Abstract
A degenerate set of PCR primers were used to clone a gene encoding a cytochrome P450 (the P450RhF gene) from Rhodococcus sp. strain NCIMB 9784 which is of unique primary structural organization. Surprisingly, analysis of the translation product revealed that the P450 is fused to a reductase domain at the C terminus which displays sequence conservation for dioxygenase reductase proteins. The reductase partner comprises flavin mononucleotide- and NADH-binding motifs and a [2Fe2S] ferredoxin-like center. The gene was engineered for heterologous expression in Escherichia coli, and conditions were found in which the enzyme was produced in a soluble form. A recombinant strain of E. coli was able to mediate the O dealkylation of 7-ethoxycoumarin in good yield, despite the absence of any recombinant redox proteins. This unprecedented finding leads us to propose that P450RhF represents the first example of a new class of cytochromes P450 in which the reducing equivalents are supplied by a novel reductase in a fused arrangement.
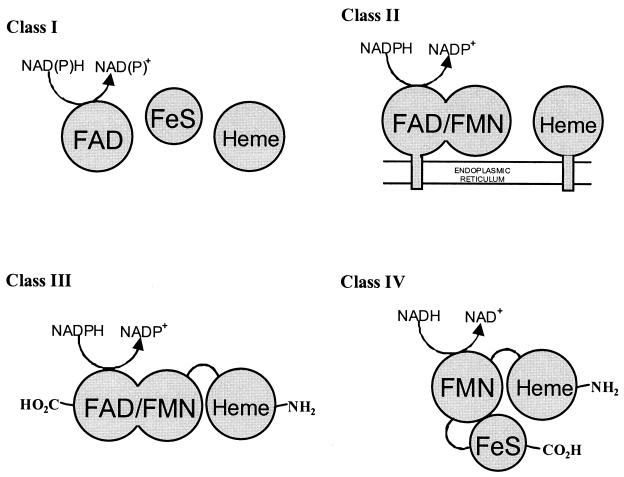
The cytochromes P450 are a diverse group of heme-containing enzymes which catalyze a wide range of oxidative reactions. They play a fundamental role in biochemistry, pharmacology, and toxicology and have been the subject of intense research ever since their discovery in 1958 (13). Substrates include natural compounds, such as steroids, fatty acids, pheromones, leukotrienes, and prostaglandins, as well as drugs and carcinogens. The primary chemical reaction catalyzed by these monooxygenases involves the activation of molecular oxygen, leading to the insertion of a single atom of oxygen into an organic substrate with concomitant reduction of the other atom to water. Electron equivalents are supplied by NAD(P)H via different redox partners. The cytochromes P450 generally fall into two broad classes, depending on the nature of the auxiliary protein(s) (23) (Fig. 1). Class I P450s, which are found on the membranes of mitochondria or in bacteria, are three-component systems comprising a flavin adenine dinucleotide (FAD)-containing reductase, an iron-sulfur protein (ferredoxin), and the P450. Class II cytochromes P450 are two-component systems made up of an FAD-containing, flavin mononucleotide (FMN)-containing NADPH-dependent cytochrome P450 reductase and a P450. These are typified by the liver microsomal enzymes in mammalian cells which are involved in both steroid metabolism and detoxification pathways.
FIG. 1.
Schematic representation of the different classes of cytochrome P450 systems. Class I systems comprise a FAD-containing flavodoxin reductase, an iron-sulfur protein (ferredoxin), and the P450. The eukaryotic class I enzymes are associated with the mitochondrial membrane. In a class II system, the P450 is partnered with a diflavin reductase, whereas in the class III system, the diflavin reductase is fused to the P450. We propose that the new class IV system is made up of an FMN-containing reductase with a ferredoxin-like center linked to a P450 in a single polypeptide. The N and C termini of the fused class III and IV systems are shown.
Probably the best characterized bacterial cytochrome P450 is P450cam from Pseudomonas putida, which catalyzes the regio- and stereospecific hydroxylation of (1R)-(+)-camphor to 5-exo-hydroxy camphor (28). The P450cam monooxygenase system consists of three soluble proteins: putidaredoxin reductase; putidaredoxin, an intermediary iron-sulfur protein; and the cytochrome P450cam. High-resolution X-ray crystal structures of P450cam have been determined for the both the substrate-free (27) and camphor-bound (29) forms. An immense amount of work has been carried out in studying the structure-function relationships of this system, and indeed, P450cam has now been established as the archetypal bacterial cytochrome P450.
Despite the identification and cloning of an enormous number of cytochromes P450 from bacterial sources, there have been very few examples which do not fall into the general class I category of enzymes. An exception is the soluble fatty acid ω-hydroxylase from Bacillus megaterium, P450BM3, which is a catalytically self-sufficient single-polypeptide enzyme containing binding sites for heme, FAD, and FMN in equimolar ratios and requiring NADPH as a source of electrons (22). This represents a fused system, made up of components of the eukaryotic class II enzyme joined by a short linker region (14). Because its primary structural organization is fundamentally different from those of the eukaryotic class II enzymes, P450BM3 is considered to constitute the first example of a class III cytochrome P450 (20). Two further P450 enzymes (CYP102A2 and CYP102A3) from Bacillus subtilis which display an overall organization similar to that of P450BM3 have been identified (16). Recently, two membrane-bound eukaryotic counterparts to P450BM3 have been cloned from Fusarium oxysporum (18) and Fusarium verticillioides (34).
