Abstract
In this work we used a new strategy designed to reduce the size of the library that needs to be explored in family shuffling to evolve new biphenyl dioxygenases (BPDOs). Instead of shuffling the whole gene, we have targeted a fragment of bphA that is critical for enzyme specificity. We also describe a new protocol to screen for more potent BPDOs that is based on the detection of catechol metabolites from chlorobiphenyls. Several BphA variants with extended potency to degrade polychlorinated biphenyls (PCBs) were obtained by shuffling critical segments of bphA genes from Burkholderia sp. strain LB400, Comamonas testosteroni B-356, and Rhodococcus globerulus P6. Unlike all parents, these variants exhibited high activity toward 2,2′-, 3,3′-, and 4,4′-dichlorobiphenyls and were able to oxygenate the very persistent 2,6-dichlorobiphenyl. The data showed that the replacement of a short segment (335TFNNIRI341) of LB400 BphA by the corresponding segment (333GINTIRT339) of B-356 BphA or P6 BphA contributes to relax the enzyme toward PCB substrates.
Biphenyl dioxygenase (BPDO) is the first enzyme of the biphenyl (BPH) catabolic pathway. BPDO comprises three components: the iron-sulfur oxygenase (ISPBPH) made up of α (Mr = 51,000) and β (Mr = 22,000) subunits, the ferredoxin (FERBPH, Mr = 12,000), and the ferredoxin reductase (REDBPH, Mr = 43,000). The encoding genes for both Burkholderia sp. strain LB400 (15, 16) and Comamonas testosteroni B-356 (35) are bphA (ISPBPH α subunit), bphE (ISPBPH β subunit), bphF (FERBPH), and bphG (REDBPH). ISPBPH catalyzes a 2,3-dihydroxylation of BPH. BPDO is of particular interest because of its potential application as biocatalyst to oxygenate priority pollutants such as polychlorinated BPHs (PCBs) or to manufacture fine chemicals.
Despite their nearly identical amino acid sequences, the LB400 (15) and Pseudomonas pseudoalcaligenes strain KF707 BPDOs (36) display distinct ranges of PCB substrate (28). LB400 BPDO oxygenates 2,2′-dichlorobiphenyl (2,2′CB) and exhibits a 3,4-oxygenation of 2,2′,5,5′CB, whereas KF707 BPDO oxygenates 4,4′CB and is poorly active toward the ortho-substituted congeners (28). C. testosteroni B-356 and Rhodococcus globerulus P6 BPDOs, which are more distantly related to KF707 and LB400 BPDOs, poorly catalyze the oxygenation of para-substituted congeners and degrade fairly well 3,3′CB (11, 19). There is 75% amino acid sequence identity between the B-356 and LB400 BphAs (35), 64% amino acid sequence identity between P6 BphA1 and LB400 BphA (2), and 65% amino acid sequence identity between P6 BphA1 and B-356 BphA (35).
Variant BPDOs that had inherited the catalytic features of both parents and exhibiting extended activities toward several tetra- and penta-substituted congeners were obtained by shuffling the very closely related LB400 bphA with KF707 bphA1 (9, 23). However, no novel BPDO has yet been described which can efficiently catalyze oxygenation of the most persistent congeners.
Family shuffling of genes of lesser homology increases the sequence diversity of the library, resulting in an accelerated rate of enzyme functional improvement (12). It is also a powerful tool to identify the major structural features of a protein family that confer a desired phenotype. However, as the sequence space to explore increases, a larger proportion of the progeny members are prone to be inactive (37), which implies that the screening assay needs to be strongly selective or based on a microarray design. In order to reduce the size of the library that needs to be explored in family shuffling of bphA of lesser homology, we targeted a portion of BphA that is critical for substrate specificity and selectivity. The current approach was based on strong evidence that structural features of the C-terminal portion of BphA influence the regioselectivity and regiospecificity of the enzyme (9, 23, 28). Furthermore, active BphA hybrids were recently obtained by replacing long stretches of LB-400 bphA encoding the C-terminal portion of the protein by the corresponding stretches of B-356 bphA (4). The purpose of this investigation was to shuffle targeted stretches of bphA genes of lesser homology in order to obtain better-performing BphA variants and to identify some of the major structural features of the C-terminal portion of BphA that contribute to relax the enzyme toward PCB congeners.
Current protocols to screen for clones expressing active BPDO rely on a trans-complementation assay involving the 2,3-dihydro-2,3-dihydroxybiphenyl 2,3-dehydrogenase (BphB) and the 2,3-dihydroxybiphenyl 2,3-dioxygenase (BphC) to transform the substrate into the yellow 2-hydroxy-6-oxo-6-phenyl-hexa-2,4-dienoic acid (HOPDA) (9, 23). However, although BphB is quite relaxed (5), BphC is much more specific (14). A more recent protocol, which does not require a meta-fission dioxygenase, relies on the Gibbs reagent to detect phenols produced from oxygenation of the aromatic substrates (21). However, the Gibbs reagent reacts poorly with bicyclic phenols (29). In the present study, we describe a new screening protocol for the detection of catechol metabolites from CBs based on nonenzymatic oxidation of catechols to dark-colored metabolites. Because the ortho-substituted PCB congeners are among the most resistant to microbial attack, the evolved BPDOs were screened for their ability to oxygenate 2,2′CB, and the evolved variants showing the highest potency toward PCB congeners were then characterized.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and general protocols.
