Abstract
Chromosome dimers in Escherichia coli are resolved at the dif locus by two recombinases, XerC and XerD, and the septum-anchored FtsK protein. Chromosome dimer resolution (CDR) is subject to strong spatiotemporal control: it takes place at the time of cell division, and it requires the dif resolution site to be located at the junction between the two polarized chromosome arms or replichores. Failure of CDR results in trapping of DNA by the septum and RecABCD recombination (terminal recombination). We had proposed that dif sites of a dimer are first moved to the septum by mechanisms based on local polarity and that normally CDR then occurs as the septum closes. To determine whether FtsK plays a role in the mobilization process, as well as in the recombination reaction, we characterized terminal recombination in an ftsK mutant. The frequency of recombination at various points in the terminus region of the chromosome was measured and compared with the recombination frequency on a xerC mutant chromosome with respect to intensity, the region affected, and response to polarity distortion. The use of a prophage excision assay, which allows variation of the site of recombination and interference with local polarity, allowed us to find that cooperating FtsK-dependent and -independent processes localize dif at the septum and that DNA mobilization by FtsK is oriented by the polarity probably due to skewed sequence motifs of the mobilized material.
Though the architecture of the bacterial chromosome remains mysterious, the early proposal that it is established symmetrically on each replichore (31) has been supported recently by studies on the repair of the most frequent accident to befall circular chromosomes: the formation of a chromosome dimer by an odd number of recombinational exchanges between nascent chromosomes. Chromosome dimer resolution (CDR) takes place through the action of three proteins, the recombinases XerC and XerD and the septum-associated protein FtsK, on the resolution site dif (28 bp), located in the chromosome terminus (5, 6, 21, 35). Resolution takes place at the time of division and requires septum formation (34). It also requires that dif be located within a small zone of the terminus region, the DAZ (for dif activity zone [10, 22]).
Failure of CDR gives rise to several phenotypes, including an increase of 50- to 100-fold in recombination in the terminus region (11, 12, 25, 26). This terminal recombination reflects the extreme fragility of the dif region after inhibition of CDR, presumably a result of engulfment of the localized dif region by the closing septum. The recombination-stimulating events are, or culminate in, double-strand breaks, since terminal recombination is largely RecBCD dependent. Consistent with this scenario, trapping by septum of DNA joining sister nucleoids and DNA degradation near dif have been detected in CDR− mutants (18, 24, 30).
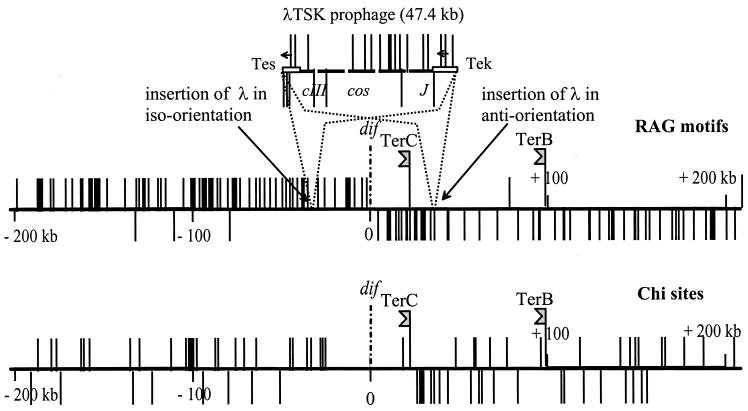
Genetic studies of the DAZ have revealed the role played by the polarity of the regions that flank dif in generating this domain. First, the “natural” orientation of ca. 30 kb on either side of dif must be maintained for dif to be active: inversions in these regions can be deleterious (29). Second, the presence near dif of the prophage λ in a certain orientation (which inverts on either sides of the site) inhibits the resolution process (12). The dif site maps in the region where the polarity of replichores generated by certain skewed oligonucleotides changes sign (8, 12). The skew of RGNAGGGS (or Rag) motifs is especially spectacular: these motifs are skewed everywhere on the chromosome (Table 1), but their skew is accentuated near dif since, along 360 kb to the left and 280 kb to the right of the site, 97% of these sequences are located on strands running 5′ to 3′ from oriC to dif (Table 1 and Fig. 1). Although there is no evidence that Rags are the determinants underlying the polarity phenomena described here, we assign them this role on a provisional basis on the grounds that they are the most promising candidates. Two arguments support this proposal: (i) the region of prophage λ responsible for the CDR inhibition effect harbors Rags skewed in a 10:1 proportion (J. Corre, unpublished results), and (ii) the regions flanking dif on the Salmonella enterica serovar Typhi or serovar Typhimurium chromosomes have only little similarity with the Escherichia coli corresponding regions, although they display the same skew of Rag motifs as in E. coli (www.sanger.ac.uk).