We are interested in characterizing novel cytochrome P450 activities from bacteria of the genus Rhodococcus, since they are known to transform a wide range of xenobiotic compounds (12). Many of these biotransformations involve the introduction of an oxygen atom at an unactivated center, a reaction often catalyzed by a cytochrome P450. We are particularly interested in isolating cytochrome P450 activities from Rhodococcus sp. strain NCIMB 9784 (previously classified as Corynebacterium sp. strain T1), which has been reported to mediate several unusual oxidative reactions, including the hydroxylation of camphor at the 6-endo position (7). In common with most other bacterial P450s, those from nocardioform actinomycetes are of the classical class I system. However, in this paper we report the cloning and preliminary characterization of P450RhF, which consists of a cytochrome P450 fused to a dioxygenase reductase-like activity. The unprecedented structural organization of P450RhF leads us to propose that this enzyme constitutes a new class of cytochrome P450. In the new class IV enzyme, electrons are relayed to the active site of the P450 through an FMN center and a [2Fe2S] ferredoxin-like component, as outlined in Fig. 1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli XL1 Blue supercompetent cells were obtained from Stratagene (La Jolla, Calif.), and E. coli BL21(DE3) was from Invitrogen Life Technologies (Carlsbad, Calif.). Both strains were routinely grown in Luria-Bertani (LB) medium at 37°C unless stated otherwise. Ampicillin was used at 100 μg/ml when required for selection. Rhodococcus sp. strains NCIMB 9784, NCIMB 12038, and NCIMB 9703 were obtained from the National Collections of Industrial, Food and Marine Bacteria (Aberdeen, Scotland, United Kingdom). Rhodococcus erythropolis NI86/21 was purchased from the National Collection of Agricultural and Industrial Microorganisms (Budapest, Hungary) and Rhodococcus fascians ATCC 35014 and Rhodococcus erythropolis ATCC 4277 were obtained from the American Type Culture Collection (Manassas, Va.). Rhodococcus sp. strain IFO 3338 was from the Institute for Fermentation, Osaka (Osaka, Japan). All the Rhodococcus strains were maintained on nutrient agar slopes at 4°C and grown in LB medium at 30°C. A kit containing the pGEM-T plasmid for the direct cloning of PCR products was purchased from Promega (Madison, Wis.). The pET3a plasmid for the heterologous expression studies was obtained from Novagen (Darmstadt, Germany). pUC18 (37) was used as a general cloning vector.
Cloning procedures.
Genomic DNA was prepared from Rhodococcus spp. as described previously (15). A PCR-based methodology described by Hyun et al. (17) was used to amplify a portion of cytochrome P450-like genes from genomic DNA by using a degenerate pair of oligonucleotide primers (Invitrogen Life Technologies) designed to the oxygen- and heme-binding motifs of these enzymes. The oligonucleotides 5′-TSCTSCTSATCGCSGGSCACGAGAC-3′, which corresponds to the oxygen-binding motif, and 5′-GCSAGGTTCTGSCCSAGGCACTGGTG-3′, which corresponds to the complementary sequence of the heme ligand pocket (S = G or C), were used to amplify cytochrome P450 genes from several Rhodococcus spp. The primers were specifically designed to accommodate the highly biased codon usage pattern found in Rhodococcus strains. Conditions for amplification were identical to those used by Hyun et al. (17). The PCR products were analyzed by agarose gel electrophoresis and purified from the gel by using standard techniques (32) and then cloned directly into pGEM-T following the manufacturer's guidelines.
The 350-bp PCR product from Rhodococcus sp. strain NCIMB 9784 was radiolabeled with [α-32P]dCTP (3,000 Ci/mmol) by random priming using standard techniques (32). The labeled DNA was used to probe Southern blots of genomic DNA from Rhodococcus sp. strain NCIMB 9784 which had been digested with a number of different restriction endonucleases. However, even using very stringent hybridization and washing conditions, multiple bands were visible in each lane following autoradiography. In order to avoid the problem of multiple hybridization, a short oligonucleotide (P45) was designed from the DNA sequence of the cloned PCR product derived from Rhodococcus sp. strain NCIMB 9784 (5′-AGAACCGCGCGGTGTGGGAGGAGA-3′) to act as a hybridization probe. The oligonucleotide was radiolabeled by a kinase reaction using T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol) under standard conditions (32) and purified using a NICK column (Amersham Pharmacia Biotech, Uppsala, Sweden). Aliquots of genomic DNA from Rhodococcus sp. strain NCIMB 9784 were digested to completion with either BclI, KpnI, or SmaI. The digests were resolved by agarose gel electrophoresis and blotted onto Hybond-N membrane (Amersham Pharmacia Biotech) by using the capillary flow method (32). DNA was fixed to the membrane by using a UV cross-linker according to the manufacturer's guidelines (Stratagene). The blot was hybridized to the radiolabeled oligonucleotide at 56°C for 48 h. The membrane was washed twice at room temperature with 300 mM NaCl and 30 μM sodium citrate containing 0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 20 min and twice at 56°C in 30 mM NaCl and 3 μM sodium citrate containing 0.1% (wt/vol) SDS for 20 min. Following autoradiography, a single cross-hybridizing band was detected in each lane. Genomic DNA digested with either BclI or SmaI was run on a preparative agarose gel, and the region corresponding to the site of hybridization was gel eluted and shotgun cloned into pUC18 vector. Positive clones were subsequently isolated after performing a colony lift procedure by hybridization to the oligonucleotide probe under conditions identical to those used for the Southern blotting. Putative positives were verified by a dot blot procedure prior to sequencing.
Heterologous gene expression in E. coli.
The cloned Rhodococcus P450RhF gene was engineered for overexpression in E. coli BL21(DE3) by using the pET3a system (Novagen). pET3a contains a unique NdeI restriction site located at an optimal distance downstream of a T7-specific promoter. Cloning at this site allows high-level expression without translational fusion to vector coding sequences. Two PCR primers (5′-CGGTGTCCAT ATGAGTGCATCAGTTC CGGCG-3′ [forward] and 5′-AGGTTGATCATT CAGAGTCGCAGGGCCAGCC-3′ [reverse]) were used to amplify the entire open reading frame (ORF) from genomic DNA and at the same time engineer an NdeI restriction site to overlap the ATG initiation codon and introduce a BclI restriction site just 3′ of the TGA stop codon. In the mutagenic primer sequences above, nucleotides differing from the genomic DNA sequence are underlined and the relevant restriction sites are shown in bold type, here and in subsequent sequences appearing in the text. The PCRs consisted of 30 cycles, with denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and elongation at 72°C for 1 min 20 s. The initial denaturation step was at 95°C for 4 min. The PCR mix included 1.5 U of Vent DNA polymerase (New England Biolabs, Beverly, Mass.), 10 mM Tris-HCl (pH 9.0 at room temperature), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 40 pmol of each primer, 10% (vol/vol) dimethyl sulfoxide, and ∼50 ng of genomic DNA in a reaction volume of 50 μl. The PCR product was purified and digested with NdeI and BclI and then cloned into the NdeI-BamHI sites of pET3a to generate the expression construct pAG03. Following cloning, the insert DNA was sequenced to ensure that no mistakes had been introduced during amplification. Plasmid pAG03 was transformed into E. coli BL21(DE3) and plated onto LB medium supplemented with 100 μg of ampicillin/ml. Cells were grown in LB medium containing 100 μg of ampicillin/ml to an optical density of 1.0. Following induction with isopropyl-β-d-thiogalactopyranoside (IPTG) at 1 mM, the cells were grown for a further 2 h. Expression of the P450RhF gene was detected initially by analyzing cell extracts by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and comparing the banding pattern with that of a negative control. The solubility of the recombinant protein was assessed by standard methodology (32). For spectrophotometric analysis of the P450, cells were resuspended in a solution containing 50 mM Tris-HCl (pH 7.4), 5 mM EDTA (pH 7.4), 1 mM dithiothreitol, and 0.3 mM phenylmethylsulfonyl fluoride. After breaking the cells by sonication, the extract was centrifuged at 12,000 × g at 4°C (15 min). The supernatant was used to obtain a difference spectrum in the presence of CO.