Escherichia coli DH11S (24) was used in this study. Several plasmids were used. pDB31[B-356-bphA], pDB31[LB400-bphA], pDB31[B-356-bphAE], and pDB31[LB-400-bphAE] in which bphA was mutated to introduce an AvrII site at positions 1354 for LB400 and 1348 for B-356 bphA were as described previously (4). pQE31[P6-bphA1] was also described previously (11). pDB31[B-356-bphAE] and pDB31[LB-400-bphAE] express His-tagged ISPBPH (ht-ISPBPH) (4, 19) carrying the His tag on BphA. pQE51[LB400-bphFGB] was obtained by deleting a 856-bp ClaI/NdeI fragment from pQE51[LB400-bphFGBC] described previously (11). DNA general protocols were done according Sambrook et al. (30). DNA was sequenced at the INRS-Institut Armand-Frappier DNA sequencing service (Laval, Quebec, Canada). Previously described procedures (5, 18) were used to express in E. coli and purify by affinity chromatography the ht-BPH catabolic enzymes. Reconstituted BPDOs comprised ht-ISPBPH plus ht-B-356 FERBPH and ht-B-356 REDBPH. 2,3-Dihydroxybiphenyl, 1,2-dihydroxynaphthalene, 3,4-dihydroxy-2,2′,5,5′CB, and 2,3-dihydroxy-2′CB were produced enzymatically according to protocols described previously (5, 11). Their identity was assessed by gas chromatography-mass spectrometry (GC-MS) analyses (11). PCB congeners, 2,3-dihydroxybenzene and 3,4-dihydroxybiphenyl, were from ULTRA Scientific, North Kingstown, R.I.
Monitoring enzyme activity and metabolite analysis.
Enzyme assays were performed in a 200-μl volume in 100 mM morpholineethanesulfonic acid buffer (pH 6.0) (18). The reactions were initiated by adding 100 nmol of substrate dissolved in acetone. The catalytic oxygenation of BPH was evaluated spectrophotometrically at 434 nm from the production of HOPDA in a coupled reaction system containing purified BPDO components plus excess amounts of purified ht-B-356 BphB and ht-B-356 BphC as described previously (18). Assays for metabolite identification were done under conditions identical to those described above except for the absence of BphB and BphC in the reaction medium. Metabolites were extracted at pH 6.0 with ethyl acetate, treated with butylboronate, and analyzed by GC-MS (5).
Site-directed mutagenesis.
Site directed-mutagenesis to replace A267 of variant II-9 by a serine was carried out by using Pharmacia-Biotech's unique site elimination mutagenses kit according to the protocol of Wang and Sul (38). The introduction of the mutation was assessed by nucleotide sequence analysis.
Preparation of DNA and PCRs for DNA shuffling and screening protocol.
A 1,037- or a 1,043-bp fragment of B-356 bphA or LB400 bphA was amplified from pDB31[bphA] by using primers 1 (5′-ACGGCTGGGCCTACGACATC-3′) and 2 (5′-CGTTGTTGCCATTGCTGCAGG-3′) external to the unique MluI-AvrII sites (Fig. 1); a 513-bp fragment of B-356 bphA (from bp 368 to the unique ScaI site) was obtained by treating the 1,037-bp fragment with ScaI; a 723-bp stretch of LB400 bphA (from bp 688 to 30 bp downstream of the stop codon in pDB31) and the corresponding 717-bp fragment of B-356 bphA were both amplified from pDB31[bphA] with primers 2 and 3 (5′-GACATGTACCACGCCGGCAC-3′); a 720-bp fragment from P6 bphA1 (from bp 664 to the stop codon) was amplified from pQE31[P6-bphA1] with primers 4 (5′-TATGTACCACGTCGGCAC-3′) and 5 (5′-GGGGTACCCCTCAGGCCGAGACATC-3′).
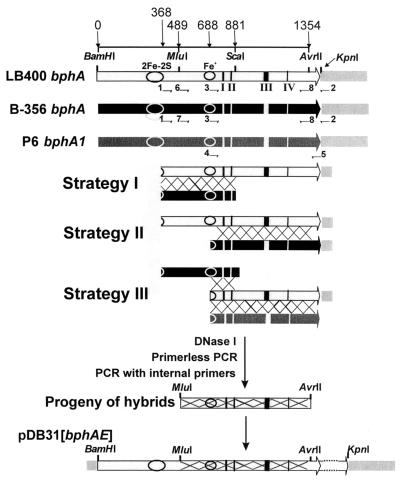
FIG. 1.
Strategies to evolve LB400 BphA. The constructions used for shuffling are drawn in the top part of the figure. These are LB400 and B-356 bphA cloned in pDB31 and P6 bphA1 cloned in pQE31. Large arrows represent genes; the gray box on the right side of the genes represents the vectors. Portions of LB400, B-356 bphA, or P6 bphA1 were PCR amplified with appropriate primers (indicated as small horizontal arrows) as described in the text. The appropriate DNA fragments for each of the three shuffling strategies were digested with DNase I and reassembled by primerless PCR. The primerless PCR products were amplified with internal primers (primers 6 or 7 and primer 8) to generate the libraries of MluI/AvrII hybrid fragments that were used to replace the corresponding fragment of LB400 bphA in pDB31[LB400-bphAE]. The numbers on top of the figure indicate the base pair positions in LB400 bphA. The indicated restriction sites are unique and common to both LB400 and B-356 bphA. Circles indicate the position of the 2Fe-2S Rieske center and the mononuclear Fe+ active center. I, II, III, and IV refer to the localization of the designated bphA regions I, II, III, and IV, respectively, of Mondello et al. (27).
