Abstract
To investigate the regulation of the Legionella pneumophila icm and dot genes required for intracellular growth, a series of nine icm::lacZ fusions were constructed. These icm::lacZ fusions were found to have different levels of expression in L. pneumophila, and five of them were more highly expressed at stationary phase than at exponential phase. When the expression of these fusions in Escherichia coli was tested, all of them were found to be expressed but three of them had dramatic changes in their levels of expression in comparison to those in L. pneumophila. Site-directed and PCR random mutagenesis with these icm::lacZ fusions was used to identify DNA regulatory elements of icm genes. Four icm genes (icmT, icmP, icmQ, and icmM) that had low levels of expression in L. pneumophila were found to contain a 6-bp sequence (TATACT) essential for their expression. This sequence was shown by primer extension to serve as their −10 promoter elements. A similar sequence, which constitutes the −10 promoter elements of the icmV, icmW, and icmR genes which had high levels of expression in L. pneumophila, was also identified. In addition, regulatory elements that probably serve as binding sites for transcription regulators were found in these genes. Altogether, 12 regulatory elements, 7 of which constitute the −10 promoter elements of the icm genes, were found. Even though all the icm and dot genes are part of one system required for L. pneumophila intracellular growth and even though their promoters are probably recognized by the vegetative sigma factor, it seems that they are subjected to different regulation mediated by several regulatory factors.
Legionella pneumophila, the causative agent of Legionnaires' disease, is a facultative intracellular pathogen. L. pneumophila is able to infect, multiply within, and kill human macrophages, as well as free-living amoebae (28, 38). The bacteria are taken up by regular phagocytosis or by a special mechanism termed “coiling” phagocytosis (9, 26). They are then found within a specialized phagosome that does not fuse with lysosomes or acidify (9, 24, 27). The specialized phagosome undergoes several recruitment events, which include association with smooth vesicles, mitochondria, and the rough endoplasmic reticulum (1, 25, 49); the bacteria multiply within the specialized phagosome until the cell eventually lyses, releasing bacteria that can start new rounds of infection (28, 38).
Two regions of genes required for human macrophage killing and intracellular multiplication have been discovered in L. pneumophila (reviewed in references 44 and 51). Region I contains 7 genes (icmV, -W, and -X and dotA, -B, -C, and -D) (8, 10, 32, 50), and region II contains 17 genes (icmT, -S , -R, -Q, -P, -O, -N, -M, -L, -K, -E, -G, -C, -D, -J, -B, and -F) (3, 35, 41, 43, 50). Complementation analysis of these genes indicated that they are probably organized as nine transcriptional units (icmTS, icmR, icmQ, icmPO, icmMLKEGCD, icmJB, icmF-tphA, icmWX, and icmV-dotA) (8, 10, 35, 41, 43, 50). Most of these genes were also shown to be required for intracellular growth in protozoan host Acanthamoeba castellanii (45). Fourteen of the Icm and Dot proteins (IcmT, -P, -O, -L, -K, -G, -C, -D, -J, and -B and DotA, -B, -C, and -D) were found to have significant sequence similarity to Tra and Trb proteins from IncI plasmids colIb-P9 and R64 (29, 46). The icm/dot system is believed to serve as a translocation system that delivers effector proteins into host cells (34, 44).
At the present time there is very little information about the regulation of L. pneumophila virulence, as well as about regulatory factors that control the expression of the 24 icm and dot genes. So far, stationary-phase sigma factor rpoS has been shown to be involved in L. pneumophila virulence: a strain with a knockout in this gene lost its ability to grow in protozoan host A. castellanii (20) and was attenuated for intracellular growth in murine bone marrow-derived macrophages (4). However, this gene was found to be dispensable for growth in HL-60-derived human macrophages and in THP-1 cells (20). Another stationary-phase-related factor whose involvement in the regulation of L. pneumophila virulence was suggested was the relA gene product (21), but this gene was found to be dispensable for intracellular growth (55). In addition, the expression of only one out of nine icm::lacZ fusions was reduced in strains containing an insertion in the rpoS or the relA gene (55), indicating that other factors control the expression of the icm and dot genes.
We were interested in identifying DNA regulatory elements that control icm and dot gene expression. To address this goal, we used random and site-directed mutagenesis on the regulatory regions of nine icm and dot genes that were fused to the lacZ reporter. We identified 12 regulatory elements in the upstream region of eight icm and dot genes. Primer extension analysis indicated that seven of these sites constitute the −10 promoter elements of the icm genes; the other sites are expected to serve as binding sites for transcription regulators.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
The L. pneumophila strain used in this work was JR32, a streptomycin-resistant, restriction-negative mutant version of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (39). The Escherichia coli strain used was MC1061 with a deletion of the lacZ gene (13). Plasmids and primers used in this work are described in Tables 1 and 2, respectively. Bacterial media, plates, and antibiotic concentrations used were as described before (43).
TABLE 1.