TABLE 1.
Distribution of Rag motifs in different chromosome regionsa
| Region (min) | No. of Rag motifs
|
No. of anti-Rags in seriesb of:
|
|||||
|---|---|---|---|---|---|---|---|
| Total | Anti | 2 | 3 | 4 | 5 | 6 | |
| 0-10 | 151 | 34 | 4 | ||||
| 10-20 | 175 | 30 | 3 | 2 | |||
| 20-30 | 184 | 23 | |||||
| 30-40c | 175 | 6 | |||||
| 40-50 | 153 | 23 | 3 | 1 | |||
| 50-60 | 159 | 25 | 5 | 1 | |||
| 60-70 | 146 | 39 | 5 | 1 | |||
| 70-80 | 149 | 36 | 11 | ||||
| 80-90d | 143 | 43 | 5 | 2 | 2 | ||
| 90-100 | 135 | 29 | 7 | 1 | |||
| Total | 1,570 | 288 | 43 | 6 | 2 | 1 | 1 |
On the sequence of the E. coli chromosome (7), a Rag motif is found every 3.3 kb on average, and the average skew is 82.4%. The skew shift is neat at dif (Fig. 1) but is less clear at oriC (not shown). For about 640 kb, 360 to the left of dif and 280 to the right (26.5 to 40.5 min on the genetic map), polarization is extreme; this region harbors 230 Rags with only 7 (3%) in antiorientation. The occurrence of anti-Rag series outside the highly polarized region is compatible with random distribution: a blind drawing of 1,340 Rags among an infinite population of iso-Rag and anti-Rag in the same proportions as in the chromosome (outside the highly polarized TER region), yields numbers of anti-Rag series close to 41 doublets, 9 triplets, and 3 tetrads. This is not very different from the actual distribution, except that the probability of occurrence of the series of 5 and 6 is low.
See Materials and Methods for discussion of the nomenclature. By “series” is meant a succession of anti-Rags uninterupted by an iso-Rag in the sequence of the region considered.
dif at 34 min.
oriC at 84 min.
FIG. 1.
Distribution of Rag and Chi elements near dif and in λTSK. The horizontal lines represent 400 kb of the TER region centered on dif. The positions of replication terminators TerC and TerB are indicated. Vertical lines indicate the positions and orientations of Rag (5′-RGNAGGGS-3′) and Chi elements (5′-GCTGGTGG-3′), based on the published sequence of E. coli (7). Chi stimulates RecBCD recombinase activity when the complex arrives at GCTGGTGG from the 3′ end (33).
One element that might play a key role in the spatiotemporal control of dimer resolution is the FtsK protein (35). FtsK is required for cell division, where its N-terminal transmembrane domain locks the protein in the septum (3, 14, 36) and serves to recruit other division proteins (9), as well as for nucleoid segregation (16, 24, 37) and for CDR (35), where its C-terminal domain is an essential participant in the XerCD-catalyzed resolution reaction (1, 2, 32). In addition to this role, we considered it possible that FtsK might play a second, prior role in CDR: that of guiding dif sites toward the ingrowing septum to ensure that synapsis can occur. Its similarity to SpoIIIE, which translocates DNA into the Bacillus subtilis prespore (17) made it an attractive candidate for such a role, and indeed the functional C-terminal fragment, FtsKc, has recently been shown also to translate on DNA (1). Our preliminary observations indicated that an ftsK mutation that eliminates the C-terminal domain results in a high rate of recombination in the terminus (12). A recent observation in this laboratory has indicated that when the normal XerCD-dif system is replaced by Cre-LoxP, efficient CDR requires not only that LoxP be located within the DAZ but also that the bacteria be FtsK+, even though FtsK is not needed for Cre-LoxP recombination (8a). FtsK must therefore be involved in the control of the regional constraints characterizing CDR.
Consequently, we have further examined the role of FtsK as a motor that drives the DNA movements needed for CDR by measuring the effect of an ftsK mutation on terminal recombination. If an ftsK mutation inhibits only Xer recombination while allowing normal dif positioning, recombinogenic lesions will occur near dif only, so that terminal recombination in this ftsK mutant will be indistinguishable from terminal recombination in a xerC mutant. If, on the other hand, FtsK is needed for positioning of dif prior to Xer recombination, the closing septum will trap DNA from a much wider region of the mutant chromosome, thus modifying the gradient of terminal recombination as a function of chromosomal position. The comparison of terminal recombination in an ftsK mutant with that in an xerC mutant indicates that two positioning processes, one FtsK dependent and one FtsK independent, cooperate to localize dif under the septum. Furthermore, because our assay can provoke interference with local polarity, these results provide evidence that FtsK acts in a polarity-sensitive process.