The DNA fragments were treated with DNase I to prepare the 50- to 100-bp fragments for DNA shuffling (39). Taq DNA polymerase was used for the primerless PCR step. Conditions were as recommended by Roche Molecular Biochemicals except that the program was as follows for 60 cycles: 94°C for 1 min, 42°C for 1 min, and 72°C for 2 min. The primerless PCR product was PCR amplified by using oligonucleotide 6 (5′-ATGGGGCCCGCTCCAGGCAC-3′) to prime LB400 bphA or oligonucleotide 7 (5′-CTGGGGGCCG TTGCAGGCAC-3′) from the MluI site to prime B-356 bphA and oligonucleotide 8 (5′-TTGAGCGTGGCCCACCTA-3′) from the AvrII site to amplify both bphA genes (Fig. 1). FastStart Taq DNA polymerase was used under the conditions recommended by Roche Molecular Biochemicals. The resulting 858-/864-bp MluI/AvrII DNA fragments were digested with MluI and AvrII and ligated to pDB31[LB400-bphAE] previously deleted from its MluI/AvrII fragment. The library was created in E. coli DH11S pQE51[LB400-bphFGB] and inoculated onto a nylon membrane on the surface of a Luria-Bertani (LB) agar plate (30). After overnight incubation at 37°C the membranes were transferred to fresh LB plates containing 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), the plates were incubated for 3 to 5 h at 37°C, and then crystals of 2,2′CB were placed into the lid of the petri dish. The plates were incubated at 37°C, and the colonies were periodically inspected visually over a period of 48 h to search for those that developed a similar or darker brown color than a control expressing LB400 BphA.
Whole-cell assays.
E. coli DH11S pQE51[LB400-bphFGB] plus pDB31[bphAE] expressing variant BphAs were tested for their ability to degrade a synthetic mix of PCBs. Log-phase cells grown with shaking at 37°C in LB broth were induced for 2 to 4 h with 0.5 mM IPTG and then washed and suspended to an optical density at 600 nm of 2.0 in M9 medium (30) containing 0.5 mM IPTG plus 0.1% (wt/vol) sodium acetate. Then, 2-ml portions of this suspension were distributed in 20-ml glass tubes covered with Teflon-lined screw caps. Each tube received 10 μl of a synthetic mix dissolved in acetone, followed by the addition of 1 μM concentrations of each of the congeners 2,6-CB, 2,4,4′CB, 2,3′,4-CB, 2,3,4′CB, 2,2′,5,5′CB, and 2,2′,3,3′CB and 5 μM concentrations of 3,3′CB and 4,4′CB. Incubation was carried out at 250 rpm at 37°C for 18 h. PCBs were extracted with two 4-ml portions of hexane. The combined extracts were dried over sodium sulfate, evaporated to a final volume of 1.5 ml, and analyzed by GC using an electron capture detector (ECD) (3). Each experiment included a set of control cultures of E. coli/pQE51bphFGB/pDB31, which were evaluated under conditions identical to those for the experimental cultures. All values reported in the present study are averages from triplicate experiments.
RESULTS
Screening protocol.
Oxidative polymerization of catechols and phenols is an important pathway in the formation of humic substances (7). The process can be catalyzed by oxidoreductive enzymes (laccases, tyrosinases, and peroxidases) (13) and by inorganic catalysts (salts and oxides of many metals and clay minerals) (8, 31). The darkening of microbial cultures that cometabolically transform aromatic xenobiotics to catechol metabolites is attributed to oxidative polymerization of catechols. Metal species or oxidative enzymes such as peroxidases and laccases present in the cultures can catalyze this reaction. However, although many investigations on the role of catechols in humic acid formation have been reported, the precise mechanism involved in culture darkening resulting from catechol accumulation is still not clear. Nevertheless, we have exploited this darkening phenomenon to introduce a new protocol to screen for clones able to convert CBs into catechols. We found that spots of catechols (2,3-dihydroxybenzene, 2,3-dihydroxybiphenyl, 1,2-dihydroxynaphthalene, 3,4-dihydroxybiphenyl, 3,4-dihydroxy-2,2′,5,5′CB and 2,3-dihydroxy-2′CB) on cellulose, nitrocellulose, or nylon membranes oxidized and darkened within minutes of exposure to ambient air when the membranes were saturated with a solution of NaOH containing a metal salt (not shown). Similarly, when an aqueous solution of any one of these catechols was dropped onto colonies of E. coli DH11S grown on a nylon membrane placed onto the surface of a LB or a M9 agar medium, the colonies darkened after 2 to 12 h of incubation at 37°C.
Based on these observations, we evaluated the capacity of IPTG-induced colonies of E. coli DH11S/pQE51[LB400-bphFGB]/pDB31[LB400-bphAE] grown on a membrane to transform BPH into a detectable colored metabolite. Several factors were considered, including the age of the cells, the temperature of incubation, the amount of IPTG, the type of membrane (cellulose, nitrocellulose, or nylon), and the mode of exposure to BPH (spraying, spreading, or sublimation). Consistent results were obtained when the protocol described in the experimental section was applied. IPTG-induced colonies of E. coli DH11S/pQE51[LB400-bphFGB]/pDB31[LB400-bphAE] grownon the surface of nylon membranes turned brownish within 2 h when exposed to BPH vapor and within 24 h when exposed to vapors of 2,2′CB or of 2,2′,5,5′CB. No change was observed when IPTG-induced colonies of E. coli DH11S/pQE51[LB400-bphFGB]/pDB31 was exposed to BPH vapor compared to a control of unexposed cells. IPTG-induced colonies of E. coli/pQE51[LB400-bphFGB]/pDB31[B-356-bphAE] darkened rapidly when exposed to BPH vapors, but their color did not change when they were exposed to vapor of 2,2′CB or of 2,2′,5,5′CB. This is consistent with the fact that LB400 but not B-356 BPDO can oxygenate 2,2′CB and 2,2′,5,5′CB.
Shuffling strategies to engineer novel PCB degrading BphAs.
