Abstract
Ferrochelatase (EC 4.99.1.1) catalyzes the last reaction in the heme biosynthetic pathway. The enzyme was studied in the bacterium Bacillus subtilis, for which the ferrochelatase three-dimensional structure is known. Two conserved amino acid residues, S54 and Q63, were changed to alanine by site-directed mutagenesis in order to detect any function they might have. The effects of these changes were studied in vivo and in vitro. S54 and Q63 are both located at helix α3. The functional group of S54 points out from the enzyme, while Q63 is located in the interior of the structure. None of these residues interact with any other amino acid residues in the ferrochelatase and their function is not understood from the three-dimensional structure. The exchange S54A, but not Q63A, reduced the growth rate of B. subtilis and resulted in the accumulation of coproporphyrin III in the growth medium. This was in contrast to the in vitro activity measurements with the purified enzymes. The ferrochelatase with the exchange S54A was as active as wild-type ferrochelatase, whereas the exchange Q63A caused a 16-fold reduction in Vmax. The function of Q63 remains unclear, but it is suggested that S54 is involved in substrate reception or delivery of the enzymatic product.
Heme is a tetrapyrrole derivative commonly used as a prosthetic group in proteins like cytochromes and catalases. The heme biosynthetic pathway from the common precursor 5-aminolevulinic acid (ALA) consists of seven enzymatic steps (6). The terminal step is catalyzed by ferrochelatase (EC 4.99.1.1; protoheme ferrolyase), which inserts a ferrous ion into protoporphyrin IX (22). It is important for the organism to keep the porphyrinogen intermediates of the pathway at a low level, since they can be oxidized to phototoxic porphyrins. Mutations in the different human heme biosynthetic genes cause inherited disorders commonly named porphyrias (31, 32). The disorder connected to the ferrochelatase gene is known as erythropoietic porphyria. The skin is one of the major organs involved in most of these diseases because porphyrin accumulation causes photosensitivity of the patient. When excited by light, porphyrins generate reactive oxygen species, which cause skin lesions. The phototoxic effect of porphyrins is, however, utilized within cancer therapy. Tumor cells are induced to accumulate porphyrins, and upon treatment with light, necrosis of these cells occurs (12, 13). A common approach to induce porphyrin accumulation is to apply the heme precursor ALA to tumor cells. The exogenous administration of ALA bypasses the heme regulatory step and results in a significant accumulation of protoporphyrin IX, which has suitable photoexcitable properties (24). The protoporphyrin IX is, however, rapidly converted to heme by ferrochelatase in the presence of iron, a process that is desirable after the light treatment but not before (39). Insight into the delivery of protoporphyrin IX and iron to ferrochelatase should help in the interpretation of the impact caused by erythropoietic porphyria mutations and could also lead to clinical improvements for photodynamic therapy of cancer.
One of the most intensively studied ferrochelatases originates from Bacillus subtilis. In this bacterium ferrochelatase is encoded by the hemH gene located in the hemEHY operon (14, 16). The monomeric enzyme has a calculated molecular mass of 35,348 Da. The structure of the B. subtilis ferrochelatase has been determined to 1.8 Å (2, 23). The ferrochelatase is folded into two similar domains, each with a four-parallel β sheet flanked by α helices. Protruding structural elements from both domains form a deep cleft, which is the active site. The structure of ferrochelatase has been solved in complex with N-methyl mesoporphyrin IX and Cu-mesoporphyrin IX (23). The former is a strong competitive inhibitor of ferrochelatase, whereas the latter is a nonphysiological enzymatic product. The structures have provided insight into the enzymatic mechanism of porphyrin metallation. The data support a mechanism in which pyrrole rings B, C, and D are fixed by ferrochelatase in a vise-like grip and force a 36° tilt on pyrrole ring A. The metal ion enters the porphyrin via the tilted pyrrole.
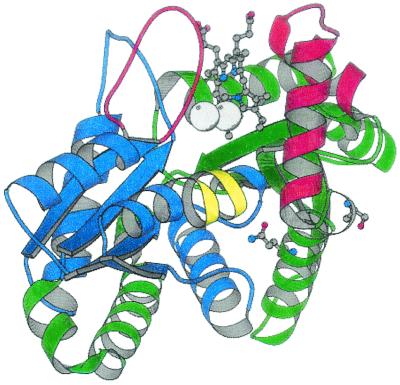
The ferrochelatases isolated from various organisms all have very similar catalytic properties, indicating that the major features of the reactions are conserved. Genes encoding ferrochelatase have been sequenced from a wide range of species including bacteria, fungi, mammals, and plants. In the alignment of the deduced polypeptides of evolutionarily distantly related ferrochelatases, less than 10% of the amino acid residues among a total of more than 300 are more or less fully conserved. These relatively few residues are most likely of fundamental importance for the function of ferrochelatase. Fatal effects on the enzyme activity might be expected if they are exchanged for any other amino acid residue. The function of most of the conserved residues can be understood from the known three-dimensional structure of ferrochelatase. Glycine and proline residues are found at the ends of helices and are of structural and steric importance. Other residues such as H183 and E264 (numbered according to the B. subtilis ferrochelatase polypeptide) play important roles in the active site. In contrast, the function of residues S54 and Q63 is not understood. Both residues are located on α-helix 3 in domain 1 more than 12 Å from the active site. The side chain of S54 is located on the outside of the ferrochelatase, whereas the Q63 side chain is internally located (Fig. 1). Neither S54 nor Q63 interacts with other amino acid residues in the ferrochelatase. Nevertheless, their function should be of vital importance to the ferrochelatase since they belong to the group of conserved residues.
