coli | 245 | 3123142 | 111 | 2506710 |
| Hin | Haemophilus influenzae | 244 | 1176150 | 109 | 1176149 |
| Hpy-A | Helicobacter pylori (J99) | 228 | 12230844 | 118 | 11387375 |
| Hpy-B | Helicobacter pylori | 228 | 3123102 | 115 | 3123101 |
| Lla | Lactococcus lactis | 235 | 12724620 | 108 | 12724619 |
| Mlo1 | Mesorhizobium loti | 242 | 13471536 | 98 | 13471535 |
| Mlo2 | Mesorhizobium loti | 244 | 13472209 | 99 | 13472208 |
| Mja | Methanococcus jannaschiic | 73 | 3219930 | 110 | 3219926 |
| Nme | Neisseria meningitidisd | 160 | 11353336 | ||
| Pmu | Pasteurella multocida | 240 | 12720674 | 109 | 12720675 |
| Pch | Pectobacterium chrysanthemi | 247 | 14970542 | 113 | 14970543 |
| Pae | Pseudomonas aeruginosa | 252 | 11349405 | 104 | 11349404 |
| Rsp | Rhodobacter sphaeroidese | 235 | 7657933 | ||
| Sme1 | Sinorhizobium meliloti | 238 | 15075182 | 102 | 15075181 |
| Sme2 | Sinorhizobium meliloti | 256 | 15074061 | 100 | 15074062 |
| Sme3 | Sinorhizobium meliloti | 248 | 15074421 | 111 | 15074422 |
| Sau | Staphylococcus aureus | 231 | 13699927 | 109 | 13699928 |
| Spn | Streptococcus pneumoniae | 218 | 14971611 | 107 | 14971612 |
| Sco | Streptomyces coelicolor | 256 | 7479559 | 102 | 7479560 |
| Vch | Vibrio cholerae | 239 | 11354432 | 105 | 11355554 |
GI, GenInfo Identifier (GenBank).
The open reading frame for this organism is not included in the current databases and was derived from the nucleic acid sequence.
The small size of the M. jannaschii BrnF homologue is due to an apparent genomic deletion. If this is a natural deletion rather than a sequencing error, the M. jannaschii homologues would be expected to be nonfunctional.
Two strains of N. meningitidis, serogroup A strain Z2491 and serogroup B strain MC58, exhibit apparently truncated BrnF homologues but no identifiable BrnE homologue. If this is due to a natural genomic deletion that occurred prior to the divergence of these two strains, these homologues would be expected to be nonfunctional pseudogenes.
No BrnE homologue could be identified for R. sphaeroides, although a full-length BrnF homologue was identified. However, the fully sequenced genome for this organism was not available at the time these studies were conducted.
Phylogenetic tree of the LIV-E family.
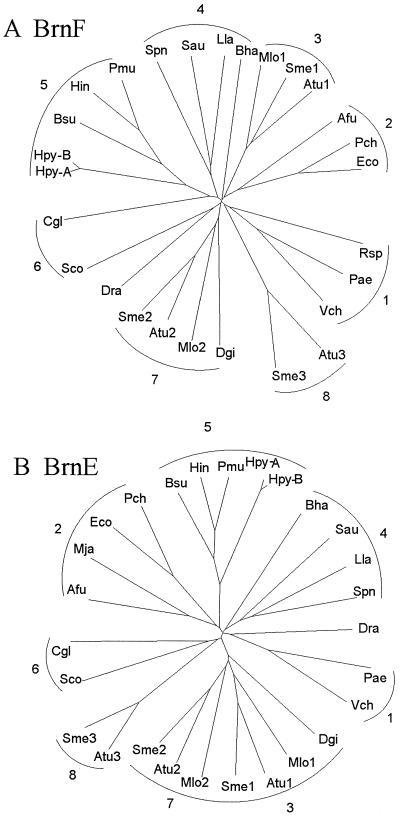
The phylogenetic trees for the BrnF and BrnE homologues are shown in Fig. 7A and B, respectively. Several features are worthy of note. First, there are two sets of orthologous α-proteobacterial proteins (Atu1, Sme1, and Mlo1; Atu2, Sme2, and Mlo2) which cluster loosely together with the Desulfovibrio gigas protein. The third set of the Atu3 and Sme3 paralogues cluster together, but they are clearly distant from the other orthologous proteins. It seems clear that the gene duplication events giving rise to these paralogues occurred early, before divergence of the species. Second, most of the gram-positive bacterial homologues cluster loosely together. In Fig. 7A, they fall into three clusters: one including the high G+C gram-positive bacterial proteins, Sco and Cgl; one including the low G+C gram-positive bacterial proteins, Spn, Lla, and Sau; and a third bearing