Plasmids used in this study
| Plasmid | Features | Reference or source |
|---|---|---|
| pGS-lac-01 | pAB-1 with a promoterless lacZ gene | 55 |
| pGS-lac-02 | pAB-1 with a promoterless lacZ gene | This study |
| pGS-Lp-34 | icmPO and their regulatory region in pUC-18 | 43 |
| pGS-Lp-35 | icmQ and its regulatory region in pUC-18 | 43 |
| pGS-Lp-36 | icmR and its regulatory region in pUC-18 | 43 |
| pGS-Lp-82 | icmV and icmW regulatory region in pUC-18 | This study |
| pGS-reg-F2 | Regulatory region of icmF in pMC1403 | 55 |
| pGS-reg-F3 | Regulatory region of icmF in pGS-lac-01 | 55 |
| pGS-reg-M2 | Regulatory region of icmM in pMC1403 | 55 |
| pGS-reg-M3 | Regulatory region of icmM in pGS-lac-01 | 55 |
| pKP-Q-21 | Regulatory region of icmQ in pMC1403 | 55 |
| pOG-J-109 | Regulatory region of icmJ in pMC1403 | 55 |
| pOG-J-122 | Regulatory region of icmJ in pGS-lac-01 | 55 |
| pOG-P-108 | Regulatory region of icmP in pMC1403 | 55 |
| pOG-P-121 | Regulatory region of icmP in pGS-lac-01 | 55 |
| pOG-Q-126 | Regulatory region of icmQ in pGS-lac-01 | 55 |
| pOG-R-125 | Regulatory region of icmR in pGS-lac-01 | 55 |
| pOG-T-107 | Regulatory region of icmT in pMC1403 | 55 |
| pOG-T-120 | Regulatory region of icmT in pGS-lac-01 | 55 |
| pOG-V-110 | Regulatory region of icmV in pMC1403 | 55 |
| pOG-V-123 | Regulatory region of icmV in pGS-lac-01 | 55 |
| pOG-W-111 | Regulatory region of icmW in pMC1403 | 55 |
| pOG-W-124 | Regulatory region of icmW in pGS-lac-01 | 55 |
| pSS-R-27 | Regulatory region of icmR in pMC1403 | 55 |
| pUC18 | oriR(ColE1) MCSa Apr | 54 |
MCS, multiple cloning sites.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| PE-icmP | GACGATCCATACTGGCGCCATG |
| PE-icmQ | TGGACCTTTTTCAATAGCATCG |
| PE-icmR | AAAAACCAAATGGATTTCGTGCACTG |
| PE-icmV | TTCATTCTGTCCCAATCGAACC |
| PE-icmW | CCAGTACTTTGCGGAGGCTTCATGGC |
| IcmF-EI | GCCGGAATTCGAACAAGGAGCAAGTATTTC |
| IcmF-Bam | CGGGGGATCCCCGTATTGCTCAGTTGTCATTATAT |
| pMC-lac | TAAGTTGGGTAACGCCAGGG |
| pMC-amp | AAGGGAATAAGGGCGACACG |
Plasmid construction.
Promoterless lacZ vector pGS-lac-01 (55) was used to construct promoterless lacZ vector pGS-lac-02. Vector pGS-lac-01 contains two BamHI sites, one in the region immediately upstream from the promoterless lacZ gene and another downstream from the lacZ gene. To use the BamHI site for cloning, pGS-lac-01 was partially digested with BamHI and self ligated after fill-in. One of the plasmids generated in which the BamHI site located upstream from the lacZ gene was left unchanged and the second site was diminished was named pGS-lac-02.
The plasmids that were used as templates for the sequencing reactions analyzed together with the primer extension reactions are listed in Table 1. Plasmid pGS-Lp-82 was constructed by cloning a HindIII-EcoRV fragment containing the icmW gene, part of the icmV and icmX genes, and the regulatory region between icmV and icmW into the pUC-18 vector.
Site-directed mutagenesis.
Site-directed mutagenesis was performed by the overlap extension PCR method (23). For each mutation, two primers that contain the mutation and that overlap one another by 20 bp were designed. The PCR template for all the mutations was the regulatory region of the gene of interest cloned in the pMC1403 vector (Table 1). PCR mutagenesis includes two steps. In the first step two PCR fragments were generated with the following pairs of primers: (i) a primer located on the vector, upstream from the regulatory region (pMC-amp), and one of the primers that contain the mutation and (ii) a primer located in the lacZ gene on the vector (pMC-lac) and the other primer that contains the same mutation on the complementary strand. The resulting two fragments were gel purified and used as templates in the second step, which includes a third PCR using the two primers located on the vector. The resulting PCR product was digested with BamHI and cloned into the pGS-lac-02 vector. All the mutations were confirmed by sequencing the whole regulatory region. The changes made were always A to C, T to G, and C to A.
PCR random mutagenesis.
Random PCR mutagenesis was performed essentially as described before (18). It was performed on the icmF regulatory region by using the icmF reverse primer containing a BamHI site (icmF-Bam) and the icmF forward primer containing an EcoRI site (icmF-EI) (Table 2). The PCR mixture included either 50 mM dATP, 0.5 mM MnCl2, and 9.5 mM MgCl2 or 50 mM dCTP, 0.5 mM MnCl2, and 5.5 mM MgCl2, along with other deoxynucleoside triphosphates at 0.2 mM, 100 ng of template (pGS-reg-F2), 25 pmol of each primer, and 1 U of Super-Therm DNA polymerase. PCR under these conditions results in 1 to 3 bp changes per 100 bp of amplicon. The PCR product was digested with EcoRI and BamHI and cloned into the pGS-lac-02 vector digested with the same enzymes. The resulting colonies were screened on MacConkey plates, suspected colonies were isolated, and the mutated regulatory regions from some of them were sequenced.
β-Galactosidase assays.