FIG. 1.
Schematic representation of the B. subtilis ferrochelatase structure showing the location of S54 and Q63 in relation to the porphyrin in the active site and the two metal binding sites. Domain 1 is colored green and domain 2 is blue. The parts of the chain in red build up the walls of the active site. The π-helix of domain 2 is in yellow. The figure was prepared using the program MOLSCRIPT (20).
In this study site-directed mutagenesis was performed on the B. subtilis ferrochelatase gene to determine the function of residues S54 and Q63. B. subtilis is genetically transformable via a multigene-controlled process involving binding, digestion, and uptake of DNA followed by recombination (7, 35). This made it possible to exchange the single wild-type hemH gene on the B. subtilis chromosome with a mutated hemH gene encoding modified S54 or Q63 residues and to perform in vivo analysis of the mutation. An expression system for the production of recombinant B. subtilis ferrochelatase in Escherichia coli exists and a straightforward purification method has been developed (17). The system was used to provide purified ferrochelatase modified at residues S54 and Q63 for activity measurements in vitro. The present study suggests that ferrochelatase is involved in substrate channeling and that the commonly used in vitro assay using Zn2+ and protoporphyrin IX solubilized in the detergent Tween 80 is highly artificial and far from the in vivo situation.
MATERIALS AND METHODS
Growth media.
B. subtilis strains were kept on tryptose blood agar base (TBAB; Difco) or agar plates containing Spizizen's minimal glucose medium (35) supplemented with required amino acids (Duchefa) at 10 mg/liter. Liquid cultures were grown in Luria-Bertani (LB) (30) medium or minimal glucose medium. For heme auxotrophs the different media were supplemented with hemin (for LB and TBAB, 2.5 mg/liter; for minimal glucose medium, 1.0 mg/liter). Bovine serum albumin (Sigma; fraction V; 500 mg/liter) was always added together with hemin. The following antibiotics were used: ampicillin (100 mg/liter) and phleomycin (1.5 mg/liter). Porphyrin-accumulating B. subtilis mutants were screened on minimal glucose medium agar plates supplemented with DEAE-Sephadex A-25 (Pharmacia Fine Chemicals; 50 g/liter). DEAE-Sephadex was suspended in water and autoclaved before it was mixed with agar.
Bacterial strains and plasmids.
B. subtilis 168Δ8 was made by transforming competent B. subtilis 168 with chromosomal DNA of B. subtilis 3G18Δ8 (16), selecting for phleomycin resistance on hemin-containing TBAB plates. For in vivo studies wild-type and mutant B. subtilis strains were used, while E. coli was used to produce the B. subtilis ferrochelatase for in vitro activity measurements. Bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| 168 | trpC2 | BGSCa |
| 3G18Δ8 | ade met trpC2 ΔhemH::ble | 16 |
| 168Δ8 | trpC2 ΔhemH::ble | This work |
| 168S54A | trpC2 hemHS54A | This work |
| 168Q63A | trpC2 hemHQ63A | This work |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) (F′ traD36 proAB+lacIqlacZΔM15) | 43 |
| BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 37 |
| Plasmids | ||
| pLUG185 | bla; 2.8-kb SphI fragment of pLUG1301 in pUC18; contains hemH flanked by 50% of hemE and 90% of hemY | 14 |
| pLUGS54Aa1 | pLUG185 derivative, contains a T-to-G point mutation in bp 3197b of hemH | This work |
| pLUGQ63Aa1 | pLUG185 derivative, contains a CA-to-GC point mutation in bp 3224 and 3225b of hemH | This work |
| pLUGT7-H | bla; B. subtilis hemH cloned downstream of the T7 promoter of pBluescript II KS(−) | 17 |
| pT7S54Ac1 | pLUGT7-H derivative, contains a T-to-G point mutation in bp 3197b of hemH | This work |
| pT7Q63Ab2 | pLUGT7-H derivative, contains a CA-to-GC point mutation in bp 3224 and 3225b of hemH | This work |
BGSC, Bacillus Genetic Stock Center, Department of Biochemistry, Ohio State University, Columbus.
Base pair numbering is according to that of a previous study (14).
Site-directed mutagenesis.
The oligonucleotides used are listed in Table 2. The point mutations were introduced using a PCR engine thermocycler (Peltier Thermal Cycler 200) in accordance with the instructions for the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, Calif.). A 50-μl reaction mixture contained 30 ng of plasmid with the cloned hemH gene, 125 ng of each oligonucleotide, 10 nmol of dNTP mixture, and 3 U of Pfu DNA polymerase (Promega). Sixteen cycles were performed as follows: 95°C, 30 s; 55°C, 60 s; 68°C, 11 min using pLUG185 as template or 9 min using pLUGT7-H as template. After the PCR was completed the template was removed by digestion with the restriction enzyme DpnI (Promega).
TABLE 2.