In a previous investigation, when an 864-bp MluI-AvrII DNA fragment (Fig. 1) encoding the C-terminal portion of LB400 BphA was replaced entirely or in part by the corresponding portions of B-356 bphA, the hybrid enzymes were active (4). Unlike the parents, some of them were active on both 2,2′CB and 3,3′CB and others were active on 4,4′CB (4). On the other hand, in a set of unpublished experiments, <1% of the progeny obtained by shuffling LB400 bphA with B-356 bphA was active toward BPH. These results prompted us to explore alternate approaches consisting of creating libraries of variant LB400 bphA obtained by family shuffling portions of the MluI-AvrII fragment. In strategy I (Fig. 1), a 1,043-bp fragment of LB400 bphA amplified with primers external to the MluI and AvrII restriction sites was digested with DNase I and reassembled by primerless PCR in the presence of a DNase I-digested 513-bp fragment of B-356 bphA. The 1,043-bp fragment was amplified from pDB31[LB400-bphA], and it stretched from bp 368 of LB400 bphA to 30 bp downstream of the stop codon (in pDB31); the 513-bp fragment was amplified from pDB31[B-356-bphA], and it extended from bp position 368 of B-356 bphA to the ScaI restriction site. In strategy II (Fig. 1), the same 1,043-bp DNA fragment of LB400 bphA was digested with DNase I and reassembled by primerless PCR in the presence of a DNase I-digested 717-bp fragment of B-356 bphA. The 717-bp fragment was amplified from pDB31[B-356-bphA], and it started at position 688 of B-356 bphA and ended 30 bp downstream of the stop codon in pDB31 (Fig. 1). In strategy III (Fig. 1), the DNase I-digested fragments used to reassemble the 1,037- to 1,043-bp fragments included a 723-bp fragment amplified from pDB31[LB400-bphA] (from position 688 of LB400-bphA to 30 bp downstream of the stop codon), a 720-bp fragment amplified from pQE31[P6-bphA1] (from position 664 of P6 bphA1 to the stop codon), and the 513-bp fragment of B-356 bphA described above. Variants generated by strategy III must differ from parental enzymes. The reassembled 1,037- to 1,043-bp fragments obtained from each of the primerless PCR were amplified with appropriate internal primers to construct the libraries of MluI-AvrII fragments that were ligated to pDB31[LB400-bphAE] previously deleted of its MluI-AvrII segment (Fig. 1). The plasmid libraries were introduced into E. coli DH11S/pQE51[LB400-bphFGB] to screen for IPTG-induced clones exhibiting a similar or darker color than a control expressing LB400 BphA when exposed to 2,2′CB vapors. Apart from the crossover events causing structural changes of the parental enzymes, the frequency of mutation occurring during shuffling was lower than 0.05%.
Characterization of the progeny from the three shuffling strategies.
From strategy I, ca. 100 clones from a library of 20,000 darkened when exposed to vapors of 2,2′CB. However, upon rescreening in the agar assay and analysis of the potency to degrade a mix of eight PCB congeners in liquid culture, none of these clones exhibited a better performance than clones carrying parental LB400 bphA. Eight randomly chosen clones active on 2,2′CB were sequenced (not shown). Two of them were identical to LB400 bphA. The others were variants resulting from recombination events in the MluI/ScaI portion of the gene. These modifications had apparently no influence on the capacity of the variant enzyme to oxygenate 2,2′CB.
From strategy II, ca. 150 IPTG-induced clones from a library of 20,000 darkened within 48 h when exposed to 2,2′CB vapors. After they were rescreened, three of them (variants II-9, II-10, and II-11) were markedly darker compared to the control expressing LB400 BphA. One more variant, II-4, showing a color intensity similar to a clone expressing LB400 BphA was retained for characterization.
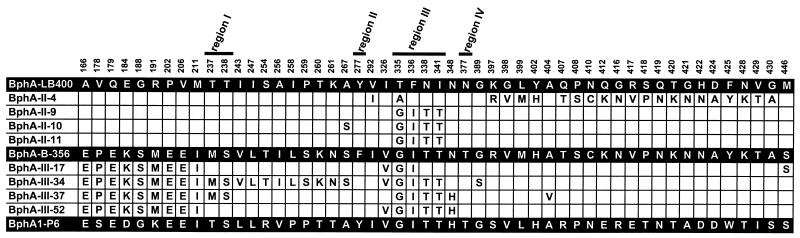
Variant II-9 was generated by the replacement of a stretch of LB400 bphA extending from bp positions 978 to 1028 by the corresponding fragment of B-356 bphA. Variant II-11 resulted from the replacement of a stretch of LB400 bphA extending from bp 978 to 1034 by the homologous portion of B-356 bphA. Variants II-9 and II-11 expressed BPDOs with identical amino acid sequences, where a short segment of LB400 BphA (335TFNNIRI341) was replaced by that of B-356 (333GINTIRT339) (Fig. 2). This segment, designated region III by Mondello et al. (28), was shown to strongly influence the potency of LB400 BphA toward PCBs. In the case of variant II-10, beside the two crossovers that allowed the replacement of region III of LB400 BphA by that of B-356 BphA, a second set of crossovers resulted in the replacement of A267 of LB400 BphA by the corresponding serine of B-356 BphA. Variant II-4 acquired a stretch of 186 bp from the distal portion of B-356 bphA; in addition, V292 of LB400 BphA was replaced by I of B-356 BphA, and a mutation at bp 1003 resulted in a change of T335 to A. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cell extracts of IPTG-induced E. coli DH11S clone expressing II-9 from pDB31 showed that equivalent amounts of both subunits of the variant BPDO were produced (results not shown). The enzyme was purified by affinity chromatography as a His-tagged protein. The UV-visible spectra of the His-tagged purified variant BPDOs were identical to that of the parents, exhibiting major peaks at 323 and 455 nm and a shoulder at 575 nm (18). The specific activities toward BPH of the His-tagged purified enzymes for II-11, II-9, and II-10 were 285, 314, and 237 nmol/min/mg, respectively. This is approximately three times higher than the value of 95 nmol/min/mg obtained for His-tagged purified LB400 BPDO and twice the value of 140 nmol/min/mg obtained for His-tagged purified B-356 BPDO. These values are close to the ones determined in previous study (19). The specific activity of II-4 (58 nmol/min/mg) was lower than that of LB400 BPDO.