only the Bacillus halodurans (Bha) protein. The single B. subtilis (Bsu) homologue does not cluster with the B. halodurans protein but instead clusters with the γ-proteobacterial proteins, Hin and Pmu, on both trees. The Bsu protein may have been acquired by horizontal transfer. Analyses of the genes encoding the B. subtilis homologues revealed significant (4%) differences in G+C content, and codon frequencies relative to these values in B. subtilis genes as a whole were also noticeably different. Finally, the γ-proteobacterial homologues (Pmu, Hin, Pca, Pae, Eco, and Vch) fall into three distant clusters on both trees, suggesting that at least three early gene duplication events occurred during evolution of the γ-proteobacterial lineage, as was evidently true for the α-proteobacteria. However, in contrast to the α-proteobacteria, with which all three paralogues were retained, at least two of the three were lost from each γ-proteobacterium.
FIG. 7.
Phylogenetic trees for the BrnF family (A) and the BrnE family (B). Protein abbreviations are as defined in Table 3. The trees were based on multiple alignments generated by using the CLUSTAL X program (36).
In silico topological analyses of BrnF and BrnE homologues.
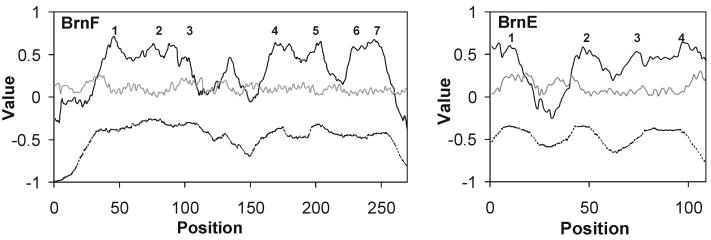
Figure 8 shows average hydropathy, amphipathicity, and similarity plots for the full-length BrnF and BrnE homologues listed in Table 3. The programs predict the presence of seven TMSs in BrnF homologues and four transmembrane spanners (TMSs) in BrnE homologues. The larger BrnF homologues are clearly better conserved than the smaller BrnE homologues.
FIG. 8.
Average hydropathy (solid line), amphipathicity (shaded line), and similarity (dashed line) for the BrnF and BrnE families. The plots were generated by using the AveHAS program (43), based on multiple alignments generated with the CLUSTAL X program.
The similar spacing of the last four BrnF peaks and the BrnE peaks suggested that these sequence dissimilar proteins arose from a common ancestor. We therefore examined BrnF homologues for regions of sequence similarity with BrnE. The last four putative TMSs in BrnF Mlo2, for example, gave a comparison score of four standard deviations with the corresponding BrnE protein, corresponding to a probability of ca. 10−5 that the sequence similarity observed occurred by chance. While suggestive of homology, this value is insufficient to establish common origin (32). We believe that the BrnFE system may have arisen by a gene triplication event from a four-TMS precursor, followed by loss of a single N-terminal TMS and extensive sequence divergence.
DISCUSSION
LysE was the first amino acid exporter characterized from C. glutamicum (39), and the use of peptides has recently resulted in the identification of the second such exporter, ThrE (34). C. glutamicum BrnFE, described here, represents the third amino acid exporter to be identified. In each of these cases, the discovery of a new large family of bacterial proteins resulted (39, 41). We suggest that all members of these novel families catalyze amino acid export, a function of general relevance that has previously been neglected.