Oligonucleotides
| Oligonucleotide | Sequence (5′ to 3′)a | Base pair no.b |
|---|---|---|
| S54Aa | ATT GGC GGC ATT GCA CCG CTT GCC CAA | 3185-3211 |
| S54Ab | TTG GGC AAG CGG TGC AAT GCC GCC AAT | 3185-3211 |
| Q63AUp:36 | CCC AAA TCA CAG AAC AGG CGG CGC ATA ATC TGG AAC | 3207-3242 |
| Q63ALo:36 | GTT CCA GAT TAT GCG CCG CCT GTT CTG TGA TTT GGG | 3207-3242 |
Oligonucleotides causing point mutations are underlined.
Base pair numbering is according to that of a previous study (14).
Transformation of bacteria.
Competent E. coli JM109 was prepared in accordance with the instructions in the manual for the Bio-Rad E. coli Pulser Transformation apparatus, while competent E. coli BL21(DE3) was prepared as previously described (25). B. subtilis 3G18Δ8 was grown to competence using a previously described method (4) using Spizizen's minimal glucose medium supplemented with hemin and bovine serum albumin. For each transformation, 0.5 ml of B. subtilis 168Δ8 culture was mixed with 1.0 μg of plasmid which had been linearized by ScaI (Boehringer, Mannheim, Germany).
DNA sequence analysis.
Plasmid and amplified chromosomal DNA were sequenced with an ABI377 sequencer (Applied Biosystems, Inc.) using Big Dye terminator supplied by the manufacturer.
General DNA techniques.
Small-scale plasmid DNA preparations were performed using the Jet Quick Miniprep kit (Genomed, Bad Oeynhausen, Germany). Large-scale preparations of plasmids were made using the Wizard Plus Midipreps DNA purification system (Promega). B. subtilis chromosomal DNA was prepared as described by Marmur (26). DNA separated on an agarose gel was purified with the Geneclean II kit (Bio101).
Growth study.
A total volume of 100 ml of minimal glucose medium in a 1-liter baffled flask was inoculated to an optical density at 600 nm (OD600) of 0.05 from overnight cultures of B. subtilis 168 (wild type) and the mutant strains. One milliliter of the cultures was removed every hour and used to measure OD600. The growth was followed for 8 h.
Preparation of soluble E. coli cell extract containing the expressed ferrochelatases.
An isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 RNA polymerase expression system was used. E. coli BL21(DE3) was transformed with the plasmid from which the hemH gene was going to be expressed. Transformants were selected on ampicillin-containing TBAB plates. A 2-liter baffled flask containing 1 liter of LB medium supplemented with ampicillin was inoculated with fresh transformants from one plate. Two flasks were used for each strain. The cells were grown at 30°C on a rotary shaker (200 rpm). At an OD600 of 0.25 IPTG was added to a final concentration of 0.5 mM. After 4 h the bacteria were harvested by centrifugation at 7,000 × g at 4°C for 10 min and the pellet was washed with 50 mM Tris-HCl buffer, pH 7.4. The pellet was resuspended in 29 ml of the 50 mM Tris-HCl buffer. Lysozyme was added to a concentration of 0.25 mg/ml and the suspension was incubated at 30°C for 20 min, followed by sonication on ice. The resulting lysate was centrifuged at 10,000 × g at 4°C for 10 min and the ferrochelatase was further purified from the supernatant according to the method of Hansson and Al-Karadaghi (17).
Ferrochelatase activity measurement.
The formation of Zn-protoporphyrin IX was measured in a fluorometric assay using a Perkin-Elmer LS50B spectrofluorometer and a 1-ml quartz cuvette. The excitation wavelength used was 420 nm and the emission wavelength was 587 nm. Excitation and emission slits were both 10 nm. A 1 mM stock solution of protoporphyrin IX was prepared in dimethyl sulfoxide. For each measurement a 1-ml reaction mixture was prepared containing 100 mM Tris-HCl (pH 7.4), 0.5, 1, or 2 μM protoporphyrin IX, 0.3 mg of Tween 80 per ml, and 5, 10, or 30 μM Zn(NO3). Reaction mixtures with different protoporphyrin concentrations, but without Zn(NO3), were prepared as cocktails. The Zn(NO3) was added at the start of each reaction. Pure ferrochelatase (5.6 μl [5.6 μg]) was added to the cuvette 1 min after the start of the reaction.
Thin-layer chromatography analysis of porphyrins.
B. subtilis mutants accumulating porphyrins were grown in 20 ml of Spizizen glucose minimal medium with required growth factors on a rotary shaker (200 rpm) overnight at 37°C. One milliliter of culture was transferred to an Eppendorf tube and centrifuged at 20,000 × g for 4 min. The supernatant was transferred to a new tube and 10 mg of DEAE-Sephadex A-25 was added to adsorb the porphyrins. The sample was centrifuged as described above and the supernatant was discarded. The porphyrins were eluted from the DEAE-Sephadex pellet with acidic acetone (12 M HCl, 13.6 M acetone; 1:4 [vol/vol]) and analyzed by thin-layer chromatography (silica gel 60; 0.25 mm thick, 10 by 20 cm; Merck) as described previously (11, 15). One to two microliters of the extract from each sample was put on the silica plate. The mobile phase was a mixture of the following organic solvents: N,N-dimethyl-formamide, methanol, ethylene glycol, acetic acid, 1-chlorobutane, and chloroform (4:35:6:0.4:18:20 [vol/vol/vol/vol/vol/vol]). Uroporphyrin III, coproporphyrin III, hematoporphyrin, and protoporphyrin IX (Porphyrin Products, Logan, Utah) dissolved in acidic acetone were used as standards.