β-Galactosidase assays were performed as described elsewhere (33). L. pneumophila strains were grown on ABCYE (ACES buffered charcoal yeast extract) plates containing chloramphenicol for 48 h. The bacteria were scraped off the plate and suspended in AYE (ACES yeast extract) broth, and bacterial optical density at 600 nm (OD600) was calibrated to 0.1 in AYE. The resulting cultures were grown on a roller drum for about 18 h until reaching an OD600 of about 3.8 (stationary phase). To test the levels of expression at exponential phase, the cultures were diluted to an OD600 of 0.1 and grown for an additional 6 to 7 h until reaching an OD600 of about 0.7 (exponential phase). The assays were done with 50 or 100 μl of culture. The substrate for lacZ hydrolysis was o-nitrophenyl-β-d-galactopyranoside.
Preparation of RNA.
RNA preparation was performed as described elsewhere (42). The L. pneumophila JR32 strain was grown on ABCYE plates for 48 h. The bacteria were scraped off the plate and suspended in AYE broth, and the bacterial OD600 was calibrated to 0.1 in AYE. The resulting cultures were grown on a roller drum for about 18 h until reaching an OD600 of about 3.8. Then the cultures were diluted to OD600 of 0.1 in AYE and grown for additional 6 to 7 h until reaching an OD600 of about 0.7. These bacteria were used for RNA preparation. The cell pellets derived from 30 ml of bacteria were suspended in 3 ml of STE buffer (10 mM Tris [pH 7], 100 mM NaCl, 1 mM EDTA), and an equal volume of aqueous 95% hot phenol (65°C) was added. This mixture was shaken vigorously at 65°C for 10 min and centrifuged for 10 min at 11,000 × g. The aqueous phase was removed, and the phenol phase was reextracted with 3 ml of STE buffer. After centrifugation, the aqueous phase was removed and combined with that already collected, and the solution was extracted twice with hot phenol, once with phenol-chloroform-isoamyl alcohol (25:24:1), and twice with STE-saturated ether. The RNA was precipitated with 2 volumes of ethanol. After approximately 12 h of incubation at −20°C, the precipitate was recovered by centrifugation at 11,000 × g for 10 min. The RNA concentration was determined by measuring OD260.
Primer extension analysis.
The primers used for the analysis are listed in Table 2. A reverse transcriptase (RT) reaction was performed in RT buffer (50 mM Tris-Cl [pH 8.3], 8 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol) containing deoxynucleoside triphosphates (1 mM concentration [each] of dATP, dCTP, dGTP, and dTTP), RNasin (10 U), 5 pmol of labeled ([γ-32P]ATP) primer, and 5 U of avian myeloblastosis virus RT (Promega, Madison, Wis.). A total of 10 μg of the RNA preparation was used in a total reaction mixture volume of 6 μl. The reaction mixture was incubated for 5 min at 45°C and then incubated at 37°C for 30 min. After the addition of 4 μl of sequencing stop buffer, the samples were heated for 5 min at 90°C before being loaded on a sequencing gel.
DNA sequencing.
The sequence was determined by the dideoxy termination method (40) using the SequiTherm cycle sequencing kit (Epicentre, Madison, Wis.). The templates for the sequencing reactions were plasmids containing the regulatory regions of the genes tested (Table 1). Sequencing was determined with the same primers used for the primer extension analysis.
RESULTS
We were interested in identifying DNA regulatory elements that control the expression of the L. pneumophila icm and dot genes required for intracellular growth. According to complementation analysis that was performed previously (10, 35, 41, 43), the icm and dot genes are organized on nine transcriptional units (icmTS, icmR, icmQ, icmPO, icmMLKEGCD, icmJB, icmF-tphA, icmWX, and icmV-dotA). The results obtained by the complementation analysis are in agreement with the organization of the genes in the two icm/dot regions. There are between 91 and 400 bp of a noncoding region upstream of genes expected to be first in a transcriptional unit and between 0 and 35 bp of a noncoding region upstream of genes that are not expected to be first in a transcriptional unit. On the basis of these data, nine icm::lacZ fusions (icmT::lacZ, icmR::lacZ, icmQ::lacZ, icmP::lacZ, icmM::lacZ, icmJ::lacZ, icmF::lacZ, icmV::lacZ, and icmW::lacZ) that contain the whole region between the gene of interest and the gene located upstream of it, with a minimum of 150 bp, were constructed.
Analysis of the expression levels of icm::lacZ fusions in L. pneumophila.
It has been suggested that L. pneumophila pathogenesis is coordinated with entry into stationary phase (4, 11, 21). To test whether icm and dot genes that were shown to be required for intracellular growth are regulated in correlation with growth phase, we compared the expression levels of the nine icm::lacZ fusions at exponential and stationary phases.