BrnFE exports the three branched-chain amino acids, l-isoleucine, l-leucine, and l-valine, at comparable rates, although it appears that l-valine is exported somewhat less efficiently than is l-leucine or l-isoleucine. One other exporter of the LIV-E family, for which limited functional data are available, is from B. subtilis (2). In this case, inactivation of the BrnE homologue (AzlD) abolishes resistance to azaleucine. It can be surmised that AzlD functions as an azaleucine exporter. Based on the available functional data, as well as on the similar structures of all family members, it can be suggested that export of hydrophobic amino acids is a characteristic of all members of the LIV-E family. However, it is not clear whether the export of amino acids observed with BrnFE is its primary physiological function or, alternatively, if there is an unrecognized function for this translocator. An argument in favor of the latter idea is that the B. subtilis genes encoding the BrnFE homologues are cotranscribed with brnQ, encoding the uptake system for branched-chain amino acids. Coexpression of these genes would be expected to create an energy-consuming futile cycle unless strictly regulated. Nevertheless, for production of l-isoleucine when the biosynthetic pathway is deregulated (10), BrnFE might significantly contribute to the efflux of this amino acid.
Based on the export properties of the C. glutamicum brnFE mutants, together with the phylogenetic data reported, we suggest that the two proteins of the system function together as an obligatory pair. The LIV-E family apparently consists of many two-component permeases where one component is predicted to have four TMSs (BrnE) and the other is predicted to have seven TMSs (BrnF), with the latter being better conserved than the former (Fig. 8). Almost all secondary carriers consist of systems with 10 to 14 TMSs, and these can exist at either one or two gene products (15, 29). In the latter case, a homo- or hetero-oligomeric system is presumed to be the active species (5). The BrnFE exporters appear to be no exception when heterodimeric structures are likely to be the active species. This uniform topological characteristic of secondary carriers may reflect the evolutionary process whereby simple channel-type proteins gave rise to carriers, often via intragenic duplication events (31). The variations on this theme, however, are remarkable (30, 32), and BrnFE, with a uniform four plus seven TMSs and two-component heterodimeric topology, represents a unique example. This suggests that the LIV-E family may exhibit unusual mechanistic features among secondary carriers.
The phylogenetic data showed that, except for α-proteobacteria, no prokaryotic organism examined has more than one member of the LIV-E family. However, the results showed that these proteins in the different bacteria are not always orthologues. Instead, early gene duplication events evidently gave rise to multiple paralogues in a primordial bacterium. These paralogues were apparently transmitted faithfully to present-day organisms only in the case of A. tumefaciens and S. meliloti, for which three paralogous systems remain. In Mesorhizobium loti, two paralogues are still present. Many other prokaryotes with completely sequenced genomes lack homologues. In a few cases, only the BrnF homologue or a truncated version of this protein was identified (see Table 3). We suggest that in these organisms these systems are inactive and may be in the process of being genetically deleted from the genomes.
In at least one organism, B. subtilis, it appears that a LIV-E family transporter was acquired by a fairly recent horizontal gene transfer event. The B. subtilis homologues cluster with those of the α-proteobacteria instead of with the low G+C gram-positive organisms, including B. halodurans (Fig. 7). Further indication of horizontal transfer of brnFE homologues results from the clustering of the encoding genes with other resistance genes in several organisms (unpublished observations). Thus, a chloramphenicol transporter maps adjacent to the E. coli brnFE genes; erythromycin resistance genes are near the brnFE genes in both Yersinia pestis and E. coli; a cadmium/zinc/cobalt resistance gene is adjacent to such genes in H. pylori, and an arsenate resistance gene is found adjacent to brnFE in C. glutamicum (Fig. 2).
Since the loss of paralogues in most proteobacteria appears to have been random (see Fig. 7), we suggest that the paralogous proteins have retained their ancestral function with little variation in substrate specificity. The differences between paralogues in the α-proteobacteria may reflect dissimilar regulatory characteristics. The fact that the phylogeny of LIV-E family members does not correlate with that for the organismal 16S rRNAs suggests that these carriers do not play roles as essential “housekeeping” proteins such as biosynthesis of essential cellular constituents or degradation of energy sources. In agreement with this suggestion is the fact that the LIV-E family carriers in both C. glutamicum and B. subtilis are apparently nonessential (2).