Other methods.
DNA was separated on 0.8% agarose gels. Proteins were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% acrylamide [wt/vol]) performed as described by Fling and Gregerson (10) using the Tris-Tricine running buffer system of Schägger and von Jagow (33). For Western blot analysis, proteins were electrotransferred after sodium dodecyl sulfate-polyacrylamide gel electrophoresis to an Immobilon P filter (Millipore) according to the method of Towbin et al. (40) using a semidry electroblotter. Polyclonal antibodies against B. subtilis ferrochelatase were from rabbit (16). Goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase was used as secondary antibody. Antigens on Immobilon P filters were visualized using a chemiluminescence detection system (Clontech Laboratory, Inc.).
RESULTS
Construction of mutant ferrochelatase.
In order to study the function of the conserved amino acid residues S54 and Q63 of ferrochelatase, these residues were changed to alanine residues by site-directed mutagenesis of the B. subtilis hemH gene. The S54A exchange was obtained using the oligonucleotides S54Aa and S54Ab as primers, and Q63A was obtained using the oligonucleotides Q63AUp:36 and Q63ALo:36 (Table 2). Plasmids pLUG185 and pLUGT7-H were used as templates. Plasmid pLUG185 is a pUC18 derivative carrying a 2.8-kb SphI DNA fragment of the B. subtilis hemEHY operon. The SphI fragment contains 540 bp of hemE, the complete hemH, and 1,238 bp of hemY (14). The pLUG185 derivatives carrying the S54A and Q63A modifications were named pLUGS54Aa1 and pLUGQ63Aa1, respectively. Plasmid pLUGT7-H is a pBluescript derivative containing 44 bp of the terminal part of hemE, the complete hemH, and 460 bp of hemY. The plasmid is used to produce recombinant B. subtilis ferrochelatase in large amounts from E. coli utilizing the T7 promoter of pBluescript (17). The pLUGT7-H derivatives carrying the S54A and Q63A modifications were named pT7S54Ac1 and pT7Q63Ab2, respectively. All genetic constructs were confirmed by DNA sequence analysis in which both strands of the complete hemH genes were sequenced.
In vivo effects of S54A and Q63A modified ferrochelatase.
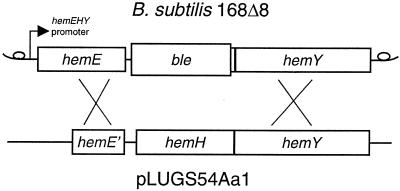
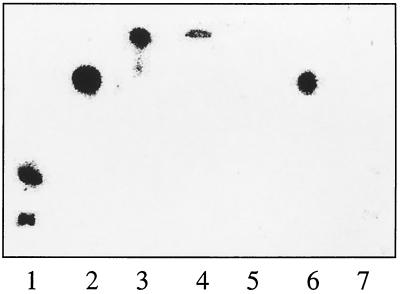
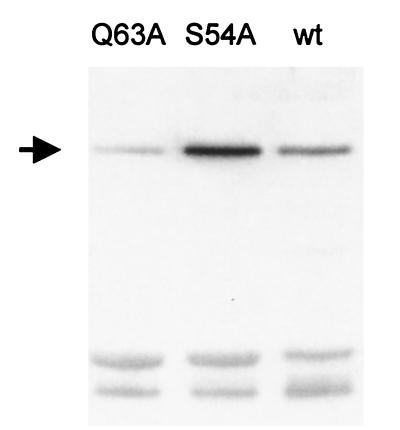
The mutated ferrochelatase gene was introduced into the B. subtilis chromosome using the B. subtilis strain 168Δ8. In B. subtilis 168Δ8 the hemH gene has been deleted and replaced by the ble gene (Fig. 2), which gives resistance to the antibiotic phleomycin. The deletion of the hemH gene results in heme auxotrophy. The ble gene was replaced by mutated hemH as follows. B. subtilis 168Δ8 was transformed with 1 μg of the plasmid pLUGS54Aa1, pLUGQ63Aa1, or pLUG185 which had been linearized by ScaI restriction enzyme cleavage (Fig. 2). Plasmid pLUG185 was used as a positive control in the experiment. Transformants were selected for heme prototrophy on TBAB plates. Fifty-one, 166, and 120 transformants were obtained upon transformation with pLUGS54Aa1, pLUGQ63Aa1, and pLUG185, respectively. Sixteen different clones from each transformation were tested for growth on two different glucose minimal media. The test media were identical except that one test plate was supplemented with hemin and phleomycin. B. subtilis 168Δ8 and the wild-type 168 strain were also tested on the plates as controls. Neither the wild type nor the transformants grew on the plates containing phleomycin and hemin. B. subtilis 168Δ8 grew well on these plates. Strain 168Δ8 could not grow on the plates which did not contain phleomycin or hemin, but the transformants and the wild type grew well. This strongly indicates that the ble gene of 168Δ8 had been replaced by the ferrochelatase genes of pLUGS54Aa1, pLUGQ63Aa1, and pLUG185 by a double crossover (Fig. 2). To confirm that no additional mutations had been introduced, the hemH and hemY genes of three mutants from each construct were amplified by PCR and sequenced. Chromosomal DNA was used as template. It was concluded that the single copy of the mutated ferrochelatase gene causing the S54A or Q63A modification was enough to support growth of the strains on minimal media. It was clear, however, that the B. subtilis strain modified with S54A, but not Q63A, was impaired by the mutation. The generation time of the wild-type 168 strain and the 168Q63A strain in minimal glucose medium was 1.6 h while it was 2.8 h for 168S54A. It was further observed that B. subtilis 168S54A accumulated a red substance in the growth medium while 168Q63A and 168 did not. The identity of the red pigment