FIG. 2.
Sequence alignment of LB400 BphA with variants obtained from shuffling portions of the distal segment of LB400 bphA with B-356 bphA and P6 bphA1. The shuffling strategies used to obtain these variants are described in Fig. 1. The alignment shows the amino acid positions of the three parental and variant BphAs for which at least one of the variant differs from LB400 BphA. In addition, the alignment shows the residues that correspond to regions I, II, III, and IV as designated by Mondello et al. (27).
It is noteworthy that in a coupled reaction with BphB and BphC, variant II-9 was able to catalyze the oxygenation of benzene, naphthalene, and toluene to produce the corresponding yellow meta-cleavage metabolite (data not shown). Variant II-10 was unable to oxygenate these substrates. Of the three substrates, LB400 BPDO can oxygenate naphthalene only (17) and B-356 BPDO can oxygenate benzene (20). Thus, variant II-9 has significantly increased the range of aromatic substrates used. In a recent report, Suenaga et al. (34) described an evolved BPDO variant, pSHF1072, with enhanced ability to oxygenate both benzene and toluene. It was found that the presence of T376 of KF707 BPDO, which corresponds to N377 of LB400 BPDO, was essential for the expression of this phenotype in pSHF1072. The fact that variant II-9 acquired N377 of LB400 BPDO suggests that other residues of the C-terminal portion of BphA influence the catalytic activity toward toluene.
Cells of E. coli carrying pQE51bphFGB and pDB31bphAE expressing the variant BPDOs were tested for their ability to degrade a mix of eight congeners in liquid culture. The IPTG-induced cells were incubated for 18 h in the presence of the PCB mix, and the percent depletion of each congener was estimated by GC-ECD. The data obtained are summarized in Table 1. E. coli clones expressing LB400 BphA degraded 2,2′,5,5′CB and 2,2′,3,3′CB significantly, and clones expressing B-356 BphA degraded 3,3′CB, 2,3′,4-CB, and 2,3,4′CB (Table 1). 2,2′CB was not included in the mix because it was not discriminated from 2,6-CB by our GC-ECD analytical settings. In the present work, levels of depletion of <10% varied markedly from one run to the other, and they were not found statistically significant when several replicate experiments were compared to controls prepared with E. coli/pQE51bphFGB/pDB31. Therefore, the levels of degradation for the poorly degraded congeners are not reported precisely in Table 1. However, based on metabolite production 2,3′,4-CB, 2,3,4′CB, 3,3′CB, and 2,4,4′CB were previously shown to be degraded at a low rate by LB400 BPDO (1, 6, 19, 32).
TABLE 1.
PCB degradative potency of BphA variants
| BPDO | % Depletiona
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 2,6-CB | 3,3′CB | 4,4′CB | 2,3′,4-CB | 2,3,4′CB | 2,4,4′CB | 2,2′,5,5′CB | 2,2′,3,3′CB | |
| LB400 | <10 | <10 | <10 | <10 | <10 | - | 94 | 76 |
| B-356 | <10 | 35 | <10 | 96 | 63 | - | - | - |
| II-4 | - | - | - | - | - | - | 84 | 47 |
| II-10 | <10 | <10 | <10 | <10 | <10 | - | 56 | 24 |
| III-17 | - | - | - | 73 | 60 | - | 20 | 27 |
| III-34 | <10 | <10 | <10 | 79 | 88 | <10 | 63 | 23 |
| II-9 | 58 | 45 | 53 | 99 | 98 | 44 | 93 | 87 |
| III-37 | 36 | 58 | 43 | 96 | 98 | 29 | 85 | 65 |
| III-52 | 57 | 51 | 52 | 97 | 99 | 50 | 92 | 83 |
That is, the percent depletion of each PCB congener of a mix of eight added to a culture of E. coli DH11S/pQE51[LB400-bphFGB]/pDB31[bphAE] expressing either of the BphA variants. The protocol is described in Materials and Methods. The values are averages of results from three separate experiments done in triplicate. The variance was <10% of the measured value in all cases. -, no degradation; the level of depletion of <10% was not statistically significant compared to the negative control and not reported precisely.
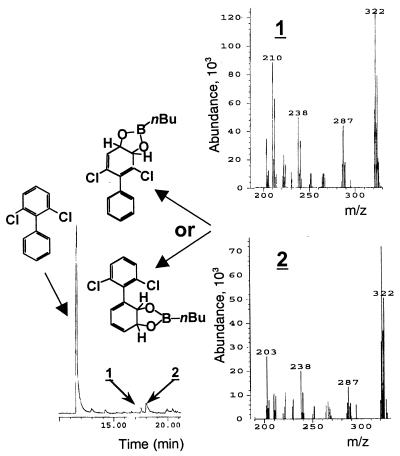
It is noteworthy that clones expressing variant II-10 showed a darker coloration than those expressing LB400 BPDO upon exposure to 2,2′CB in the screening assay, but in the liquid culture assay they exhibited a degradation potency toward the mix of eight PCBs that was similar to that of LB400 BPDO. We have no explanation at this time for this observation. On the other hand, the eight congeners of the mix were all depleted significantly by variant II-9, including the very persistent 2,6-CB (Table 1). As expected from its structure, variant II-11 showed a pattern of degradation identical to that of II-9 (not shown). The His-tagged purified II-9 BPDO metabolized 2,6-CB into two dichlorinated dihydrodiol metabolites. They were identified by comparing the GC-MS spectra of their butylboronate derivatives to published spectra of similar butylboronate-derived PCB metabolites (19, 26). The mass spectra for both metabolites exhibited a molecular ion at m/z 322 and significant ions at m/z = 287 (M-35, Cl), m/z = 238 (M-84, nBuBO), and m/z = 222 (M-100, nBuBO2) (Fig. 3). The detection of these metabolites confirms the potency of II-9 to transform 2,6-CB.