Acknowledgments
We thank M. Wessel for the lrp deletion mutant, and DEGUSSA AG for providing genome information of C. glutamicum. We also thank Mary Beth Hiller and Annelie Förstel for their assistance in the preparation of the manuscript. Salar Partovi and Yu-Feng Zhai provided assistance with the computer analyses.
This work was supported by NIH grants GM64368 and GM55434 to M. H. Saier, Jr.; grant 525/01/0916 from the Grant Agency of the Czech Republic to M. Pátek; and a grant from the Bundesland Nordrhein-Westfalen, Germany, to H. Sahm.
REFERENCES
- 1.Ankri, S., I. Serebrijski, O. Reyes, and G. Leblon. 1996. Mutations in the Corynebacterium glutamicum proline biosynthetic pathway: a natural bypass of the proA step. J. Bacteriol. 178:4412-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., M. C. Gustafsson, A. L. Sonenshein, and C. von Wachenfeldt. 1997. An Lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 179:5548-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellmann, A., M. Vrljic, M. Pátek, H. Sahm, R. Krämer, and L. Eggeling. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765-1774. [DOI] [PubMed] [Google Scholar]
- 4.Bost, S., F. Silva, and D. Belin. 1999. Transcriptional activation of ydeA, which encodes a member of the major facilitator superfamily, interferes with arabinose accumulation and induction of the Escherichia coli arabinose P-BAD promoter. J. Bacteriol. 181:2185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, Y. J., and M. H. Saier, Jr. 2001. SMR-type multidrug resistance pumps. Curr. Opin. Drug Dis. Dev. 4:237-245. [PubMed] [Google Scholar]
- 6.Condemine, G. 2000. Characterization of SotA and SotB, two Erwinia chrysanthemi proteins which modify isopropyl-β-d-thiogalactopyranoside and lactose induction of the Escherichia coli lac promoter. J. Bacteriol. 182:1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cremer, J., L. Eggeling, and H. Sahm. 1990. Cloning of the dapA dapB cluster of the lysine-secreting bacterium Corynebacterium glutamicum. Mol. Gen. Genet. 220:478-480. [Google Scholar]
- 8.Dassler, T., T. Maier, C. Winterhalter, and A. Böck. 2000. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 36:1101-1112. [DOI] [PubMed] [Google Scholar]
- 9.Ebbighausen, H., B. Weil, and R. Krämer. 1989. Transport of branched-chain amino acids in Corynebacterium glutamicum. Arch. Microbiol. 151:238-244. [DOI] [PubMed] [Google Scholar]
- 10.Eggeling, L., S. Morbach, and H. Sahm. 1997. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J. Biotechnol. 56:167-182. [Google Scholar]
- 11.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. Lüdtke, and H. Sahm. 1994. Nucleotid sequence, expression and transcriptional analysis of gltA gene encoding citrate synthase. J. Gen. Microbiol. 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 12.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haney, S. A., J. Platko, D. L. Oxender, and J. M. Calvo. 1992. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J. Bacteriol. 174:108-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann, T., and R. Krämer. 1996. Mechanism and regulation of isoleucine excretion in Corynebacterium glutamicum. Appl. Environ. Microbiol. 62:3238-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack, D. L., N. M. Yang, and M. H. Saier, Jr. 2001. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268:3620-3639. [DOI] [PubMed] [Google Scholar]
- 16.Juillard, V., D. LeBars, E. R. S. Kunji, W. N. Konings, J. Gripon, and J. Richard. 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth on milk. Appl. Environ. Microbiol. 61:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klingenberg, M., and E. Pfaff. 1977. Means of terminating reactions. Methods Enzymol. 10:680-684. [Google Scholar]
- 18.Krämer, R. 1994. Secretion of amino acids by bacteria: physiology and mechanism. FEMS Microbiol. Rev. 13:75-94. [Google Scholar]
- 19.Liebl, W., A. Bayerl, U. Stillner, and K. H. Schleifer. 1989. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol. Lett. 65:299-304. [DOI] [PubMed] [Google Scholar]