MATERIALS AND METHODS
Polarity nomenclature.
The distribution of Rag elements is taken as an example. When a Rag element displays the most frequent orientation on the replichore (the sequence 5′-RGNAGGGS-3′ running in the oriC-to-dif direction), it is said to be in iso-orientation (iso-Rag). When it displays the opposite orientation, it is said to be in antiorientation (anti-Rag). A larger DNA segment harboring several Rags with a majority in a certain orientation, such as an integrated λTSK prophage, is in iso-orientation when its own skew merges with the ambient skew and in antiorientation in the opposite situation. This is illustrated Fig. 1.
Bacterial strains, bacteriophages, and genetic procedures.
All strains analyzed for terminal recombination derive from CB0129 (W1485 F− leu thyA thi deoB or deoC supD [4]) and harbor tet (tetracycline resistance [Tcr]) or Tn10 insertions previously described (11, 12, 25). The ftsK1::cat (chloramphenicol resistance) is described elsewhere (14). The xerC2::Apr (ampicillin resistance) allele was described by Louarn et al. (26). The Δ(recB recC)::Apr mutation was as described previously (11). These mutations were transferred by P1 transduction by standard protocols (27). Bacteriophage λTSK is described by Corre et al. (12). Cultures were routinely made in Luria-Bertani (LB) medium (27).
Prophage excision assay.
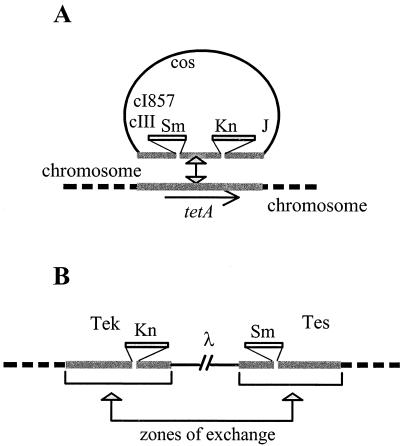
Our routine test for indigenous recombination is a prophage excision assay (12, 25). It is performed on strains made lysogens for a phage λ derivative (λTSK) that is proficient for lyzogenization but carries tet sequences coming from Tn10 transposon in place of the att int region (Fig. 2). This prophage can integrate by homologous recombination into a chromosome that carries a resident Tn10 or tet sequence and then becomes flanked by direct repeats. Since the phage repressor is thermosensitive, bacteria cured of the prophage can be scored easily as CFU at 42°C. Curing results preferentially from excisive homologous recombination between the flanking repeats. Routinely, frequencies of cured bacteria were determined as follows: eight 24-h-old individual colonies grown on LB agar medium at 30°C were each resuspended in 0.5 ml of LB broth and grown overnight at 30°C. These clones were then mixed as two groups of four clones; each group was plated on LB agar (containing 8 × 10−3 M sodium citrate to avoid λ reinfection when the strains were λ sensitive) and then incubated at 30 or 42°C. Replicate determinations displayed significant variability, as expected from the stochastic nature of the excision event, but in general this did not exceed ±20% of the reported average value. When CDR is inhibited by an ftsK mutation, the difference in selective values of the lysogen versus the nonlysogen parent is minor and can be neglected: growth competition experiments have indicated that there is no, or only limited, selective advantage for lysogens versus nonlysogens after 25 generations or after a 5-day incubation in stationary phase (data not shown). Since bacterial curing frequencies are measured after the same number of generations (ca. 30) for all strains, the variations observed are solely imputable to variations in excision frequencies per cell generation.
FIG. 2.
Prophage excision assay. (A) System for monitoring indigenous recombination. Phage λTSK carries a 2.8-kb tetAR (Tcr) fragment from Tn10 interrupted within tetA by two insertions providing kanamycin resistance (Knr) and streptomycin-spectinomycin resistance (Smr). It can recombine with a resident tetAR sequence and inactivate the Tcr character to form a Tcs lysogen. Owing to the cI857 mutation, the prophage is repressed only at a low temperature (30°C). It is induced and kills its host at 42°C. (B) Prophage loss after excisive recombination between the flanking repeats restores the ability to form colonies at 42°C, so that the number of temperature-resistant bacteria present in a clone derived from a single lysogen is a measure of prophage excision frequency when lysogens and nonlysogens display the same generation time, which is the case in CDR− mutants.
RESULTS
How DNA lesions due to CDR inactivation lead to terminal recombination.