FIG. 3.
(Left) GC spectra of butylboronate-derived metabolites produced from 2,6-CB by variant II-9 BPDO. (Right) Mass spectra of metabolites 1 and 2. The structures of the metabolites are shown on the left and are tentatively identified as the 2,6-dichloro-3,4-dihydro-3,4-dihydroxybiphenyl and 2,6-dichloro-2′,3′-dihydro-2′,3′-dihydroxybiphenyl.
The inability of II-10 to oxygenate most of the congeners of the mix is noteworthy (Table 1) since it indicates a significant contribution of A267 of LB400 BphA to the selectivity or recognition of some PCB substrates. To confirm the importance of this residue by site-directed mutagenesis, we introduced a mutation at position 267 of variant II-9 to change A267 by S as in B-356 BPDO. The resulting mutant (II-9-267) was cloned into E. coli DH11S to evaluate its PCB-degrading potency. As with variant II-10, when IPTG-induced cells expressing II-9-267 were grown in the presence of the synthetic mix of eight congeners for 18 h, no significant depletion of 3,3′CB and 4,4′CB was recorded and, respectively, 59 and 23% depletion was obtained for 2,2′,5,5′CB and 2,2′,3,3′CB. Thus, position 267 strongly influences the range of PCB substrate used by the enzyme.
Sixty clones able to convert 2,2′CB were recovered out of 15,000 analyzed when strategy III was applied. Four clones (III-17, III-34, III-37, and III-52) were retained based on their activity toward 2,2′CB in the agar plate assays and on their pattern of activity when tested for their ability to degrade the synthetic mix of eight congeners in liquid medium. Variants III-52 and III-37 exhibited the highest potency. Their activity toward the PCB mix was similar to that of variant II-9 (Table 1), and the specific activity of purified preparations toward BPH was in the range of 350 nmol/min/mg. The pattern of PCB congeners degraded by variants III-17 and III-34 was narrower than that degraded by variants III-37 and III-52, and the specific activity of purified enzyme preparation was in the range of 100 nmol/min/mg. Purified BPDO variants from strategy III exhibited UV-visible spectra identical to parent enzymes (not shown). Except for III-17, all other variants had inherited the entire region III of P6 BphA (333GINTIRT339). However, the portion of the α subunit inherited from B-356 BphA differed for each of them (Fig. 2), which indicates an influence of residues outside region III on the range of PCB substrates that BPDOs can oxygenate.
DISCUSSION
In the present study, we used a new screening assay to identify evolved BPDOs with desired phenotypes. The assay is based on the darkening of catechol metabolites derived from PCBs. While this study was under way, Meyer et al. (27) reported that a similar assay was used to change the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. This assay was qualitative only. Furthermore, as shown by Meyer et al. (27), in addition to true mutations affecting enzyme specificity, the increase of intensity of coloration over time can be influenced by factors such as enzyme stability and solubility inside the cell. These considerations might explain why, even though variant II-10 was less active than LB400 BPDO toward all PCB congeners tested, colonies expressing this variant were darker than those expressing LB400 BPDO when they were exposed to vapors of 2,2′CB. Nevertheless, this assay was found to be very useful for identifying novel enzymes exhibiting improved PCB-transforming activities among a library of evolved enzymes. Our data and those of Meyer et al. (27) clearly show that it is possible to rely on the time of apparition of the coloration and on the intensity of coloration compared to control colonies expressing a less-active enzyme to screen for enzyme exhibiting phenotypes of interest. Because it does not require the participation of BphC, the assay can potentially be applied to congeners containing a higher level of chlorine substitution. In the present study, 2,2′CB was preferred as the screening agent to other, more persistent congeners such as 2,6-CB. Because the assay was used for the first time, E. coli clones expressing LB4000 BPDO could conveniently be used as positive controls when assayed on 2,2′CB, whereas no colored metabolites were generated from 2,6-CB. Furthermore, we expected that the frequency of variants exhibiting a higher activity than the positive controls on 2,2′CB would be higher than the frequency of variants exhibiting a catalytic activity that neither of the parents could perform.
Interesting variants were obtained from two of the three shuffling strategies examined during this work. Data obtained with variants from strategy II are consistent with those obtained by Mondello et al. (28) from the examination of variants obtained by site-directed mutagenesis. These authors found that the most potent variants were those in which the entire region III of LB400 BphA was replaced by that of KF707 BphA (AINTIRT) (28). Substitution of T335 by an alanine, as in KF707 BphA, did not significantly broaden the range of PCB substrate used. This is also the case of variant II-4. Noticeably, a single round of family shuffling between targeted portions of bphA was sufficient to generate variants able to efficiently oxygenate 2,2′CB, 3,3′CB, and 4,4′CB. In addition, these variants could degrade the very persistent 2,6-CB, which is a major metabolite of the anaerobic PCB dechlorination process (25). Most potent variants were only recovered from the libraries generated by strategies II and III, wherein it was possible to replace the 335TFNNIRI pattern of LB400 BphA by the GINTIRT pattern of B-356 or P6 BphAs. The data show the power of the in vitro-directed evolution process to unravel the enzymes' structural features associated with their biochemical properties. Thus, the data clearly demonstrate the influence of region III (Mondello et al. [28]) on the range of PCB substrate that the enzyme can oxygenate. On the other hand, the sequence comparison of variants III-37, III-52, and II-9 shows that many sets of amino acid combinations of the C-terminal portion of BphA can confer the ability to oxygenate a broad range of PCB congeners. The only requirement is that the sequence pattern of region III be GINTIRT instead of TFNNIRI as in LB400 BphA.