Biotransformation.
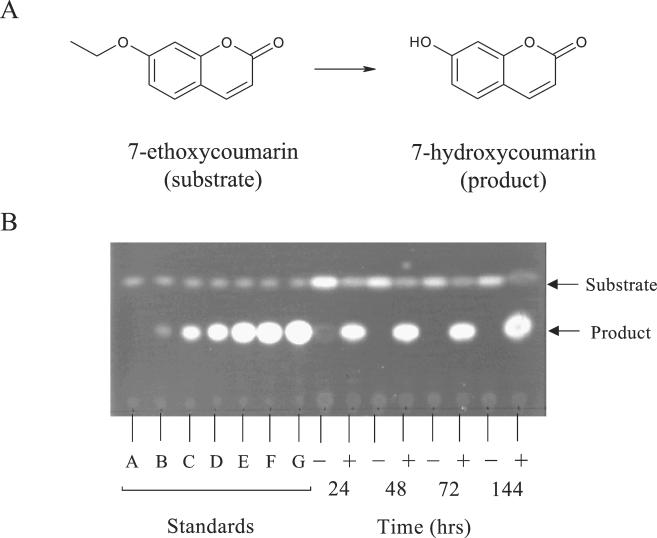
The E. coli expression strain [BL21(DE3)pAG03] was grown in a 25-ml culture of LB medium containing 100 μg of ampicillin/ml at 30°C. Strains containing the same plasmid without a DNA insert [BL21(DE3)pET3a] were used as a negative control. Typically, the substrate 7-ethoxycoumarin was present in the culture medium at the time of inoculation at a concentration of 0.5 mM. Aliquots (0.5 ml) of culture broth were analyzed after 24, 48, 72, and 144 h. Reactions were stopped by adding ethyl acetate and vortexing. The reaction products were extracted with ethyl acetate and separated from the starting material by silica gel thin-layer chromatography (TLC) using a solvent system of hexane-ethyl acetate (3:2, vol/vol). Both the substrate and product are fluorescent and could be readily detected by exposing the plates to UV light (300 to 320 nm). To act as standards, different concentrations of 7-hydroxycoumarin were prepared in the culture medium and extracted into ethyl acetate. The percentage conversion was estimated by normalizing against known concentrations of 7-hydroxycoumarin by using the ImageMaster TotalLab (version 1.0) software package (Amersham Pharmacia Biotech). The product of the biotransformation was isolated by preparative TLC for further analytical analysis.
DNA sequencing and analysis.
The dideoxy chain termination method (33) of DNA sequencing was performed throughout, and the reactions were resolved on an automated DNA sequencer (ABI PRISM 377; Perkin-Elmer Life Sciences). All sequencing was carried out on both strands. DNA sequences were determined using a primer-hopping strategy, with synthetic oligonucleotides as primers. Nucleotide sequence data were assembled and analyzed using the DNAStrider and MacVector software packages. Database homology searches (SWISSPROT release 40 protein database) were carried out using the BLASTP (2) program. Multiple sequence alignments were made with ClustalW, version 1.7 (36), using the default parameters.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been deposited at EMBL and GenBank with accession number AF459424.
RESULTS
PCR amplification of cytochrome P450-like gene fragments from Rhodococcus spp.
Initially we used a PCR-based (17) approach to amplify the gene(s) from genomic DNA encoding cytochrome P450 enzyme(s). This method relies on using a pair of degenerate PCR primers designed to the oxygen-binding motif and a second primer corresponding to the complementary sequence of the heme ligand pocket. The expected size of the PCR product, based on the average number of amino acid residues between the two motifs in this family of enzymes, was 350 bp. In addition to Rhodococcus sp. strain NCIMB 9784, we screened a further six different Rhodococcus strains, which acted as convenient controls. These were Rhodococcus sp. strain NCIMB 12038, Rhodococcus sp. strain NCIMB 9703, Rhodococcus erythropolis NI86/21, Rhodococcus fascians ATCC 35014, Rhodococcus erythropolis ATCC 4277, and Rhodococcus sp. strain IFO 3338. PCR products of the expected size were obtained from Rhodococcus sp. strain NCIMB 9784, Rhodococcus sp. strain NCIMB 12038, and Rhodococcus sp. strain IFO 3338. The PCR products were cloned into pGEM-T and sequenced. Translation of the sequences revealed the all three products displayed convincing homology to authentic cytochrome P450 enzymes in the SWISSPROT database (results not shown). However, as judged from analysis of 12 individual clones from each experiment, it appeared that only one cytochrome P450-like sequence was amplified from a given strain. Genome sequencing studies have shown that Mycobacterium tuberculosis CDC1551 (The Institute for Genome Research; www.tigr.org) encodes 20 cytochromes P450, and 18 P450 ORFs have been identified from the recently completed genomic sequence of Streptomyces coelicolor A3 (Sanger Institute Sequencing Project; www.sanger.ac.uk). Furthermore, four out of the seven strains failed to give a product, including Rhodococcus erythropolis NI86/21, which is known to encode at least one cytochrome P450 (21). Given the relatively low level of sequence identity between members of the cytochrome P450 family of enzymes, it appears that only a subset of cytochrome P450 genes are amplified using this pair of degenerate primers. Nevertheless, the technique was useful in the amplification of new and potentially interesting cytochrome P450 ORFs.
Cloning of the complete cytochrome P450 gene.