was coproporphyrin III, as determined with a silica gel thin-layer chromatography system using uroporphyrin III, coproporphyrin III, hematoporphyrin IX, and protoporphyrin IX as references (Fig. 3). The concentration of coproporphyrin III in the medium of an overnight culture of 168S54A was 6 μM, as determined spectrophotometrically. The slower growth of B. subtilis 168S54A compared to the other two strains could not be explained by a reduced amount of ferrochelatase in this strain. Rather, 168S54A had five times more ferrochelatase than the wild-type 168 strain as determined by Western blot analysis in which equal amounts of total cellular protein were loaded (Fig. 4). The overproduction is probably a response to lack of heme and is the first observation demonstrating that the amount of ferrochelatase varies in B. subtilis cells.
FIG. 2.
Map of the hemEHY operon in the plasmid pLUGS54Aa1 linearized by ScaI cleavage and in the B. subtilis 168Δ8 chromosome. In strain 168Δ8, the hemH gene (bp 2992-3874) has been deleted and replaced by a phleomycin-resistance gene, ble. The mutated hemH gene in pLUGS54Aa1 causing a serine-to-alanine modification at the protein level could be integrated into the B. subtilis chromosome by a double crossover as indicated. The numbering of base pairs follows that used in a previous study (14).
FIG. 3.
Porphyrins in growth cultures of three different B. subtilis strains analyzed by thin-layer chromatography. Lanes 1 to 4 are reference samples, while lanes 5 to 7 are samples from different growth cultures. Lane 1, uroporphyrin III; lane 2, coproporphyrin III; lane 3, hematoporphyrin IX; lane 4, protoporphyrin IX; lane 5, B. subtilis 168; lane 6, B. subtilis 168S54A; lane 7, B. subtilis 168Q63A.
FIG. 4.
Western blot analysis of ferrochelatase in three B. subtilis strains. The strains were 168Q63A (lane Q63A), 168S54A (S54A), and wild-type 168 (wt). The arrow indicates the ferrochelatase antigen. Five micrograms of total cellular protein was loaded in each lane.
If the B. subtilis 168S54A strain was left on a TBAB growth plate for 2 weeks at room temperature, it was observed that faster growing secondary mutants accumulated in the colonies of 168S54A. The second site mutants were isolated by streaking the cells for single colonies on minimal glucose medium agar plates supplemented with DEAE-Sephadex A-25. On this medium, colonies accumulating coproporphyrin III exhibited a bright orange fluorescent halo under UV light. Most second site mutants were leaky, as they still accumulated coproporphyrin III. Four nonleaky second site mutants could be isolated. No coproporphyrin accumulation could be detected in the nonleaky strains. Chromosomal DNA was isolated from nine of the many leaky second site mutants and from the four nonleaky second site mutants. The hemH and hemY genes of the extracted chromosomal DNA was amplified. The entire PCR products were analyzed by DNA sequencing. All leaky second site mutants remained S54A and no other changes in the DNA sequence could be found. The nonleaky strains were true revertants, as they had changed back to a serine residue at position 54.
Activity of mutant ferrochelatase in vitro.
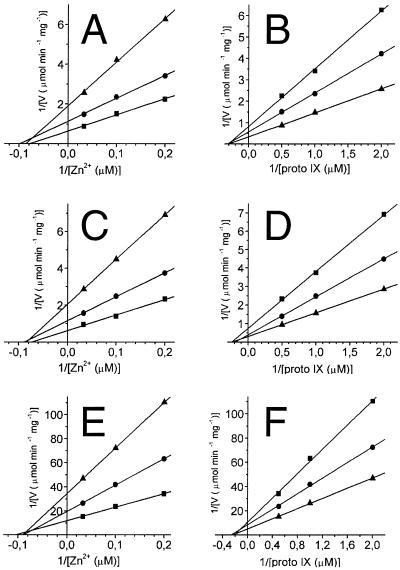
In order to analyze the effects of the mutations in vitro, the modified ferrochelatases were produced in E. coli BL21(DE3), transformed with pLUGT7-H, pT7S54Ac1, or pT7Q63Ab2, and purified to homogeneity as described previously (17). Plasmids pT7S54Ac1 and pT7Q63Ab2 have the desired mutations in hemH resulting in the exchanges S54A and Q63A, respectively. During the purification procedure the wild-type ferrochelatase and the two modified proteins behaved similarly. Activity measurements were made with 5.6 μg of purified ferrochelatase. The incorporation of Zn2+ into protoporphyrin IX was monitored in a fluorometric assay. Nine pairs of different concentrations of Zn2+ and protoporphyrin IX were used. The concentrations of Zn2+ were 5, 10, and 30 μM and those of protoporphyrin IX were 0.5, 1, and 2 μM. Vmax, Km, and kcat of the wild type and the two modified ferrochelatases were determined from Lineweaver-Burk plots (Fig. 5 and Table 3). The values determined for the ferrochelatase carrying the S54A modification were very similar to those of the wild-type enzyme. In contrast, the catalytic activity of ferrochelatase carrying the Q63A modification was considerably lower, although the Km values were only slightly affected. These results were totally contradictory to the in vivo observations, where the growth rates were affected by the S54A modification but not by Q63A.