A BLASTP search was made to compare the sequence patterns for region III of the α subunits of all BPH, benzene, toluene, isopropylbenzene, and ethylbenzene dioxygenases recorded in the sequence data banks. These oxygenases were selected on the basis of the structural analogy of their substrates and based on the fact that, like BphA, they are Rieske-type proteins that require electron equivalents provided by a two-component ferredoxin and ferredoxin reductase system. It is noteworthy that the BLASTP analysis showed that the sequence pattern for region III of most of these enzymes is very similar to the GINTIRT pattern of B-356 and the P6 region III (data not shown). LB400 BphA (28) and Alcaligenes eutrophus H850 BphA (28), which are the two most versatile naturally occurring BphAs, are also the only ones to exhibit a very distinct sequence pattern for region III. Given the observation that our most potent evolved BphAs had acquired the broadly dispersed GINTIRT pattern, its replacement in nature in LB400 and in H850 BphA by the less-efficient TFNNIRI pattern needs to be clarified.
Based on sequence alignment with the related naphthalene dioxygenase (NDO), for which the tridimensional structure is known (10, 22), the amino acid residues of region III of BphA (corresponding to 310CSGVFKV316 of NDO) would be located inside the substrate-binding pocket, which explains their influence on substrate binding and recognition. However, amino acid residues outside region III can influence substrate specificity or substrate recognition, as evidenced by the fact that the range of substrates oxygenated by variant II-10 is much narrower than those oxygenated by variants II-9 and II-11. The single difference between these variants is the acquisition by II-10 of S267 of B-356 BphA to replace A267 of LB400 BphA. In a recent report, the neighbor residue A268 was found to influence LB400 BphA potency to catalyze the oxygenation of benzene, toluene, and alkylbenzenes (34). Based on NDO structural information (22), V252 of NDO, which corresponds to S267 of B-356 BphA, is located in the vicinity of the mononuclear iron center, which is believed to be the active site. Therefore, this position appears to play a crucial role in substrate recognition and/or binding, or it strongly interacts with some of the residues of region III.
In a previous study (5), when region III of LB400 BPDO was changed to that of KF707 BPDO, changing N377 of LB400 BphA to a T, as in KF707 BphA, caused a loss of activity toward the ortho-substituted congeners. The important contribution of residue 377 to the enzyme specificity was confirmed by the observation that a recombinant in which T376 of KF707 BphA was changed to N acquired 3,4-dioxygenase activity on 2,2′,5,5′CB (33). However, changing N377 of LB400 BphA to a T when the original region III of LB400 was not altered did not affect the substrate specificity of the variant enzyme (28). These observations suggest that the structural space that needs to be explored to unveil sets of structural arrangements likely to enhance BPDO potency is tremendous. A rational approach is useful for identifying residues affecting enzyme specificity; however, a more empirical approach will be necessary to create libraries of enhanced BPDO variants able to oxygenate the most persistent congeners. The in vitro-directed evolution process appears to be an appropriate strategy for this purpose. However, successful application of this approach is probably dependent on the diversity of the combinatorial libraries obtained from the shuffling of DNA molecules. The data presented here suggest that an approach consisting of family shuffling of a targeted region of bphA can greatly reduce the size of BphA library required to find more potent BPDOs.
Acknowledgments
This work was supported by grants STP224153 and OGP0039579 from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Arnett, C. M., J. V. Parales, and J. D. Haddock. 2000. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the BPDO of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 66:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asturias, J. A., E. Diaz, and K. N. Timmis. 1995. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene 156:11-18. [DOI] [PubMed] [Google Scholar]
- 3.Barriault, D., and M. Sylvestre. 1993. Factors affecting PCB degradation by an implanted bacterial strain in soil microcosms. Can J. Microbiol. 39:594-602. [DOI] [PubMed] [Google Scholar]
- 4.Barriault, D., C. Simard, H. Chatel, and, M. Sylvestre. 2001. Characterization of hybrid biphenyl dioxygenases obtained by recombining Burkholderia sp. strain LB400 bphA with the homologous gene of Comamonas testosteroni strain B-356. Can. J. Microbiol. 47:1025-1032. [PubMed] [Google Scholar]
- 5.Barriault, D., M. Vedadi, J. Powlowski, and M. Sylvestre. 1999. cis-2,3-Dihydro-2,3-dihydroxybiphenyl dehydrogenase and cis-1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase catalyze dehydrogenation of the same range of substrates. Biochem. Biophys. Res. Commun. 260:181-187. [DOI] [PubMed] [Google Scholar]
- 6.Bedard, D. L., and M. L. Haberl. 1990. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb. Ecol. 20:87-102. [DOI] [PubMed] [Google Scholar]
- 7.Bollag, J. M. 1992. Decontaminating soil with enzymes. Environ. Sci. Technol. 26:1876-1881. [Google Scholar]
- 8.Borraccino, R., M. Kharoune, R. Giot, S. N. Agathos, E. J. Nyns, H. P. Naveau, and A. Pauss. 2001. Abiotic transformation of catechol and 1-naphthol in aqueous solution: influence of environmental factors. Water Res. 35:3329-3373. [DOI] [PubMed] [Google Scholar]