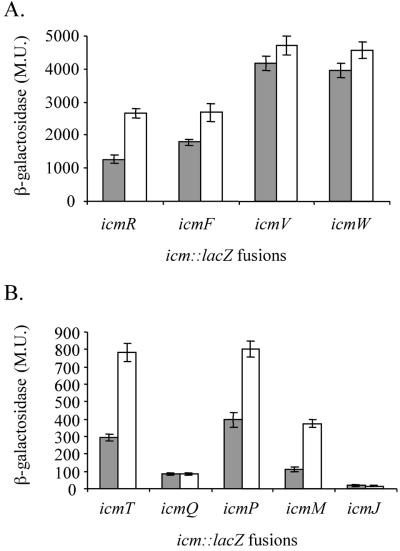
Even though all the icm and dot genes were shown to be required for intracellular growth and host cell killing, their expression levels were found to be different from one another. As can be seen in Fig. 1, the icm::lacZ fusions can be divided into two groups according to their expression levels in the wild-type L. pneumophila JR32 strain. Four icm::lacZ fusions (icmR::lacZ, icmF::lacZ, icmV::lacZ, and icmW::lacZ) were found to have high levels of β-galactosidase expression (above 1,000 Miller units [MU]), and two of them had higher levels of expression at stationary phase than at exponential phase (Fig. 1A). In contrast, five icm::lacZ fusions (icmT::lacZ, icmP::lacZ, icmQ::lacZ, icmM::lacZ, and icmJ::lacZ) were found to have low levels of β-galactosidase expression (below 1,000 MU), and three of these fusions had higher levels of expression at stationary phase than at exponential phase (Fig. 1B). Only one of these fusions (icmP::lacZ) was shown to have lower levels of expression in strains containing an insertion in the genes coding for RpoS or RelA than in wild-type strains (55); the other fusions (icmT::lacZ, icmM::lacZ, icmR::lacZ, and icmF::lacZ), which had higher levels of expression at stationary phase than at exponential phase, are probably regulated by other factors.
FIG. 1.
Expression of icm::lacZ fusions in L. pneumophila at exponential and stationary phases. The expression of the nine icm::lacZ fusions (for the genes icmT, icmR, icmQ, icmP, icmM, icmJ, icmF, icmW, and icmV) in wild-type strain JR32 at exponential (gray) and stationary phases (white) was examined. β-Galactosidase activity was measured as described in Materials and Methods. Four of the icm::lacZ fusions were found to have high β-galactosidase activities (A), and five were found to have low activities (B). The results (Miller units [M.U.]) are the averages ± standard deviations (error bars) of at least three different experiments.
Analysis of the expression levels of icm::lacZ fusions in E. coli.
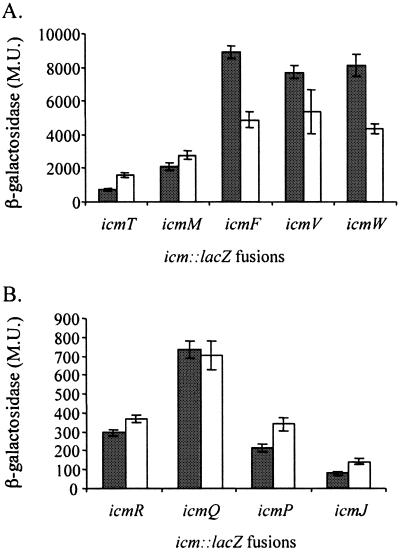
One way to get an indication of whether unique L. pneumophila regulatory factors participate in icm gene expression is to compare their expression in L. pneumophila with that in E. coli. When we compared the levels of expression of the icm::lacZ fusions described above in E. coli and L. pneumophila, several interesting differences were observed (compare Fig. 1 and 2). Three icm::lacZ fusions (icmF::lacZ, icmV::lacZ, and icmW::lacZ) that had high levels of expression in L. pneumophila were also found to have high levels of expression in E. coli (compare Fig. 1A and 2A). In contrast, the icmR::lacZ fusion, which was highly expressed in L. pneumophila, had a very low level of expression in E. coli (fourfold reduction at exponential phase and sevenfold reduction at stationary phase; compare Fig. 1A and 2B). Two other fusions (icmQ::lacZ and icmM::lacZ) had the opposite result: their levels of expression were higher in E. coli than in L. pneumophila, showing an increase of at least sevenfold both at exponential and stationary phases (compare Fig. 1 and 2). Four out of the five icm::lacZ fusions that had higher levels of expression at stationary phase than at exponential phase in L. pneumophila (icmT::lacZ, icmP::lacZ, icmM::lacZ, and icmR::lacZ) showed increased levels of β-galactosidase at stationary phase in E. coli as well, but the increase was less pronounced.
FIG. 2.
Expression of icm::lacZ fusions in E. coli at exponential and stationary phases. The expression of the nine icm::lacZ fusions (for the genes icmT, icmR, icmQ, icmP, icmM, icmJ, icmF, icmW, and icmV) in E. coli strain MC1061 at exponential (gray) and stationary phases (white) was examined. β-Galactosidase activity was measured as described in Materials and Methods. Five of the icm::lacZ fusions were found to have high β-galactosidase activities (A), and four were found to have low activities (B). The results (Miller units [M.U.]) are the averages ± standard deviations (error bars) of at least three different experiments.
We think that these results clearly indicate that the promoters of the L. pneumophila icm and dot virulence genes are recognized in E. coli and are probably transcribed by a sigma factor present in both bacteria. However, it is most likely that there are other regulatory factors (a repressor for icmQ and icmM and an activator for icmR) present in L. pneumophila that are absent in E. coli.
Identification of −10 promoter elements of icm genes that have low expression levels.