FIG. 5.
Lineweaver-Burk plots of activities obtained with wild-type ferrochelatase (A, B) and S54A (C, D) and Q63A (E, F) modified ferrochelatase. (A, C, E) Three different fixed concentrations of protoporphyrin IX were used (0.5, 1, and 2 μM). (B, D, F) Three different fixed concentrations of Zn2+ were used (5, 10, and 30 μM). Vmax and Km values were calculated from secondary plots of panels A to F (see Table 3).
TABLE 3.
Catalytic properties of modified ferrochelatases compared to the wild-type enzymea
| Modification | Vmaxb |
Km (μM)
|
kcat (min−1) | |
|---|---|---|---|---|
| Zn2+ | Protoporphyrin IX | |||
| Wild type | 800 | 11 | 3.3 | 28 |
| S54A | 750 | 11 | 4.0 | 27 |
| Q63A | 49 | 12 | 5.4 | 1.7 |
The data were obtained from secondary plots of the graphs in Fig. 5.
Vmax is expressed as nanomoles of Zn-protoporphyrin IX min−1 mg of protein−1.
DISCUSSION
The function of the conserved amino acid residues S54 and Q63 in the B. subtilis ferrochelatase is not understood from their locations in the protein structure. Modification of these residues to alanine had opposite effects on the ferrochelatase activity studied in vivo and in vitro. The exchange Q63A caused a 16-fold reduction in Vmax when measured in vitro but had no detectable effects in vivo when monitored as growth rates. The catalytic activity of B. subtilis ferrochelatase in vivo and in vitro has been analyzed before (16). The activity in vivo was calculated from the rate of heme formation during exponential growth in relation to the amount of ferrochelatase present in these cells. The apparent turnover was determined to be 0.2 min−1 (16). This apparent turnover in vivo is 100 times lower than the measured turnover in vitro (28 min−1 in this study, 24 min−1 in reference 16). It therefore seems possible that a Q63A modified ferrochelatase with a 16-fold lower Vmax than the wild type is still capable of supporting synthesis of wild-type amounts of heme molecules for exponentially growing cells. This provides an explanation for the difference in the in vivo and in vitro results for Q63A, but the function of Q63 in the reaction mechanism of ferrochelatase still remains unclear. Our kinetic analysis shows, however, that the Q63A modified ferrochelatase primarily affects the Vmax of the enzyme, whereas the Km values for Zn2+ and protoporphyrin IX are similar to those of the wild-type enzyme.
The S54A modified ferrochelatase behaved differently from Q63A since it had no effect on the ferrochelatase activity in vitro but caused cells to grow slower in vivo. An upregulation of the amount of ferrochelatase in the mutant cells was not enough to compensate for the mutation. In addition, the mutant cells accumulated coproporphyrin III. Cells blocked in one or several later steps in heme biosynthesis often accumulate and excrete porphyrins, which are considered to be the autooxidized products of the corresponding porphyrinogens (15, 27). Interestingly, iron-deficient B. subtilis has also been reported to accumulate coproporphyrin (28). The location of the conserved S54 residue on the surface of the ferrochelatase structure suggests that S54 is part of a docking site, presumably of another protein. We suggest that the docking protein could either deliver protoporphyrin IX or Fe2+ or retrieve the heme product. A conserved S54 residue of such a docking site should be of fundamental importance for the function of the chelatase reaction and a disturbed interaction could be expected to reduce growth and cause accumulation of substrates. Such behavior was demonstrated by our B. subtilis 168S54A strain in vivo. In vitro, the porphyrin substrate as well as the metalloporphyrin product are solubilized by the detergent Tween 80. In such an artificial environment the docking site of ferrochelatase is not used and an S54A modification does not affect the ability of ferrochelatase to catalyze the reaction. These results suggest that the in vitro assay for ferrochelatase activity measurements is highly artificial, not only because unphysiological substrates are often used. In the cell the transport of heme and heme biosynthetic intermediates is still to a large extent unknown, but albumin and lipoproteins are suggested as important transporter proteins (34). No direct transfer of heme from ferrochelatase to heme utilizing proteins has been found. Among possible candidates in B. subtilis are CtaB, which uses heme as a substrate in the pathway of a-type heme biosynthesis (38), and CydC and CydD, which are essential components for the correct assembly of cytochrome bd oxidase (41).