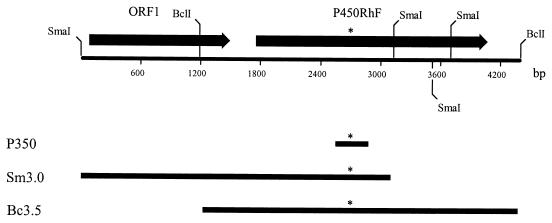
In order to clone the complete cytochrome P450 gene from Rhodococcus sp. strain NCIMB 9784, we initially used the corresponding 350-bp PCR product as a hybridization probe to screen genomic DNA by Southern blotting. Despite using very stringent hybridization and washing conditions, multiple cross-reacting bands were detected, indicating that similar sequences exist within the genome. In an attempt to overcome the problem of multiple hybridization, an oligonucleotide probe (P45) was synthesized based on the sequence of a portion of the PCR product. In designing the P45 hybridization probe, we were careful not to include sequence elements which could cause a reduction in the efficacy of probe-target interaction, such as an oligomer with the potential to form a stable hairpin structure. Southern blotting experiments using this probe resulted in a single specific cross-hybridizing band in each lane. Two positively hybridizing fragments of DNA, a 3.0-kbp SmaI and a 3.5-kbp BclI fragment, were subsequently cloned into pUC18 to give clones Sm3.0 and Bc3.5, respectively. Sequence analysis showed that together, the partially overlapping clones covered just over 4.4 kbp of chromosomal DNA (Fig. 2). The overall G+C content of the sequenced fragment was 69.7%, which is entirely consistent with the high G+C content of Rhodococcus spp. (12). A sequence similarity search using the BLAST program in the SWISSPROT database assisted in identifying two ORFs.
FIG. 2.
Restriction map of a 4.4-kbp segment of chromosomal DNA from Rhodococcus sp. strain NCIMB 9784 containing the P450RhF gene. ORFs are denoted by arrows, which indicate the direction of transcription. The relative positions of the three overlapping clones are shown by the thick bars below the restriction map. P350 represents the PCR product obtained by using a degenerate set of primers designed from the oxygen- and heme-binding motifs of cytochrome P450 enzymes. The asterisks denote the site from which an oligonucleotide hybridization probe (P45) was designed. This probe was then used to clone both Sm3.0 and Bc3.5 from a sublibrary of genomic DNA. Only the restriction sites mentioned in the text are shown.
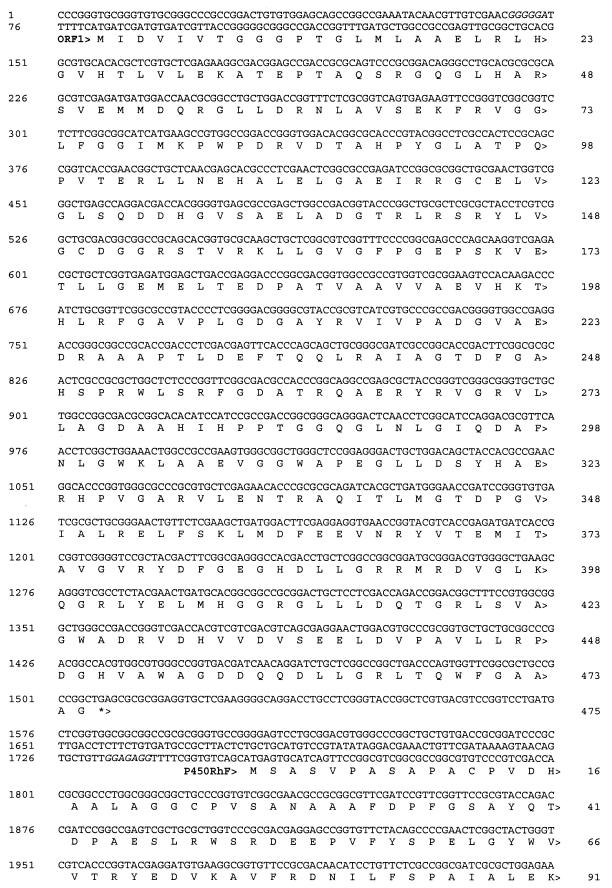
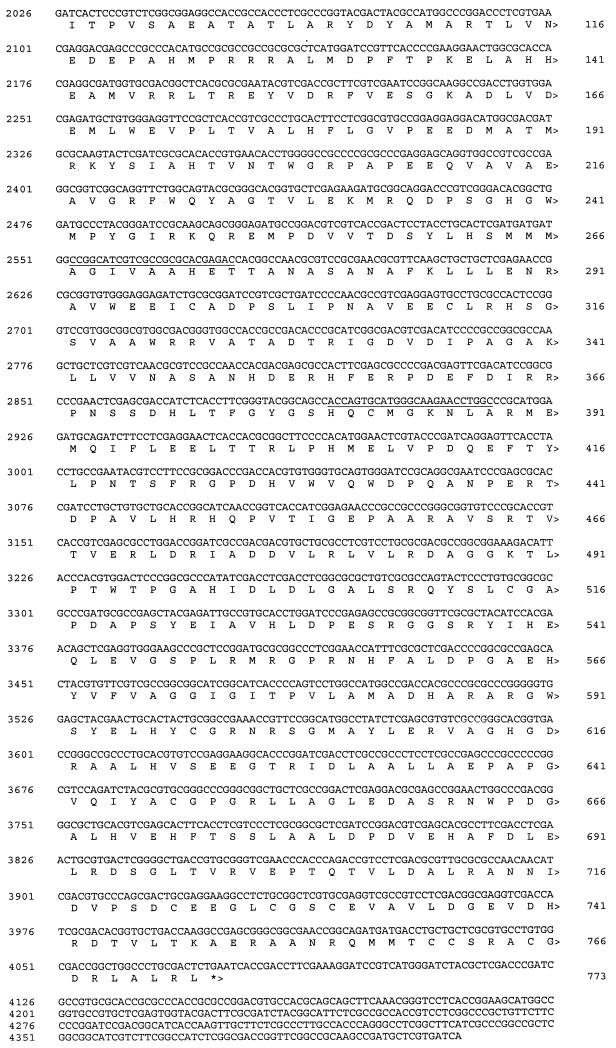
The site of hybridization of the P45 probe was identified by sequence analysis and is indicated by asterisks in Fig. 2. This forms part of an ORF (called P450RhF) which encodes a protein of 773 amino acids (Fig. 3) . The deduced ATG start codon is preceded by a plausible Shine-Dalgarno sequence (GGAGAGG) at a distance of 14 nucleotides. The distance between the start site and the ribosome-binding site is greater than that generally observed, but no other reasonable start codons could be found. The GTG codon for valine 25 was considered as a possible candidate, since it is located three nucleotides downstream of a purine-rich segment of the sequence. However, this was rejected, since analysis of both the codon usage pattern upstream of the potential start point and the sequence similarity of the translated product indicated that this was the coding sequence.
FIG. 3.