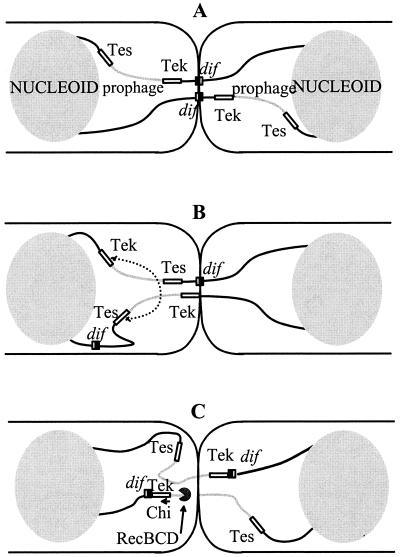
Terminal recombination is not a normal consequence of failed CDR but rather is an imposed consequence of the assay system that we use to detect fragility in the terminus region. When CDR is inactive, the septum traps a chromosome dimer and isolates the nucleoids from each other. This results, by mechanisms currently unknown, in DNA lesions that can stimulate recombination. Detection of these events by genetic means implies that a circular monomer chromosome is regenerated by homologous recombination. This seldom occurs on normal chromosomes since xer mutants that harbor an unresolved dimer usually are doomed (18). The observation suggests that the monomer units of chromosome dimers are normally distributed evenly about the dif sites located at the septum, so that absence of terminal diploidy in either sister cell precludes repair recombination (Fig. 3A). A small overlap restricted to the region devoid of Chi sites near dif (Fig. 1) may occur, however, as suggested by the observation that a recD mutation improves viability of a xerC mutant (30). Apart from this minor contribution, we only detect terminal recombination because the λ prophage of our assay induces uneven distribution of the terminus DNA between sister cells (Fig. 3B). This facilitates homology searching, thus helping cells to recover viability.
FIG. 3.
Model for prophage excision and recovery of circular chromosome monomers from a dimer in a xerC mutant. (A) The strain harbors an iso-oriented prophage in the terminus region, with no associated perturbation of the positioning of dif sites at division. Trapping by the septum immobilizes the dif regions and leads to generation of double-strand ends. The absence of overlap leaves no possibility for recombination to yield a circular monomer in either of the daughter cells. Both cells are doomed. (B) A pseudo-DAZ generated at the junction of an antioriented prophage and the chromosome can result in septal trapping of DNA distant from dif, whereas the positioning of the second thread is correct. The resulting overlap of terminus sequences can be used to generate a prophage-cured circular monomer by homologous recombination between the indicated Tes and Tek elements in the cell to the left, while the cell to the right is doomed. (C) When the prophage maps at or near dif, positioning operates to place prophage DNA under the septum. In the example depicted (chosen for simplicity), breakage in the trapped zone results in simultaneous prophage excision and chromosome repair, provided the RecBCD enzyme, responsible for the repair, encounters the stimulating Chi site present in the tet sequence. Both cells can recover a viable monomer chromosome.
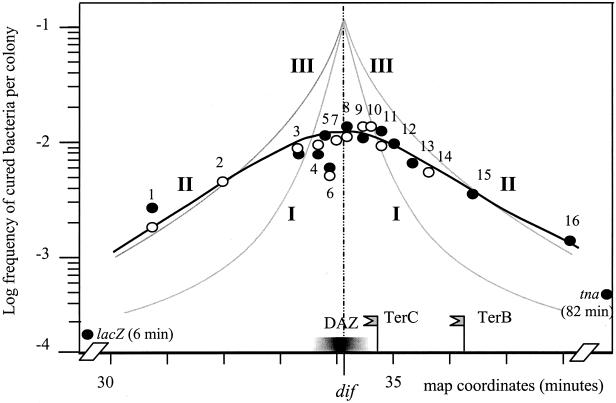
The polar effect of the prophage is easily detected in xer mutants (12) and is recalled in Fig. 4, in which curve I shows the frequency of terminal recombination in strains carrying iso-oriented prophages and curve III shows the equivalent data for antioriented prophages. Although the frequency of chromosome dimer trapping and of resulting DNA lesions is presumably the same in all xerC2 mutant strains, clearly, the ability to repair these lesions depends on prophage orientation and position since curve I is always below curve III, except at dif. This can be explained on the basis of a polarity-dependent positioning mechanism. When the prophage maps at or near dif, the positioning mechanism sets the prophage under the septum, so that the situation depicted in Fig. 3C is generated: recombination-stimulating events occur within the prophage, and the chromosomes can then be repaired in either or both cells by recombination between prophage flanking regions, with loss of the prophage and generation of the temperature-resistant cells that we select. This occurs irrespective of prophage orientation: both curves rise to the same maximum of terminal recombination when the prophage is inserted at dif (Fig. 4). If the prophage is displaced away from dif, the probability of chromosome repair becomes dependent on prophage orientation, in a way consistent with the models of Fig. 3A and B. It remains elevated when prophages are antioriented but decreases rapidly when prophages are iso-oriented (compare curves I and III in Fig. 4). Terminal recombination is detectable with iso-oriented prophages located some distance from dif (curve I; see also Table 2), probably because neither the prophage nor the flanking repeats are as polarized as the terminus (Fig. 1) so that chromosome termini in these strains may not be as evenly distributed as depicted in Fig. 3A. Nevertheless, the prophage excision assay is a system of choice for detecting alteration of the polarity dependence of dif positioning.