- 20.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, J. Y., P. F. Miller, M. Gosink, and E. R. Olson. 1999. The identification of a new family of sugar efflux pumps in Escherichia coli. Mol. Microbiol. 31:1845-1851. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J. Y., P. F. Miller, J. Willard, and E. R. Olson. 1999. Functional and biochemical characterization of Escherichia coli sugar efflux pumps. J. Biol. Chem. 274:22977-22984. [DOI] [PubMed] [Google Scholar]
- 23.Menkel, E., G. Thierbach, L. Eggeling, and H. Sahm. 1989. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ. Microbiol. 55:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milner, J. L., B. Vink, and J. M. Wood. 1987. Transmembrane amino acid flux in bacterial cells. CRC Crit. Rev. Biotechnol. 5:1-48. [DOI] [PubMed] [Google Scholar]
- 25.Morbach, S., C. Junger, H. Sahm, and L. Eggeling. 2000. Attenuation control of ilvBNC in Corynebacterium glutamicum: evidence of leader peptide formation without the presence of a ribosome binding site. J. Biosci. Bioeng. 90:501-507. [DOI] [PubMed] [Google Scholar]
- 26.Morbach, S., H. Sahm, and L. Eggeling. 1996. l-Isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl. Environ. Microbiol. 62:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli: a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Pátek, M., B. J. Eikmanns, J. Pátek, and H. Sahm. 1996. Promoters from Corynebacterium glutamicum: cloning, molecular analysis and search for a consensus motif. Microbiology 142:1297-1309. [DOI] [PubMed] [Google Scholar]
- 29.Saier, M. H., Jr. 2000. Vectorial metabolism and the evolution of transport systems. J. Bacteriol. 182:5029-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saier, M. H., Jr. 2001. Evolution of transport proteins, p. 1-10. In J. K. Setlow (ed.) Genetic engineering: principles and methods, vol. 23. Kluwer Academic/Plenum Publishers, New York, N.Y. [DOI] [PubMed]
- 31.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saier, M. H., Jr. and T.-T. Tseng. 1999. Evolutionary origins of transmembrane transport systems, p. 252-274. In J. K. Broome-Smith, S. Baumberg, C. J. Stirling, and F. B. Ward (ed.), Transport of molecules across microbial membranes. Symposium 58: Society for General Microbiology. Cambridge University Press, Cambridge, United Kingdom.
- 33.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 34.Simic, P., H. Sahm, and L. Eggeling. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tauch, A., T. Hermann, A. Burkovski, R. Krämer, A. Pühler, and J. Kalinowski. 1998. Isoleucine uptake in Corynebacterium glutamicum ATCC 13032 is directed by the brnQ gene product. Arch. Microbiol. 169:303-312. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniakm, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 38.Vasicová, P., Z. Abrhámová, J. Nesvera, M. Pátek, H. Sahm, and B. Eikmanns. 1998. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnol. Tech. 12:743-746. [Google Scholar]
- 39.Vrljic, M., H. Sahm, and L. Eggeling. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22:815-826. [DOI] [PubMed] [Google Scholar]
- 40.Vrljic, M., J. Garg, A. Bellmann, S. Wachi, R. Freudl, M. J. Malecki, H. Sahm, V. J. Kozina, L. Eggeling, and M. H. Saier, Jr. 1999. The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme for a novel superfamily of transmembrane solute translocators. J. Mol. Microbiol. Biotechnol. 1:327-336. [PubMed] [Google Scholar]
- 41.Yen, M.-R., Y.-H. Tseng, P. Simic, H. Sahm, L. Eggeling, and M. H. Saier, Jr. 2002. The ubiquitous ThrE family of putative transmembrane amino acid efflux transporters. Res. Microbiol. 153:19-25. [DOI] [PubMed] [Google Scholar]
- 42.Zakataeva, N. P., V. V. Aleshin, I. L. Tokmakova, P. V. Troshin, and V. A. Livshits. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228-232. [DOI] [PubMed] [Google Scholar]
- 43.Zhai, Y., and M. H. Saier, Jr. 2001. A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J. Mol. Microbiol. Biotechnol. 3:285-286. [PubMed] [Google Scholar]
- 44.Zittrich, S., and R. Krämer. 1994. Quantitative discrimination of carrier-mediated excretion of isoleucine from uptake and diffusion in Corynebacterium glutamicum. J. Bacteriol. 176:6892-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]