DNA sequence of the 4,411-bp SmaI-BclI region encoding the cytochrome P450 (P450RhF) from Rhodococcus sp. strain NCIMB 9784. The deduced ORFs are labeled, and the direction of transcription is indicated by arrowheads. Stop codons are denoted by asterisks, and potential ribosome binding sites are shown in italics. The sites of annealing of the PCR primers used to amplify the segment of DNA between the oxygen- and heme-binding sites are underlined.
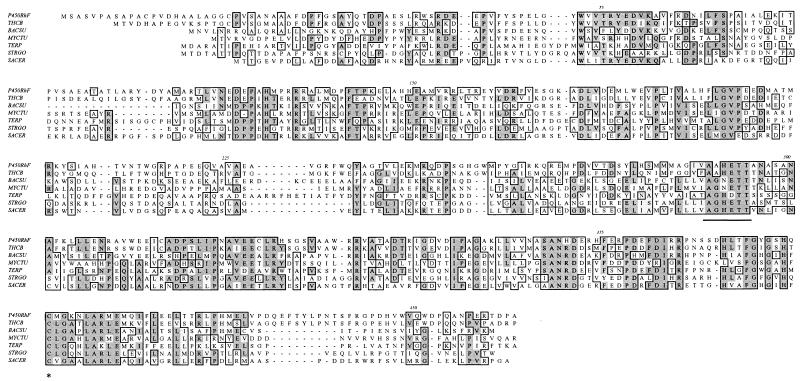
Comparison of the translated sequence with those of the SWISSPROT database shows that the N-terminal region (residues 1 to 444) displays convincing homology to authentic cytochrome P450 enzymes (Fig. 4). A high level of homology to the thiocarbamate-inducible cytochrome P450 (CYP116) from Rhodococcus erythropolis NI86/21 (21) was found (55% identity). The sequence 375FGYGSHQCMG matches the PROSITE consensus motif ([F/W][S/G/N/H]X[G/D]X[R/H/P/T]XC[L/I/V/M/F/A/P][G/A/D]) for cytochrome P450 enzymes, spanning the hydrophobic heme-ligand pocket. The region around the putative oxygen-binding site (271AAHETT) is also very highly conserved.
FIG. 4.
Amino acid sequence alignment of the N-terminal portion of P450RhF with those of other cytochrome P450 enzymes. The cytochrome P450-like region within P450RhF encompasses residues 1 to 444, the residues which have been used in this alignment. THCB, thiocarbamate-inducible cytochrome P450 from Rhodococcus erythropolis NI86/21 (21); BACSU, cytochrome P450 from Bacillus subtilis (1); MYCTU, putative cytochrome P450 from Mycobacterium tuberculosis H37Rv (8); TERP, terpene-inducible cytochrome P450 from Pseudomonas spp. (26); STRGO, cytochrome P450-SU2 from Streptomyces griseolus (24); SACER, cytochrome P450 107A1 from Saccharopolyspora erythraea (3). Regions of sequence conservation are highlighted in the boxed areas. The conserved oxygen-binding site is underlined. The conserved cysteine residue which provides the sixth ligand to the heme iron is highlighted with an asterisk.
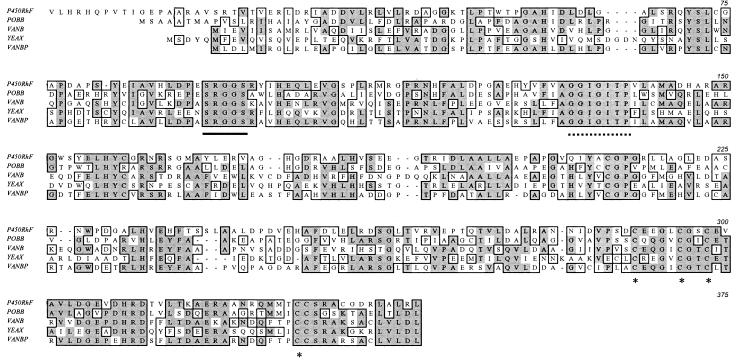
The C-terminal portion of the translation product of the P450RhF gene displays good homology to a number of oxygenase reductase proteins. An alignment of this sequence with those of four representative reductase subunits of oxygenases from various sources is shown in Fig. 5. For clarity, we have defined the C-terminal region of the protein as covering the section from residue 445 (i.e., the end of the cytochrome P450 homology) to residue 773. By homology to genes encoding proteins of known function, the C-terminal region can be subdivided into three distinct functional parts, an FMN-binding domain, a NAD+-binding domain, and a plant ferredoxin-type [2Fe2S] domain. The sequence 532SRGGS conforms to the consensus motif [G/S]RGGS, for binding the phosphate group of FMN. The fingerprint sequence GXGXXP for NADH binding can also be identified [571AGGIGITP] in a region of sequence that is highly conserved within the representative members of this class of enzyme. The crystal structure of the closely related phthalate dioxygenase reductase indicates that the proline residue in this sequence contacts the bound nicotinamide (9). At the C terminus, there are four highly conserved cysteine residues, which are highlighted in Fig. 5. The three cysteines which are clustered together in the primary amino acid sequence [722CEEGLCGSC] conform to the PROSITE consensus motif CXX[GA]XC[GAST]XC for a [2Fe2S] ferredoxin.
FIG. 5.
Amino acid sequence alignment of the C-terminal portion of P450RhF with various dioxygenase reductase subunits. For the purposes of this alignment, we have defined the C-terminal region of P450RhF as extending from residue 445 to residue 773. POBB, phenoxybenzoate dioxygenase beta subunit from Pseudomonas pseudoalcaligenes (11); VANB, vanillate O demethylase oxidoreductase from Pseudomonas sp. strain HR199 (30); YEAX, putative dioxygenase beta subunit from E. coli (5); VANBP, vanillate O demethylase oxidoreductase from Pseudomonas ATCC 19151 (6). Regions of sequence conservation are highlighted in the boxed areas. The FMN-binding motif is underlined, and the NADH-binding motif is indicated by a dotted line. The four cysteine residues which are involved in binding the iron-sulfur cluster are highlighted with asterisks.
A second ORF just upstream of P450RhF was identified which encodes a protein of 475 amino acids (Fig. 3). The deduced ATG start codon is positioned 6 bp 3′ of a purine-rich segment [GGGGGA] which may act as a ribosome-binding site. When the translated sequence was screened against the SWISSPROT database, it was found to display significant resemblance to those of several flavin-type bacterial hydroxylases, such as the pentachlorophenol-4-monooxygenase from a Flavobacterium sp. (25) (31% identity) or the hydroxylase TcmG of the tetracenomycin C biosynthetic gene cluster from Streptomyces glaucescens (10) (25% overall identity; similarity confined to the N terminus) (data not shown). Alignment of the N-terminal region of the protein revealed a sequence [8GGGPTGLMLAAELRLHG] which corresponds to the canonical motif [GXGXXGXXXAXXXXXXG] for flavin nucleotide binding (19).