FIG. 4.
Frequency of prophage loss from various positions in the TER region of the ftsK1::cat mutant. The frequency of cured bacteria is plotted against the position of the tetAR integration locus. Symbols: •, prophages in antiorientation; ○, prophages in iso-orientation. Curve II, based on the present data obtained in the ftsK1 mutant, takes into account both orientations. The other curves are extracted from previously published work (12): curve I, xerC2 mutant, prophage iso-oriented; curve III, xerC2 mutant, prophage antioriented. The positions of relevant loci (dif, TerC, TerB) are indicated. The extent of the DAZ, the zone in which dif gains progressively resolution activity as it is moved toward its natural site (29), is shown by a graded bar. The positions tested (the distances in kilobases from dif are in parentheses, negative to the left and positive to the right): 1, zda192 (−151); 2, trg (−100); 3, zdc310 (−35.3); 4, zdc330 (−15.8); 5, Δ(zdc330-zdc338) (−15.8/−7.0); 6, zdc338 (−7.0); 7, Δ(zdc338-hipA) (−7.0/+1); 8, hipA (+1); 9, zdd355 (+8.9); 10, zdd365 (+17.8); 11, zdd370 (+23.2); 12, zde381 (+34); 13, zde395 (+48); 14, zde406 (+58); 15, zdf237 (+100); and 16, zdh57 (+180). Both prophage orientations were assayed at positions 1, 3, 4, 6, 8, 9, and 11. Curing frequencies of prophages inserted at positions external to the terminus, lacZ (6 min), and tna (82 min) are also indicated. They were as low in the FtsK− background, 1.5 × 10−4 and 3.5 × 10−4, respectively, as in wild-type bacteria at the same positions (25).
TABLE 2.
Genetic controls on terminal recombination at two positions
| Relevant genotype | Tr frequencya at:
|
|||
|---|---|---|---|---|
|
zdc310
|
zdd355
|
|||
| Probe anti | Probe iso | Probe anti | Probe iso | |
| Wild type | 5.1 × 10−2 | 1.4 × 10−3 | 5.7 × 10−2 | 1.4 × 10−3 |
| xerC2 | 1.2 × 10−2 | 3.2 × 10−3 | 3.8 × 10−2 | 1.0 × 10−2 |
| ftsK1::cat | 8.1 × 10−3 | 8.6 × 10−3 | 1.2 × 10−2 | 1.2 × 10−2 |
| ftsK1 xerC2 | 6.2 × 10−3 | 8.3 × 10−3 | 1.4 × 10−2 | 1.4 × 10−2 |
| ftsK1 Δ(recB recC) | ND | 5.0 × 10−4 | ND | 1.1 × 10−4 |
The frequencies of temperature-resistant (Tr) cured bacteria were determined about 30 generations after the initial single-cell isolation. Average values are presented. Probe anti, the prophage λTSK is inserted in antiorientation; Probe iso, the prophage λTSK is inserted in iso-orientation. ND, not determined.
Inactivation of the FtsK C-terminal domain alters terminus positioning and suppresses polar effects.
The ftsK1::cat allele encodes an N-terminal domain that is functional in cell division and part of the linker domain but not the C-terminal domain which acts in CDR (14). It was introduced by phage P1 transduction into an isogenic family of CDR+ strains carrying iso-oriented and antioriented prophages. This introduction always resulted in filamentation and/or chain formation of a fraction of the bacteria, confirming inactivation of CDR by the mutation. The frequency of prophage excision in the resulting strains was then measured by assaying for thermoresistant derivatives (see Materials and Methods). The results (curve II in Fig. 3) show that excisive recombination, while at least 10-fold higher in the terminus than at the control positions outside this region (lacZ and tna) is lower by an order of magnitude than in the xerC2 mutant, at least near dif. The drawing of curve II (Fig. 3) does not take into account the fact that excision frequencies observed at position zdc338 were two- to threefold lower than at neighbor positions. This phenomenon was not observed in deletion mutants of this region and is unrelated to dimer resolution, since the whole region (except dif) is dispensable for CDR (10, 22). It has not been yet further analyzed. The ftsK1 mutation also resulted in elevated levels of prophage excision over an extended region (ca. 300 kb) in contrast to the narrow zone affected with iso-oriented prophage in the xerC2 mutant. Moreover, frequencies of cured derivatives were independent of prophage orientation, including for several positions at which both prophage orientations were assayed. These observations show that, in the absence of the cytoplasmic domain of FtsK, (i) the terminus is submitted to elevated recombination activity, thus confirming our previous report (12), and (ii) terminus fragility affects an expanded domain without regard to polarity. This implies that FtsK+ controls the size of the region submitted to trapping and imposes the polarity constraints.