It is obvious that substrate channeling of tetrapyrrole intermediates between the enzymes of the heme biosynthetic pathway would be a solution to avoid accumulation of porphyrins. The observation that coproporphyrin III, and not protoporphyrin IX, the substrate of ferrochelatase, accumulates in the S54A ferrochelatase mutant is intriguing and could support the idea that the last three enzymes in the heme biosynthetic pathway are involved in substrate channeling. In order to study this, a better understanding of the metabolism of coproporphyrinogen III and protoporphyrinogen IX in B. subtilis has to be obtained. At present one protoporphyrinogen IX oxidase and two coproporphyrinogen III oxidases have been found in this bacterium (15, 18, 19). Substrate channeling of tetrapyrrole intermediates between the last two enzymes of the heme pathway has been suggested before (9), but the hypothesis was abandoned as no coordination of the expression of the corresponding genes in mice was found (5). Other data, obtained by Straka et al. (36), support the idea that ferrochelatase interacts with a protein. In situ experiments found the functional unit of bovine ferrochelatase to be 82 kDa. It could not be determined if the protein accompanying the 40-kDa ferrochelatase was an unknown protein or another ferrochelatase molecule. The latter interpretation is supported by the recent structure of the human ferrochelatase, which forms a dimer (42). Residues from helices α11 and α12 and strand β6 are found in the interface between the dimers. These secondary elements correspond to α9, α10, and β6 in the monomeric B. subtilis ferrochelatase structure. It is excluded that S54 and Q63, in the focus of this study, could participate in the stabilization of a dimer, since they are located in α3 and not in α9, α10, or β6.
The iron used as a substrate by ferrochelatase is in the form of a ferrous ion, not a ferric ion (22). Ferrous iron is easily oxidized to ferric iron, but it has been suggested that the intracellular environment should be reducing enough to keep iron in its reduced form (29). Bacterioferritin and ferritin are two distantly related iron-storage proteins (3). The uptake of iron is an oxidation process in which ferrous iron is oxidized and stored as a ferric-hydroxyphosphate core. The release of iron from bacterioferritin and ferritin is less understood but is likely to involve a reduction of ferric to ferrous iron (8), which thus would provide iron in a suitable form for ferrochelatase. It is doubtful, however, that bacterioferritin or ferritin is a ubiquitous direct supplier of ferrous ions for ferrochelatase of different species, since many organisms, including B. subtilis, apparently lack these proteins judging from their genome sequences (21). In addition, no phenotype has been determined for E. coli bacterioferritin mutants (1).
As the S54 residue is conserved in all ferrochelatases studied so far, it is reasonable to believe that the putative protein interacting with ferrochelatase is a protein common to all organisms. To trace this protein we will try an approach using the second site mutants of the B. subtilis 168S54A strain obtained in this study. A second site mutant which does not accumulate coproporphyrin III will hopefully be found in which the S54A modification still remains. Such a strain could harbor a compensatory mutation in the gene encoding the protein interacting with ferrochelatase. A DNA library would be constructed from that second site mutant and used to retrieve the gene by complementation of the original mutant selecting for non-porphyrin-accumulating bacteria. In such work, leaky second site mutants cannot be used as their phenotype is too close to that of the primary mutants.
Acknowledgments
We are grateful to Simon Gough for fruitful discussions.
This work was made possible thanks to generous support from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) and the Carl Tryggers Foundation.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Karadaghi, S., M. Hansson, S. Nikonov, B. Jönsson, and L. Hederstedt. 1997. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure 5:1501-1510. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 4.Arwert, F., and G. Venema. 1973. Transformation in Bacillus subtilis. Fate of newly introduced transforming DNA. Mol. Gen. Genet. 123:185-198. [DOI] [PubMed] [Google Scholar]
- 5.Conder, L. H., S. I. Woodard, and H. A. Dailey. 1991. Multiple mechanisms for the regulation of haem synthesis during erythroid cell differentiation. Biochem. J. 275:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dailey, H. A. 1990. Conversion of coproporphyrinogen to protoheme in higher eukaryotes and bacteria: terminal three enzymes, p. 123-161. In H. A. Dailey (ed.), Biosynthesis of heme and chlorophylls. McGraw-Hill Publishing Co., New York, N.Y.