- 9.Brühlmann, F., and W. Chen. 1999. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol. Bioeng. 63:544-551. [DOI] [PubMed] [Google Scholar]
- 10.Carredano, E., A. Karlsson, B. Kauppi, D. Choudhury, R. E. Parales, J. V. Parales, K. Lee, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2000. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J. Mol. Biol. 296:701-712. [DOI] [PubMed] [Google Scholar]
- 11.Chebrou, H., Y. Hurtubise, D. Barriault, and M. Sylvestre. 1999. Heterologous expression and characterization of the purified oxygenase component of Rhodococcus globerulus P6 biphenyl dioxygenase and of chimeras derived from it. J. Bacteriol. 181:4805-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crameri, A., S. A. Raillard, E. Bermudez, and W. P. Stemmer. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391:288-291. [DOI] [PubMed] [Google Scholar]
- 13.Dec, J., and J. M. Bollag. 1994. Dehalogenation of chlorinated phenols during oxidative coupling. Environ. Sci. Technol. 28:484-490. [DOI] [PubMed] [Google Scholar]
- 14.Eltis, L. D., B. Hofmann, H. J. Hecht, H. Lunsdorf, and K. N. Timmis. 1993. Purification and crystallization of 2,3-dihydroxybiphenyl 1,2-dioxygenase. J. Biol. Chem. 268:2727-2732. [PubMed] [Google Scholar]
- 15.Erickson, B. D., and F. J. Mondello. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fain, M. G., and J. D. Haddock. 2001. Phenotypic and phylogenetic characterization of Burkholderia (Pseudomonas) sp. strain LB400. Curr. Microbiol. 42:269-275. [DOI] [PubMed] [Google Scholar]
- 17.Haddock, J. D., and D. T. Gibson. 1995. Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1996. Characterization of active recombinant His-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J. Biol. Chem. 271:8152-8156. [DOI] [PubMed] [Google Scholar]
- 19.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1998. Involvement of the terminal oxygenase beta subunit in the biphenyl dioxygenase reactivity pattern toward chlorobiphenyls. J. Bacteriol. 180:5828-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurtubise, Y., D. Barriault, J. Powlowski, and M. Sylvestre. 1995. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J. Bacteriol. 177:6610-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joern, J. M., T. Sakamoto, A. A. Arisawa, and F. H. Arnold. 2001. A versatile high throughput screen for dioxygenase activity using solid-phase digital imaging. J. Biomol. Screen. 6:219-223. [DOI] [PubMed] [Google Scholar]
- 22.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 23.Kumamaru, T., H. Suenaga, M. Mitsuoka, T. Watanabe, and K. Furukawa. 1998. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol. 16:663-666. [DOI] [PubMed] [Google Scholar]
- 24.Lin, J. J., M. Smith, J. Jessee, and F. Bloom. 1992. DH11S: an Escherichia coli strain for reparation of single-stranded DNA from phagemid vectors. BioTechniques 12:718-721. [PubMed] [Google Scholar]
- 25.Maltseva, O. V., T. V. Tsoi, J. F. Quensen, M. Fukuda, and J. M. Tiedje. 1999. Degradation of anaerobic reductive dechlorination products of Aroclor 1242 by four aerobic bacteria. Biodegradation 10:363-371. [DOI] [PubMed] [Google Scholar]
- 26.Massé, R., F. Messier, C. Ayotte, M.-F. Lévesque, and M. Sylvestre. 1989. A comprehensive gas chromatographic/mass spectrometric analysis of 4-chlorobiphenyl bacterial degradation products. Biomed. Environ. Mass Spectrom. 18:27-47. [Google Scholar]
- 27.Meyer, A., A. Schmid, M. Held, A. H. Westphal, M. Rothlisberger, H. P. E. Kohler, W. J. H. van Berkel, and B. Witholt. 2002. Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J. Biol. Chem. 277:5575-5582. [DOI] [PubMed] [Google Scholar]
- 28.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quintana, M. G., C. Didion, and H. Dalton. 1997. Colorimetric method for a rapid detection of oxygenated aromatic biotransformation products. Biotechnol. Tech. 11:585-587. [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schweigert, N., A. J. B. Zehnder, and R. I. L. Eggen. 2001. Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ. Microbiol. 3:81-91. [DOI] [PubMed] [Google Scholar]
- 32.Seeger, M., M. Zielinski, K. N. Timmis, and B. Hofer. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suenaga, H., A. Nishi, T. Watanabe, M. Sakai, and K. Furukawa. 1999. Engineering a hybrid pseudomonad to acquire 3,4-dioxygenase activity for polychlorinated biphenyls. J. Biosci. Bioeng. 87:430-435. [DOI] [PubMed] [Google Scholar]
- 34.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sylvestre, M., M. Sirois, Y. Hurtubise, J. Bergeron, D. Ahmad, F. Shareck, A. Larose, D. Barriault, I. Guillemette, and J. M. Juteau. 1996. Sequencing of Comamonas testosteroni strain B-356-biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among gram-negative bacterial biphenyl dioxygenases. Gene 174:195-202. [DOI] [PubMed] [Google Scholar]
- 36.Taira, K., J. Hirose, S. Hayashida, and K. Furukawa. 1992. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 267:4844-4853. [PubMed] [Google Scholar]
- 37.Voigt, C. A., S. Kauffman, and Z. G. Wang. 2001. Rational evolutionary design: the theory of in vitro protein evolution. Adv. Protein Chem. 55:79-160. [DOI] [PubMed] [Google Scholar]
- 38.Wang, D., and H. S. Sul. 1996. Site-directed mutagenesis for large insertions by oligonucleotide primers in optimized molar ratios. BioTechniques 22:70-72. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, H., and F. H. Arnold. 1997. Optimization of DNA shuffling for high-fidelity recombination. Nucleic Acids Res. 25:1307-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]