Alteration of the same process triggers terminal recombination in ftsK and in xerC mutants, and repair requires RecBC.
Fragility due to inactivation of FtsK and that due to absence of XerC should be related, since both factors are needed for CDR. To examine this prediction, we constructed xerC2 ftsK1 double mutants and measured their capacity for prophage excision at two positions. Excision frequencies of the double mutants were about the same as those of the equivalent ftsK1 single mutants (Table 2), supporting the view that both genes are involved in the same pathway. Furthermore, with antioriented prophages, the double mutant displayed lower excision rates than the single xerC2 mutant. The ftsK1 mutation is thus epistatic on a xerC2 mutation, indicating that FtsK has, in addition to its role in Xer recombination, other functions in the resolution pathway and that these functions operate before Xer recombination.
RecBCD is an actor in all processes of hyperrecombination in the terminus described thus far (11, 12, 19). This is also the case for ftsK1-induced fragility, since the frequency of prophage curing is strongly decreased when recB and recC genes are deleted (Table 2). Thus, the recombination-stimulating events occurring in the ftsK1 mutant produce double-strand ends, as in other situations leading to terminus fragility. These double-strand ends may result from guillotining (the trapped DNA is broken) or garroting (the next replication forks arriving in the trapped region are stopped, and subsequently nascent strands are extruded and anneal).
DISCUSSION
Our major observations are as follows: (i) inactivation of the ftsK C-terminal domain makes the terminus fragile; (ii) terminus fragility due to ftsK mutation has the same cause as that of xer mutants, most probably the processing of unresolved dimer chromosomes; and (iii) the role of replichore polarity in terminus fragility seen in FtsK+ cells is not observed in the absence of the ftsK C-terminal domain.
When terminus positioning is near normal but CDR is inactivated—the situation corresponding to curve I of Fig. 4—trapping occurs at or near dif, so that the consequences of aborted CDR are detected only in the vicinity of dif. The lower but much broader profile of terminal recombination in the ftsK1 mutant (curve II in Fig. 4) suggests that the fragile region is much less accurately determined in this mutant, although clearly still belonging to the terminus. A straightforward interpretation is that positioning of the terminus is the result of two successive processes. The first is a coarse positioning independent of FtsK, reflected by curve II, that accompanies or follows postreplication reconstruction of nucleoid structure. This positioning is not precise enough to allow efficient resolution at dif, but it is sufficient to situate the DNA that links the nucleoids in the terminus region. The second mechanism is a fine-tuning involving FtsK that positions the dif sites close to each other under the septum, so that CDR can take place or, if CDR fails, so that recombinational rescue is restricted to a limited region near dif.
Coarse positioning might be passive, a mere consequence of the terminus being replicated by a machinery anchored in the center of the cell where the septum will eventually form (13, 23), accompanied by sequential compaction of nascent DNA into two nucleoids. In this model, the links between nucleoids would “naturally” be confined to the terminus. Alternatively, the retention of a terminus macrodomain (TER region; 20 to 30% of the chromosome) in the vicinity of the future septum may be determined actively and specifically. This possibility is supported by cytological analyses of the terminus (20, 28). Experiments now in progress that are aimed at determining whether relocation of the region where replication forks collide causes a corresponding displacement of the region of elevated excision should shed light on this issue.
A remarkable finding is that terminus fragility is indifferent to prophage orientation in ftsK1 mutants, in contrast to the observation that in xerC mutants terminal recombination is a function of prophage orientation (Fig. 4 and Table 2). The conclusion is that the polarity-sensitive positioning mechanism must be FtsK dependent. How FtsK reads and uses DNA polarity furnished by Rags, or other similarly skewed elements, to mobilize DNA is unknown. Polar elements may act as inhibitors or as activators of the sliding of DNA through the FtsK-based edifice built at the septum. In either case, the direction of DNA movement under the closing septum would be dictated by the mobilized material itself.