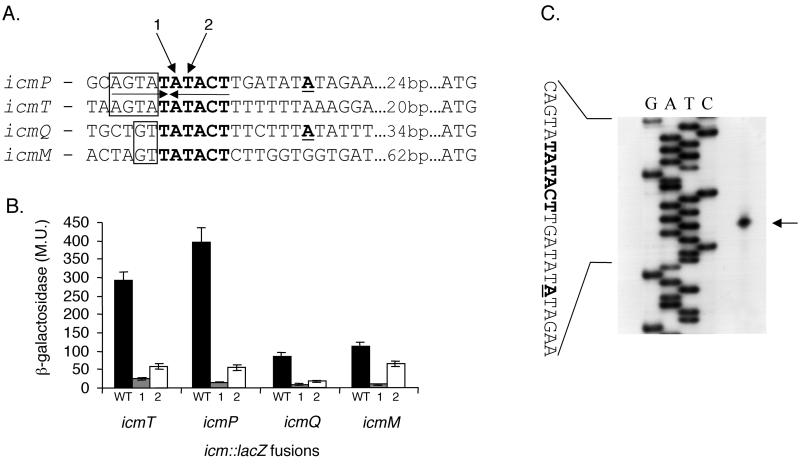
Careful analysis of the upstream regulatory regions of four icm genes (icmT, icmP, icmQ, and icmM) that were found to have low levels of expression in L. pneumophila, revealed a 6-bp (TATACT) putative consensus sequence (Fig. 3A). The distances of this sequence from the first ATG codons of the corresponding genes (32 to 74 bp), as well as the sequence itself, suggest that it might serve as the −10 promoter element for these genes. This sequence differs by only 1 nucleotide from the −10 promoter element recognized by E. coli vegetative sigma factor RpoD (TATAAT) (53). To examine the importance of this sequence in the expression of these icm::lacZ fusions, 2 nucleotides in each of these sequences were mutated and the effect of the change was examined. As can be seen in Fig. 3B, the change in either of these two positions (the second or third nucleotide of the putative consensus sequence) caused a dramatic reduction in the levels of expression of these icm::lacZ fusions. The mutation in the second position of the putative consensus (mutation 1 in Fig. 3) almost eliminated the expression of the lacZ reporter, and the levels of β-galactosidase observed were close to the expression level of the promoterless lacZ vector, the negative control (pGS-lac-02) (data not shown).
FIG. 3.
Analysis of the regulatory regions of icm::lacZ fusions that have low levels of expression. (A) Sequences of the regulatory regions of icmP, icmT, icmQ, and icmM are shown. The putative consensus sequence is in boldface, and the distances to the first ATG are indicated. The nucleotides representing the 5′ end of the mRNA are in boldface and underlined. Additional sequences that form potential inverted repeats with the consensus found are boxed. Arrows 1 and 2, nucleotides mutated. The changes made were always A to C and T to G. (B) Expression of icmT::lacZ, icmP::lacZ, icmQ::lacZ, and icmM::lacZ fusions and of two mutant versions of each fusion in L. pneumophila at exponential phase was determined. For each of the fusions, results for the wild-type (WT) regulatory region and for regulatory regions containing mutations in positions 1 and 2 are shown. β-Galactosidase activity was measured as described in Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. (C) Mapping of the 5′ end of the icmP transcript by primer extension. Primer extension was performed as described in Materials and Methods with a primer complementary to the 5′ end of the icmP gene. G, A, T, and C (top), products of the sequencing reaction obtained by using the same primer. The sequence presented is that of the sense strand. Arrow, nucleotide representing the 5′ end of the mRNA. Boldface and underlining are as described for panel A.
As indicated in the two previous sections, icmT and icmP have similar expression levels and expression profiles in L. pneumophila (higher expression at stationary phase than at exponential phase). In addition, they contain the same 4 nucleotides upstream from their TATACT consensus sequences; these 4 nucleotides might form an inverted repeat with the consensus found (Fig. 3A). On the other hand, icmM and icmQ also have similar levels of expression in L. pneumophila and they were both found to have higher levels of expression in E. coli than in L. pneumophila (sevenfold increase at both stationary and exponential phases). Due to these results, the transcription start sites for one of the genes in each pair (icmP and icmQ) were determined (Fig. 3A), and the primer extension result for the icmP gene is presented in Fig. 3C. As can be seen clearly from the primer extension analysis, the TATACT site constitutes the −10 promoter element of these genes, as was expected from the distance of the consensus from the first ATG of the genes, the sequence itself, and the mutagenesis results.
All the positions in the TATACT consensus are required for maximal expression.
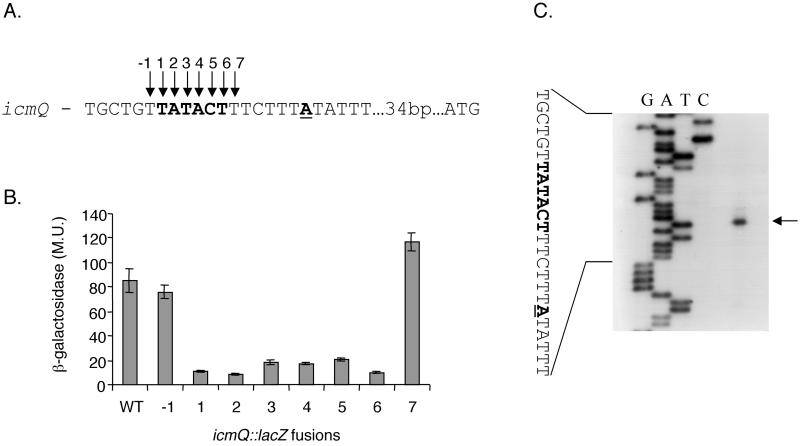
To determine if all the positions in the TATACT consensus are required for maximal expression, we performed a careful analysis of the icmQ consensus sequence (Fig. 4). Eight mutations in the icmQ consensus sequence were constructed (Fig. 4A): the 6 nucleotides of the TATACT consensus as well as 1 nucleotide on both sides of it were mutated. As can be seen very clearly in Fig. 4B, all six positions that are expected to be part of the consensus sequence were found to be required for maximal expression of the icmQ::lacZ fusion. The mutations upstream and downstream from the consensus sequence did not reduce the expression of the icmQ::lacZ fusion, and they determine the boundaries of the consensus.