Heterologous expression of the P450RhF gene.
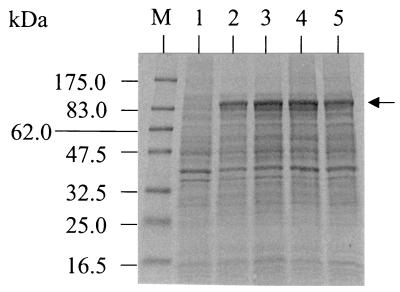
The expression construct (pAG03) was transformed into E. coli BL21(DE3), and the cell extract was analyzed by SDS-PAGE. A protein of the expected size (∼85 kDa) was readily detected on a Coomassie-stained gel from an extract of E. coli BL21(DE3)pAG03 which was not observed in the negative control of the same strain harboring pET3a (Fig. 6). No detectable difference in the observed levels of heterologous expression was seen between IPTG-induced and noninduced cells, which indicates a degree of leakiness within the system. For this reason, all subsequent expression experiments were carried out on uninduced cell cultures. When the cells were grown at a temperature of 37°C, most of the P450RhF appeared in the insoluble fraction, whereas the protein was largely soluble in cells grown at 30°C.
FIG. 6.
SDS-PAGE of cell extracts showing the expression of the P450RhF gene in E. coli. Coomassie-stained 10 to 20% gradient polyacrylamide gel of whole-cell extracts. Two expression clones, both E. coli BL21(DE3)pAG03, were analyzed following growth at 30°C in the absence of IPTG (lanes 2 and 4). There was no significant difference in the banding profiles following induction with IPTG (lanes 3 and 5, respectively). Lane 1 shows the results for the negative control, E. coli BL21(DE3)pET3a, following incubation with IPTG. The arrow indicates the position of the recombinant protein. Molecular mass markers are shown in lane M.
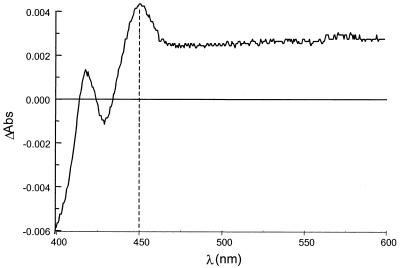
Spectrophotometric evidence for the presence of a P450 enzyme in a crude cell extract of E. coli BL21(DE3)pAG03 was obtained from a CO-induced difference spectrum (Fig. 7). A peak at 450 nm was observed only for cells harboring pAG03, which is characteristic of a cytochrome P450 enzyme.
FIG. 7.
Difference spectrum in the presence of CO for a crude cell extract of E. coli BL21(DE3)pAG03. The peak at 450 nm is highlighted.
Whole-cell biotransformation.
Several reports in the literature have cited the ability of cytochrome P450 activities to mediate the O dealkylation of 7-ethoxycoumarin (3). This reaction has the advantage that both the substrate and expected product are strongly fluorescent and can be readily separated by TLC. Since the expression system was leaky, cells were grown in the presence of 7-ethoxycoumarin (0.5 mM). The results we observed using the expression strain [BL21(DE3)pAG03] were typically of a 40 to 50% conversion of 7-ethoxycoumarin to 7-hydroxycoumarin within 144 h (Fig. 8). There was no detectable formation of product in the negative control [BL21(DE3)pET3a] over the same period of time, although there was a gradual loss of substrate, probably caused by evaporation during the course of the fermentation. The identity of the product of the biotransformation was subsequently confirmed to be 7-hydroxycoumarin by electrospray mass spectrometry (data not shown). Similar experiments using Rhodococcus sp. strain NCIMB 9784 showed that the cells were capable of performing the biotransformation, but the yield of product was considerably lower than in the recombinant system (data not shown).
FIG. 8.
Biotransformation of 7-ethoxycoumarin by a recombinant strain of E. coli expressing the P450RhF gene. (A) Dealkylation of 7-ethoxycoumarin to form 7-hydroxycoumarin. (B) TLC analysis of ethyl acetate extracts from the culture broth of either the expression strain [BL21(DE3)pAG03] (marked with a “+”) or the negative control [BL21(DE3)pET3a] (marked “−”). The time values indicate the number of hours following inoculation. Substrate (0.5 mM) was added to the medium at the time of inoculation. Different concentrations of 7-hydroxycoumarin were prepared in the culture medium and then extracted into ethyl acetate to act as standards. These were normalized to allow estimation of the percentage of conversion of substrate to product. The concentration (mM) of each standard was as follows: A, blank; B, 0.01; C, 0.05; D, 0.10; E, 0.25; F, 0.50; and G, 1.00.
Since the gene was cloned without any prior knowledge of its protein product, it is not immediately apparent which compounds may act as suitable substrates. It is known that Rhodococcus sp. strain NCIMB 9784 is able to utilize camphor as a carbon source and that the first step in its degradation is a hydroxylation at the 6-endo position (7, 15). However, we could not detect any biotransformation of camphor using the E. coli expression strain (A. Celik, personal communication).
DISCUSSION
We have used a generic PCR-based methodology to clone genes encoding cytochrome P450 activities from Rhodococcus spp. Our results using one pair of degenerate PCR primers suggest that only a subset of target sequences are amplified. This result presumably arises from the diversity that exists in the sequence of the target DNA even when care is taken to identify regions of the P450 gene with the greatest degree of conservation and the least codon redundancy. Nevertheless, this technique allowed the rapid cloning of novel cytochrome P450 genes which may otherwise not have been identified using a classical enzyme activity screen, especially where the basal level of expression is very low. In this study, one gene in particular (the P450RhF gene), from Rhodococcus sp. strain NCIMB 9784, was of immense interest due to its unprecedented structural organization.
Cytochrome P450 activities have been isolated from a wide range of bacterial sources, but to our knowledge, P450BM3 (22) from Bacillus megaterium and its homologs CYP102A2 and CYP102A3 from Bacillus subtilis (16) are presently the only examples of self-sufficient cytochromes P450. These enzymes do not require the participation of additional proteins to transfer electrons from the reduced pyridine nucleotide to the active site. In this paper we describe the cloning and heterologous expression in E. coli of the P450RhF gene, encoding a cytochrome P450 which is fused to a unique reductase activity. There are several noteworthy features of this novel enzyme.