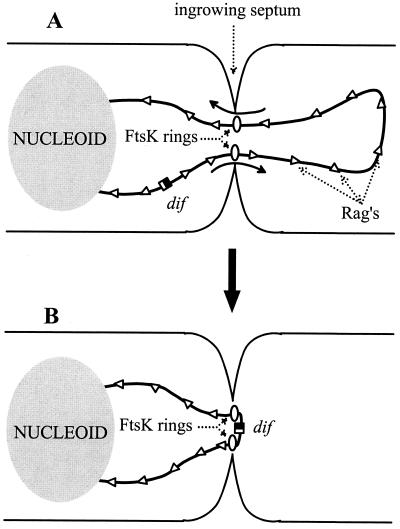
Does FtsK have a role beyond ensuring the resolution of dimers? For example, the FtsK-dependent positioning process may help preclude accidental trapping of DNA by the septum. This could happen when a DNA loop is extruded past the septum into the opposite cell compartment. An FtsK ring might form on each thread of DNA passing through the septum and move the intruding loop in the direction dictated by the skewed elements. As shown in Fig. 5, the final result would be elimination of the loop and positioning of dif under the septum (an epiphenomenon in the case of a monomer chromosome). This again illustrates the interest for the cell in possessing a mobilization system in which the direction of DNA movement is dictated only by elements belonging to the mobilized material. The 14-min region from 26.5 to 40.5 min is characterized by a very low frequency of anti-Rags (Table 1). The strong polarity of Rag motifs in the TER region has perhaps evolved to combat the trapping of material that is forced by the chromosome or cell architecture to remain close to the septum. Outside the strongly polarized TER region, intrusion loops, if formed, might also be destroyed by the FtsK-dependent mobilization process. In this case, anti-Rag clusters would have the task of limiting the journey of a loop toward dif. The occurrence of such clusters is, however, consistent with random choice, suggesting that they have not been favored by selection (Table 1).
FIG. 5.
How FtsK might eliminate intrusion loops and position dif sites under the septum. (A) A DNA loop extruded from the nucleoid on the left wanders in the cytoplasm of the future sister cell to the right. When the septum is closing, FtsK forms ring structures around the two DNA threads. FtsK mobilizes these threads in the directions (indicated by arrows) dictated by Rags or similar DNA polar elements. (B) When one FtsK ring meets a region polarized in antiorientation (here, the DAZ at the replichore junction near dif), it stops mobilizing DNA; the other ring keeps acting, so that the size of the loop is gradually reduced. When the second ring meets the DAZ, dif is located under the septum and the loop is eliminated.
In the relatively short time that has elapsed since the discovery of FtsK, studies of this protein have proved remarkably fruitful, as highlighted by a recent review (15). The protein constitutes a keystone of the cell division process. Its ability to coordinate chromosome segregation and cell division bears witness to a versatility that is underscored by its skill in processing DNA: it translocates DNA, it modulates the synapse between dif sites and, as shown here, it even reads DNA and interprets its polarity. Further insights may come from isolation of ftsK mutations specifically affected either in the positioning step or in Xer recombination.
Acknowledgments
This work was supported by recurrent funding from CNRS and MENRT, by contract 9823 from the Association de la Recherche contre le Cancer (ARC), and by contract 99N60/0211 from PRFMMIP.
We thank François Cornet for numerous discussions. We are most grateful to David Lane for his interest, critical reading, and many improvements to the manuscript.