FIG. 4.
Analysis of the regulatory region of the icmQ gene. (A) The sequence of the regulatory region of icmQ is shown (boldface, putative consensus sequence). The nucleotide representing the 5′ end of the mRNA is in boldface and underlined. Arrows −1 to 7, nucleotides mutated. The changes made were always A to C, T to G, and C to A. (B) The expression of the icmQ::lacZ fusion (WT) and of the eight mutants from panel A in L. pneumophila at exponential phase was measured. β-Galactosidase activity was measured as described in Materials and Methods. The results are the averages ± standard deviations (error bars) of at least three different experiments. (C) Mapping of the 5′ end of the icmQ transcript by primer extension. Primer extension was performed as described in Materials and Methods with a primer complementary to the 5′ end of the icmQ gene. G, A, T, and C (top), products of the sequencing reaction obtained by using the same primer. The sequence presented is that of the sense strand. Arrow, nucleotide representing the 5′ end of the mRNA. Boldface and underlining are as described for panel A.
A primer extension analysis of this region determined the transcription start site of the icmQ transcript (Fig. 4C). The transcription start site was located 6 bp downstream from the last nucleotide of the consensus, indicating that the TATACT consensus constitutes the −10 promoter element of this gene. It is important to note that the changes at positions 1, 2, and 6 caused the strongest reduction in the expression level of the icmQ::lacZ fusion whereas the changes in the three other positions of the consensus (positions 3, 4, and 5) had a clear but less dramatic effect on the expression of this fusion. These results are in agreement with the known data about E. coli rpoD promoters showing that the first, second, and sixth nucleotide of its −10 promoter element are the most conserved (31).
icmR contains at least three regulatory elements.
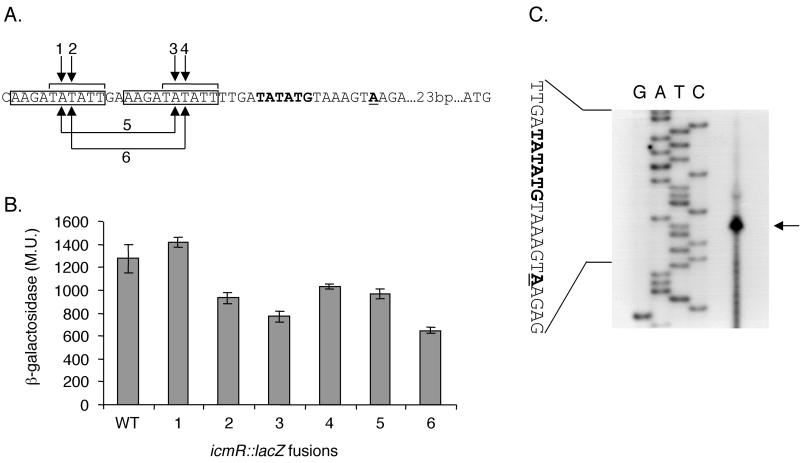
Analysis of the icmR regulatory region identified two identical sequences (AAGATATATT) located next to each other. Because the 6 bp (TATATT) located at the 5′ end of this sequence differ by only 1 nucleotide from the −10 promoter element described above (TATACT), we thought that these two sites might serve as regulatory elements of the icmR gene. To test this hypothesis, single as well as double mutations in the icmR regulatory region were constructed (Fig. 5A). As can be seen in Fig. 5B, the effect of the mutations in the icmR regulatory region was less pronounced than the effect of mutations in icmT, icmP, icmQ, and icmM. However, the combination of the two mutations at the third position (mutation 6 in Fig. 5B) caused a reduction of about 50% in the expression of the icmR::lacZ fusion, which was a more severe reduction than that due to each individual mutation at this position (mutations 2 and 4 in Fig. 5B).
FIG. 5.
Analysis of the regulatory region of the icmR gene. (A). The sequence of the regulatory region of icmR is shown, and the distance from the first ATG is indicated. The two putative consensus sequences are boxed, and the part of the consensus sequence (TATATT) similar to the consensus sequences found in the icmT, icmP, icmQ, and icmM genes is also indicated. The nucleotide representing the 5′ end of the mRNA is in boldface and underlined, and the −10 promoter element is in boldface. Arrows 1 to 6, nucleotides mutated. The changes made were always A to C and T to G. (B) The expression of the icmR::lacZ fusion (WT) and of the six mutants from panel A in L. pneumophila at exponential phase was measured. β-Galactosidase activity was measured as described in Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments. (C) Mapping of the 5′ end of the icmR transcript by primer extension. Primer extension was performed as described in Materials and Methods with a primer complementary to the 5′ end of the icmR gene. G, A, T, and C (top), products of the sequencing reaction obtained by using the same primer. The sequence presented is that of the sense strand. Arrow, nucleotide representing the 5′ end of the mRNA. Boldface and underlining are as described for panel A.