In common with P450BM3, the cytochrome P450 activity of P450RhF is located at the N-terminal region. An alignment of this portion of P450RhF with the database indicates significant homology to authentic cytochromes P450 and in particular to the class I thiocarbamate-inducible P450 (CYP116) from Rhodococcus erythropolis NI86/21 (21). This leads us to propose that P450RhF constitutes a new member of the CYP116 family of enzymes. The high degree of sequence conservation is difficult to rationalize at this time but may be due to the close evolutionary relationship of these two organisms within the same taxon. The C-terminal reductase portion of P450RhF comprises an FMN-binding, NADH-binding, and [2Fe2S] ferredoxin center and closely resembles a dioxygenase reductase. This portion of P450RhF differs markedly from that of P450BM3, which lacks an iron-sulfur component and is generally recognized as being a fused class II enzyme system. In contrast, P450RhF is more analogous to a fused version of the classical prokaryotic cytochrome P450 system, although in this case the electrons are presumably transferred through the FMN semiquinone rather than FAD. The reductase component is significantly smaller than its BM3 counterpart, which means that there is a large difference between the overall size of the polypeptide P450RhF (85 kDa) and that of self-sufficient P450BM3 (119 kDa).
Examination of the sequence of P450RhF indicates that there is only a short segment of about 16 amino acids (residues 445 to 460) between the end of the cytochrome P450-like sequence and the start of the reductase-like sequence. This length is similar to that of the linker region separating the N-terminal P450 domain from the C-terminal reductase portion of P450BM3 (14), although there is no apparent sequence homology.
Nothing is known of the natural function of this cytochrome P450 activity in the organism or of its likely substrate specificity. Sequence analysis of the cloned segment of the DNA containing the P450RhF gene shows that immediately upstream is an ORF encoding a flavin-type monooxygenase. Comparison with related sequences from other organisms suggests that the gene product could be involved in the metabolism of aromatic or possibly polychlorinated aromatic compounds, although this may not be related to the role of P450RhF in the cell.
To investigate the activity of the gene product, we engineered the P450RhF gene into an expression system in E. coli. Using the recombinant system, a number of potential substrates were tested in whole-cell biotransformations, most of which gave negative results (A. Celik, personal communication). However, a strain of E. coli expressing the P450RhF gene was able to mediate the O dealkylation of 7-ethoxycoumarin in good yield (∼40%) in the absence of any recombinant ferredoxin or ferredoxin reductase activity. Control cultures of cells lacking the P450RhF gene gave no detectable product. This seems to suggest that P450RhF is a self-sufficient cytochrome P450 and analogous in this respect to P450BM3, although we cannot rule out the possibility that P450RhF may obtain reducing equivalents from endogenous electron transport proteins within E. coli. Further studies are required to establish whether the enzyme is able to mediate catalysis without the participation of other redox partners. We are presently purifying the recombinant enzyme to allow for a complete biochemical characterization.
Interestingly, a class I P450 has been purified from Rhodococcus sp. strain NCIMB 9784 which catalyzes the hydroxylation of camphor at the 6-endo position (G. Grogan, personal communication). Furthermore, a gene which appears to encode a ferredoxin reductase activity has been cloned from this organism (15). Therefore, within the same host both a class I and the new class IV P450 system coexist.
One intriguing implication of this work is the possibility that class I P450 enzymes may furnish electrons from sources other than the previously recognized redox partners (i.e., the ferredoxin/ferredoxin reductase couple). Proteins homologous in sequence to the C-terminal reductase portion of P450RhF are found in a diverse range of microorganisms. For example, a hypothetical oxidoreductase from M. tuberculosis CDC1551 (accession number AAK47165), identified as a result of a whole-genome sequencing project, displays 34% identity to the C-terminal region (residues 445 to 773) of P450RhF (alignment not shown). In the present paper, an oxidoreductase of this type has been implicated in functioning as a redox partner to a P450 by virtue of a natural gene fusion. We therefore speculate that in M. tuberculosis CDC1551, the electron transfer protein may act as redox partner to one or more of the 20 known P450 enzymes from this strain. Indeed, this could constitute yet another class of cytochrome P450 system in which the P450 and P450RhF reductase-like elements are encoded by discrete genes.
Within the area of biocatalysis, there is enormous interest in generating efficient P450 enzymes with tailored substrate specificities. A major obstacle to achieving this goal has been the complexity of an enzyme system involving auxiliary redox proteins for delivering electrons to the catalytic center. Efforts to construct artificial self-sufficient P450 catalytic systems have mainly focused on membrane-bound enzymes (31, 38), although there are examples of class I triple fusions (4, 35). Despite being of immense interest, this approach has not proved successful in producing enzymes with catalytic utility. It would be intriguing to investigate the possibility of generating chimeric proteins by genetically fusing different P450 enzymes to the reductase portion of P450RhF. If successful, this could provide a means of generating novel enzymes with potentially high catalytic activity due to the fused arrangement of redox partners. A similar approach, in which chimeric proteins were formed by fusing a P450 to the diflavin reductase of P450BM3, has generally been unsuccessful, since in most cases the pathway of electron relay failed to function effectively (S.K. Chapman, personal communication). However, the interaction of the P450RhF reductase with its P450 partner may be naturally more malleable to manipulation, enabling the P450 domain to be swapped without necessarily hindering the electron transfer process.
Acknowledgments
We gratefully acknowledge the Biotechnology and Biological Sciences Research Council (BBSRC) and Department of Trade and Industry (DTI) for funding under the aegis of a LINK programme. We also thank the Edinburgh Protein Interaction Centre (Wellcome Trust) and the European Community Socrates Programme for additional funding. The project, which is in collaboration with University College London, is also financially supported by GlaxoSmithKline (GSK), Pfizer, and Ultrafine Chemicals.
The DNA sequencing was carried out by Anne White at DNASHEF Technologies (Edinburgh Royal Infirmary, Edinburgh, Scotland). We thank Steve Chapman, Tobe Ost, Ayhan Celik (all from University of Edinburgh), and Mike Dawson (GSK) for their valuable comments and assistance during the preparation of the manuscript.