REFERENCES
- 1.Aussel, L., F. X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108:195-205. [DOI] [PubMed] [Google Scholar]
- 2.Barre, F. X., M. Aroyo, S. D. Colloms, A. Helfrich, F. Cornet, and D. J. Sherratt. 2000. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 14:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene. ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, R. E., J. Louarn, J. Martuscelli, and L. Caro. 1972. Origin and sequence of chromosome replication in Escherichia coli. J. Mol. Biol. 70:549-566. [DOI] [PubMed] [Google Scholar]
- 5.Blakely, G., S. Colloms, G. May, M. Burke, and D. Sherratt. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789-798. [PubMed] [Google Scholar]
- 6.Blakely, G., G. May, R. McCulloch, L. K. Arciszewska, M. Burke, S. T. Lovett, and D. J. Sherratt. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K-12. Cell 75:351-361. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Capiaux, H., F. Cornet, J. Corre, M. I. Guijo, K. Perals, J. E. Rebollo, and J. M. Louarn. 2001. Polarization of the Escherichia coli chromosome: a view from the terminus. Biochimie 83:161-170. [DOI] [PubMed] [Google Scholar]
- 8a.Capiaux, H., C. Lesterlin, K. Pérals, J.-M. Louarn, and F. Cornet. A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep., in press. [DOI] [PMC free article] [PubMed]
- 9.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI requires FtsK, but not FtsN, for colocalization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 10.Cornet, F., J. Louarn, J. Patte, and J. M. Louarn. 1996. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 10:1152-1161. [DOI] [PubMed] [Google Scholar]
- 11.Corre, J., F. Cornet, J. Patte, and J. M. Louarn. 1997. Unraveling a region-specific hyper-recombination phenomenon: genetic control and modalities of terminal recombination in Escherichia coli. Genetics 147:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corre, J., J. Patte, and J. M. Louarn. 2000. Prophage lambda induces terminal recombination in Escherichia coli by inhibiting chromosome dimer resolution: an orientation-dependent cis-effect lending support to bipolarization of the terminus. Genetics 154:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Den Blaauwen, T., N. Buddelmeijer, M. E. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diez, A. A., A. Farewell, U. Nannmark, and T. Nystrom. 1997. A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J. Bacteriol. 179:5878-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donachie, W. D. 2002. FtsK: Maxwell's demon? Mol. Cell 9:206-207. [DOI] [PubMed] [Google Scholar]
- 16.Draper, G. C., N. McLennan, K. Begg, M. Masters, and W. D. Donachie. 1998. Only the N-terminal domain of FtsK functions in cell division. J. Bacteriol. 180:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Errington, J., J. Bath, and L. J. Wu. 2001. DNA transport in bacteria. Nat. Rev. Mol. Cell. Biol. 2:538-545. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks, E. C., H. Szerlong, T. Hill, and P. Kuempel. 2000. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC, and xerD) of Escherichia coli. Mol. Microbiol. 36:973-981. [DOI] [PubMed] [Google Scholar]
- 19.Horiuchi, T., Y. Fujimura, H. Nishitani, T. Kobayashi, and M. Hidaka. 1994. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J. Bacteriol. 176:4656-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-703. [DOI] [PubMed] [Google Scholar]
- 21.Kuempel, P. L., J. M. Henson, L. Dircks, M. Tecklenburg, and D. F. Lim. 1991. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 3:799-811. [PubMed] [Google Scholar]
- 22.Kuempel, P., A. Hogaard, M. Nielsen, O. Nagappan, and M. Tecklenburg. 1996. Use of a transposon (Tndif) to obtain suppressing and nonsuppressing insertions of the dif resolvase site of Escherichia coli. Genes Dev. 10:1162-1171. [DOI] [PubMed] [Google Scholar]
- 23.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 24.Liu, G., G. C. Draper, and W. D. Donachie. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol. 29:893-903. [DOI] [PubMed] [Google Scholar]
- 25.Louarn, J. M., J. Louarn, V. François, and J. Patte. 1991. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J. Bacteriol. 173:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louarn, J., F. Cornet, V. François, J. Patte, and J. M. Louarn. 1994. Hyperrecombination in the terminus region of the Escherichia coli chromosome: possible relation to nucleoid organization. J. Bacteriol. 176:7524-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Niki, H., Y. Yamaichi, and S. Hiraga. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 14:212-223. [PMC free article] [PubMed] [Google Scholar]
- 29.Perals, K., F. Cornet, Y. Merlet, I. Delon, and J. M. Louarn. 2000. Functional polarization of the Escherichia coli chromosome terminus: the dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol. Microbiol. 36:33-43. [DOI] [PubMed] [Google Scholar]
- 30.Prikryl, J., E. C. Hendricks, and P. L. Kuempel. 2001. DNA degradation in the terminus region of resolvase mutants of Escherichia coli, and suppression of this degradation and the Dif phenotype by recD. Biochimie 83:171-176. [DOI] [PubMed] [Google Scholar]
- 31.Rebollo, J. E., V. Francois, and J. M. Louarn. 1988. Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 85:9391-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recchia, G. D., M. Aroyo, D. Wolf, G. Blakely, and D. J. Sherratt. 1999. FtsK-dependent and-independent pathways of Xer site-specific recombination. EMBO J. 18:5724-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahl, F. W., M. M. Stahl, R. E. Malone, and J. M. Crasemann. 1980. Directionality and nonreciprocality of chi-stimulated recombination in phage lambda. Genetics 94:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner, W. W., and P. L. Kuempel. 1998. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol. Microbiol. 27:257-268. [DOI] [PubMed] [Google Scholar]
- 35.Steiner, W., G. Liu, W. D. Donachie, and P. Kuempel. 1999. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol. Microbiol. 31:579-583. [DOI] [PubMed] [Google Scholar]
- 36.Wang, L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731-740. [DOI] [PubMed] [Google Scholar]
- 37.Yu, X. C., E. K. Weihe, and W. Margolin. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 180:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]