To determine if either of these two sites serves as a −10 promoter element of the icmR gene, the start site of its transcript was determined (Fig. 5C). The transcription start site clearly indicates that the two sites mutated are not the −10 promoter element of the icmR gene. The sequence of its −10 promoter element was found to be TATATG, which is only 2 nucleotides different from the −10 promoter elements identified for icmT, icmP, icmQ, and icmM. We concluded that the AAGATATATT consensus plays a significant regulatory role in the expression of the icmR gene, probably as a binding site for a regulatory protein, and that the TATATG site constitutes the −10 promoter element of this gene.
Identification of a regulatory elements of icm genes that have high expression levels.
icmW, icmV, and icmF are the only three genes that were found to have high levels of expression in both L. pneumophila and E. coli (Fig. 1A and 2A). To find regulatory elements that are involved in the expression of these genes, the regulatory region of the icmF transcriptional unit was randomly mutated by PCR. Four clones that each contain a single mutation were identified, and all mutations mapped close to one another. Three mutations (from three independent PCRs) were found at the same position, and a fourth mutation was found five nucleotides downstream from it (Fig. 6A). These mutations caused a severe reduction in the level of expression of the icmF::lacZ fusion (data not shown).
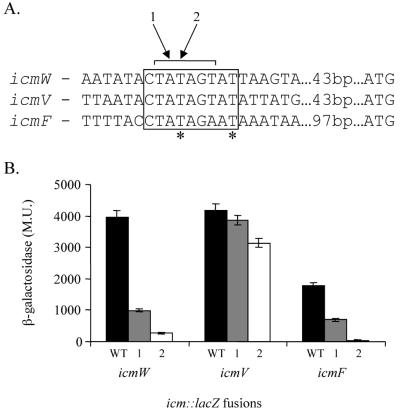
FIG. 6.
Analysis of the regulatory region of icm::lacZ fusions that have high expression levels. (A) Sequences of the regulatory regions of icmW, icmV, and icmF. The distances to the first ATG are indicated. The region of sequence similarity is boxed, and the sequence (TATAGT) similar to those of the −10 promoter elements of the icmT, icmP, icmQ, and icmM genes is indicated. ∗, nucleotides in the icmF regulatory region in which random mutations were identified; arrows 1 and 2, nucleotides mutated by site-directed mutagenesis. The changes made were always A to C and T to G. (B) The expression of icmW::lacZ, icmV::lacZ, and icmF::lacZ fusions as well as of two mutant versions of each of these fusions in L. pneumophila at exponential phase was measured. For each of the genes, the wild-type (WT) regulatory region and the regulatory regions containing mutations in positions 1 and 2 are shown. β-Galactosidase activity was measured as described in Materials and Methods. The data are the averages ± standard deviations (error bars) of at least three different experiments.
From a comparison of the randomly mutated site in the icmF regulatory region to the regulatory regions of icmV and icmW (these two genes are transcribed back to back and their regulatory regions overlap) a 9-bp putative consensus sequence (CTATAGTAT) was identified (Fig. 6A). To determine the importance of this site in the expression of icmV, icmW, and icmF, 2 nucleotides in each of these putative regulatory elements were mutated (Fig. 6A). As can be seen in Fig. 6B, the effect of these mutations on the icmW::lacZ and icmF::lacZ fusions was dramatic. The expression of these lacZ fusions was reduced about 10-fold when the fourth position of the consensus was mutated (mutation 2). In constructs, the effect on the icmV::lacZ fusion was minor, and only one of the mutations resulted in a 20% reduction in expression. Because of this result and because icmV and icmW are transcribed back to back and the mutated sites overlap one another (Fig. 7; the region is shown as double stranded), we decided to analyze their regulatory regions further, as described below.
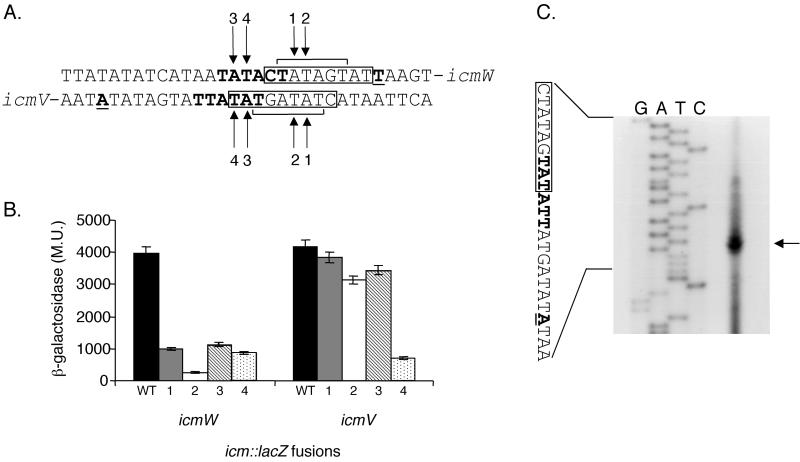
FIG. 7.