REFERENCES
- 1.Ahn, K. S., and R. G. Wake. 1991. Variations and coding features of the sequence spanning the replication terminus of Bacillus subtilis 168 and W23 chromosomes. Gene 98:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. F., and C. R. Hutchinson. 1992. Characterization of Saccharopolyspora erythraea cytochrome P450 genes and enzymes, including 6-deoxyerythronolide B hydroxylase. J. Bacteriol. 174:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, S. M., J. A. Harikrishna, G. D. Szklarz, and W. L. Miller. 1994. The mitochondrial environment is required for activity of the cholesterol side-chain cleavage enzyme, cytochrome P450scc. Proc. Natl. Acad. Sci. USA 91:7247-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Brunel, F., and J. Davison. 1988. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J. Bacteriol. 170:4924-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, P. J., G. Meerman, I. C. Gunsalus, R. Srinivasan, and K. L. Rinehart, Jr. 1966. New acyclic acid metabolite in camphor oxidation. J. Am. Chem. Soc. 88:618-619. [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Correll, C. C., C. J. Batie, D. P. Ballou, and M. L. Ludwig. 1992. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 258:1604-1610. [DOI] [PubMed] [Google Scholar]
- 10.Decker, H., H. Motamedi, and C. R. Hutchinson. 1993. Nucleotide sequences and heterologous expression of tcmG and tcmP, biosynthetic genes for tetracenomycin C in Streptomyces glaucescens. J. Bacteriol. 175:3876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehmel, U., K. H. Engesser, K. N. Timmis, and D. F. Dwyer. 1995. Cloning, nucleotide sequence, and expression of the gene encoding a novel dioxygenase involved in metabolism of carboxydiphenyl ethers in Pseudomonas pseudoalcaligenes POB310. Arch. Microbiol. 163:35-41. [DOI] [PubMed] [Google Scholar]
- 12.Finnerty, W. R. 1992. The biology and genetics of the genus Rhodococcus. Annu. Rev. Microbiol. 46:193-218. [DOI] [PubMed] [Google Scholar]
- 13.Garfinkel, D. 1958. Pig-liver microsomes. I. Enzymic and pigment composition of different microsomal fractions. Arch. Biochem. Biophys. 77:493-509. [DOI] [PubMed] [Google Scholar]
- 14.Govindaraj, S., and T. L. Poulos. 1996. Probing the structure of the linker connecting the reductase and heme domains of cytochrome P450BM-3 using site-directed mutagenesis. Protein Sci. 5:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grogan, G., G. A. Roberts, D. Bougioukou, N. J. Turner, and S. L. Flitsch. 2001. The desymmetrization of bicyclic beta-diketones by an enzymatic retro-claisen reaction. A new reaction of the crotonase superfamily. J. Biol. Chem. 276:12565-12572. [DOI] [PubMed] [Google Scholar]
- 16.Gustafsson, M. C. U. 2000. Ph.D. thesis. Bacterial cytochromes P450: studies on cytochrome P450 102A2 and P450 102A3 of Bacillus subtilis. Lund University, Lund, Sweden.
- 17.Hyun, C. G., J. M. Kim, S. K. Hong, and J. W. Suh. 1998. An efficient approach for cloning P450 hydroxylase genes from actinomycetes. J. Microbiol. Biotechnol. 8:295-299. [Google Scholar]
- 18.Kitazume, T., N. Takaya, N. Nakayama, and H. Shoun. 2000. Fusarium oxysporum fatty-acid subterminal hydroxylase (CYP505) is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J. Biol. Chem. 275:39734-39740. [DOI] [PubMed] [Google Scholar]
- 19.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 20.Miles, C. S., T. W. B. Ost, M. A. Noble, A. W. Munro, and S. K. Chapman. 2000. Protein engineering of cytochrome P450. Biochim. Biophys. Acta 1543:383-407. [DOI] [PubMed] [Google Scholar]
- 21.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. De Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narhi, L. O., and A. J. Fulco. 1986. Characterization of a catalytically self-sufficient 119,000 dalton cytochrome P450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 261:7160-7169. [PubMed] [Google Scholar]
- 23.Nebert, D. W., and F. J. Gonzalez. 1987. P450 genes: structure, evolution and regulation. Annu. Rev. Biochem. 56:945-993. [DOI] [PubMed] [Google Scholar]
- 24.Omer, C. A., R. Lenstra, P. J. Litle, C. Dean, J. M. Tepperman, K. J. Leto, J. A. Romesser, and D. P. O'Keefe. 1990. Genes for two herbicide-inducible cytochromes P450 from Streptomyces griseolus. J. Bacteriol. 172:3335-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orser, C. S., C. C. Lange, L. Xun, T. C. Zarht, and B. J. Schneider. 1993. Cloning, sequence analysis and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson, J. A., J. Y. Lu, J. Geisselsoder, S. Graham-Lorence, C. Carmona, F. Witney, and M. C. Lorence. 1992. Cytochrome P450terp. Isolation and purification of the protein and cloning and sequencing of its operon. J. Biol. Chem. 267:14193-14203. [PubMed] [Google Scholar]
- 27.Poulos, T. L., B. C. Finzel, and A. J. Howard. 1986. Crystal structure of substrate-free Pseudomonas putida cytochrome P450. Biochemistry 25:5314-5322. [DOI] [PubMed] [Google Scholar]
- 28.Poulos, T. L., B. C. Finzel, and A. J. Howard. 1987. High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 195:687-700. [DOI] [PubMed] [Google Scholar]
- 29.Poulos, T. L., B. C. Finzel, I. C. Gunsalus, G. C. Wagner, and J. Kraut. 1985. The 2.6 Å crystal structure of Pseudomonas putida P450. J. Biol. Chem. 260:16122-16130. [PubMed] [Google Scholar]
- 30.Priefert, H., J. Rabenhorst, and A. Steinbuchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakaki, T., S. Kominami, S. Takemori, H. Ohkawa, M. Akiyoshi-Shibata, and Y. Yabusaki. 1994. Kinetic studies on a genetically engineered fused enzyme between rat cytochrome P4501A1 and yeast NADPH-P450 reductase. Biochemistry 33:4933-4939. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo, J.-A., R. H. Proctor, and R. D. Plattner. 2001. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 34:155-165. [DOI] [PubMed] [Google Scholar]
- 35.Sibbesen, O., J. J. De Voss, and P. R. Ortiz de Montellano. 1996. Putidaredoxin reductase-putidaredoxin-cytochrome P450cam triple fusion protein. Construction of a self-sufficient Escherichia coli catalytic system. J. Biol. Chem. 271:22462-22469. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 38.Wada, A., P. A. Mathew, H. J. Barnes, D. Sanders, R. W. Estabrook, and M. R. Waterman. 1991. Expression of functional bovine cholesterol side chain cleavage cytochrome P450 (P450scc) in Escherichia coli. Arch. Biochem. Biophys. 290:376-380. [DOI] [PubMed] [Google Scholar]