- 7.Dubnau, D. 1993. Genetic exchange and homologous recombination, p. 555-584. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria; biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 8.Fatemi, S. J. A., F. H. A. Kadir, D. J. Williamson, and G. R. Moore. 1991. The uptake, storage, and mobilization of iron and aluminum in biology. Adv. Inorg. Chem. 36:409-448. [Google Scholar]
- 9.Ferreira, G. C., T. L. Andrew, S. W. Karr, and H. A. Dailey. 1988. Organization of the terminal two enzymes of the heme biosynthetic pathway. J. Biol. Chem. 263:3835-3839. [PubMed] [Google Scholar]
- 10.Fling, S. P., and D. S. Gregerson. 1986. Peptide and protein molecular weight determination by electrophoresis using a high-molarity Tris buffer system without urea. Anal. Biochem. 155:83-88. [DOI] [PubMed] [Google Scholar]
- 11.Friedmann, H. C., and E. T. Baldwin. 1984. Reverse-phase purification and silica gel thin-layer chromatography of porphyrin carboxylic acids. Anal. Biochem. 137:473-480. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, J., S. Weber, and R. Kaufmann. 2000. Genotoxic potential of porphyrin type photosensitizers with particular emphasis on 5-aminolevulinic acid: implications for clinical photodynamic therapy. Free Radic. Biol. Med. 15:537-548. [DOI] [PubMed] [Google Scholar]
- 13.Granville, D. J., B. M. McManus, and D. W. Hunt. 2001. Photodynamic therapy: shedding light on the biochemical pathways regulating porphyrin-mediated cell death. Histol. Histopathol. 16:309-317. [DOI] [PubMed] [Google Scholar]
- 14.Hansson, M., and L. Hederstedt. 1992. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J. Bacteriol. 174:8081-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansson, M., and L. Hederstedt. 1994. Bacillus subtilis HemY is a peripheral membrane protein essential for protoheme IX synthesis which can oxidize coproporphyrinogen III and protoporphyrinogen IX. J. Bacteriol. 176:5962-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansson, M., and L. Hederstedt. 1994. Purification and characterization of a water-soluble ferrochelatase from Bacillus subtilis. Eur. J. Biochem. 220:201-208. [DOI] [PubMed] [Google Scholar]
- 17.Hansson, M., and S. Al-Karadaghi. 1995. Purification, crystallization, and preliminary X-ray analysis of Bacillus subtilis ferrochelatase. Proteins 23:607-610. [DOI] [PubMed] [Google Scholar]
- 18.Hippler, B., G. Homuth, T. Hoffmann, C. Hungerer, W. Schumann, and D. Jahn. 1997. Characterization of Bacillus subtilis hemN. J. Bacteriol. 179:7181-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homuth, G., A. Rompf, W. Schumann, and D. Jahn. 1999. Transcriptional control of Bacillus subtilis hemN and hemZ. J. Bacteriol. 181:5922-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraulis, P. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 21.Kunst, F., et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Labbe-Bois, R., and J.-M. Camadro. 1994. Ferrochelatase in Saccharomyces cerevisiae, p. 413-453. In G. Winkelmann and D. R. Winge (ed.), Metal ions in fungi. Marcel Dekker Inc., New York, N.Y.
- 23.Lecerof, D., M. Fodje, A. Hansson, M. Hansson, and S. Al-Karadaghi. 2000. Structural and mechanistic basis of porphyrin metallation by ferrochelatase. J. Mol. Biol. 297:221-232. [DOI] [PubMed] [Google Scholar]
- 24.Malik, Z., and H. Lugaci. 1987. Destruction of erythroleukaemic cells by photoactivation of endogenous porphyrins. Br. J. Cancer 56:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel, M., and A. Higa. 1979. Calcium-dependent bacteriophage DNA infections. J. Mol. Biol. 53:159-162. [DOI] [PubMed] [Google Scholar]
- 26.Marmur, J. 1961. A procedure for isolation of deoxyribonucleic acid from bacterias. J. Mol. Biol. 3:208-218. [Google Scholar]
- 27.Miczák, A., I. Berek, and G. Ivánovics. 1976. Mapping the uroporphyrinogen decarboxylase, coproporphyrinogen oxidase and ferrochelatase loci in Bacillus subtilis. Mol. Gen. Genet. 146:85-87. [DOI] [PubMed] [Google Scholar]
- 28.Peters, W. J., and R. A. Warren. 1970. The mechanism of iron uptake in Bacillus subtilis. Can. J. Microbiol. 16:1179-1185. [DOI] [PubMed] [Google Scholar]
- 29.Porra, R. J., and O. T. G. Jones. 1963. Studies on ferrochelatase. 1. Assay and properties of ferrochelatase from a pig-liver mitochondrial extract. Biochemistry 87:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sarkany, R. P. 1999. Porphyria. From Sir Walter Raleigh to molecular biology. Adv. Exp. Med. Biol. 455:235-241. [PubMed] [Google Scholar]
- 32.Sassa, S. 2000. Hematologic aspects of the porphyrias. Int. J. Hematol. 71:1-17. [PubMed] [Google Scholar]
- 33.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 34.Smith, A. 1990. Transport of tetrapyrroles: mechanisms and biological and regulatory consequences, p. 435-490. In H. A. Dailey (ed.), Biosynthesis of heme and chlorophylls. McGraw-Hill Publishing Co., New York, N.Y.
- 35.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straka, J. G., J. R. Bloomer, and E. S. Kempner. 1991. The functional size of ferrochelatase determined in situ by radiation inactivation. J. Biol. Chem. 266:24637-24641. [PubMed] [Google Scholar]
- 37.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 38.Svensson, B., M. Lübben, and L. Hederstedt. 1993. Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Mol. Microbiol. 10:193-201. [DOI] [PubMed] [Google Scholar]
- 39.Tan, W. C., N. Krasner, and M. Lombard. 1997. Enhancement of photodynamic therapy in gastric cancer cells by removal of iron. Gut 41:14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winstedt, L., K.-I. Yoshida, Y. Fujita, and C. von Wachenfeldt. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, C.-K., H. A. Dailey, J. P. Rose, A. Burden, V. M. Sellers, and B.-C. Wang. 2001. The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat. Struct. Biol. 8:156-160. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]