Analysis of the icmW and icmV regulatory regions. (A) The sequences of the regulatory regions of icmW and icmV are shown. These two genes are transcribed back to back, and the region is shown as double stranded. The 9-bp consensus sequence is boxed, and the internal 6-bp sequence (TATAGT) similar to the promoter elements found in the icmT, icmP, icmQ, and icmM genes is indicated. The nucleotides representing the 5′ end of the mRNA are in boldface and underlined, and the −10 promoter elements are in boldface. Arrows 1 to 4, nucleotides mutated. The changes made were always A to C and T to G. (B) The expression of icmW::lacZ, and icmV::lacZ fusions as well as four mutant versions of each of these fusions in L. pneumophila at exponential phase was measured. For each of the genes, the wild-type (WT) regulatory region and the regulatory regions containing mutations in positions 1, 2, 3, and 4 are shown. β-Galactosidase activity was measured as described in Materials and Methods. The data are the averages of at least three different experiments. (C) Mapping of the 5′ end of the icmV transcript by primer extension. Primer extension was performed as described in Materials and Methods with a primer complementary to the 5′ end of the icmV gene. G, A, T, and C (top), products of the sequencing reaction obtained by using the same primer. The sequence presented is that of the sense strand. Arrow, nucleotide representing the 5′ end of the mRNA. Boldface and underlining are as described for panel A.
icmV and icmW have overlapping regulatory regions.
Careful analysis of the icmV and icmW overlapping regulatory regions revealed an interesting organization of several overlapping putative consensus sequences. The two sites examined in the previous section (CTATAGTAT; one in the icmV regulatory region and one in the icmW regulatory region; Fig. 7A) overlap one another by 6 bp, and both of them also overlap another possible regulatory element (Fig. 7A). In the icmW regulatory region, upstream from the CTATAGTAT site (overlapping by 2 nucleotides), there is a TATACT site (a sequence identical to those of the −10 promoter elements found in icmT, icmP, icmQ, and icmM). Moreover, in the icmV regulatory region, downstream from the CTATAGTAT site (overlapping by three nucleotides), there is a TATATT site (a sequence identical to part of the consensus sequence found in icmR). The organization of these sites in relation to one another is presented in Fig. 7A.
To determine the involvement of the sites mentioned above in the expression of icmV and icmW, four additional mutations in this region were constructed and their effect on the expression of these fusions was determined (Fig. 7B). In icmW, mutations in both sites (CTATAGTAT and TATACT) result in dramatic reduction in β-galactosidase activity. In icmV, the mutations in the CTATAGTAT putative consensus had a minor effect on the expression of the lacZ gene but the change in the third position of the TATATT putative consensus reduced the expression to less then 20%.
To determine which of these sites, if any, constitute the −10 promoter elements of the icmV and icmW genes, the transcription start sites of both genes were determined, and the primer extension analysis of the icmV gene is presented in Fig. 7C. This analysis clearly indicates that TATACT in icmW and TATATT in icmV constitute the −10 promoter elements of these genes. The 9-bp consensus sequence (CTATAGTAT) identified probably serves as a binding site for a transcription regulator.
DISCUSSION
L. pneumophila is an intracellular pathogen that inhibits phagosome-lysosome fusion and that grows inside human macrophages and amoebae (28, 38). Thus far, 24 icm and dot genes required for intracellular growth have been found and characterized (44, 51). Even though these genes have been known for some time, there is no information concerning DNA regulatory elements and regulatory factors that control their expression.
It has been suggested that two major stationary-phase-related factors (RpoS and RelA) are involved with L. pneumophila pathogenicity (4, 11, 21). However, the RpoS sigma factor was shown to be dispensable for intracellular growth in HL-60-derived macrophages and THP-1 cells (20), and the RelA regulatory factor was shown to be dispensable for intracellular growth in both amoebae and HL-60-derived human macrophages (55). In contrast, the icm and dot genes were shown to be required for intracellular growth in all the hosts examined (HL-60- and U937-derived human macrophages [41, 52], murine bone marrow-derived macrophages [50], and protozoan hosts A. castellanii [45] and Dictyostelium discoideum [47]). In addition, analysis of the effect of a strain containing an insertion in the rpoS gene (20) or the relA gene (55) on the expression of the nine icm::lacZ fusions described revealed that the expression of only one fusion (icmP::lacZ) was moderately reduced (55). This might explain the increase in β-galactosidase activity observed with the icmP::lacZ fusion at stationary phase (Fig. 1), but it cannot explain the results for the icmT::lacZ, icmR::lacZ, icmM::lacZ, and icmF::lacZ fusions, in which higher levels of β-galactosidase at stationary phase than at exponential phase were observed as well (Fig. 1).
One approach to identify regulatory factors that control the expression of a certain gene is to identify the DNA regulatory elements of the gene of interest and then to find regulatory factors that interact with these sites. To gain information about the DNA regulatory elements that control icm and dot gene expression, we analyzed their upstream regulatory regions by extensive site-directed and random mutagenesis as well as primer extension analysis. We were able to identify 12 DNA regulatory elements that are involved in the regulation of eight icm genes. Seven of these sites constitute the −10 promoter elements of the icm genes, and the other five sites (two in the icmR regulatory region and one each in the icmV, icmW, and icmF regulatory regions) probably serve as binding sites for regulatory proteins. The sequences of all these regulatory elements are summarized in Fig. 8.
FIG. 8.
Sequence alignment of the icm regulatory regions. The sequences are aligned according to the −10 promoter elements (boldface). The transcription start sites are in boldface and are underlined. ∗, mutations (29 in all). In icmP, icmT, icmQ, and icmM, sequence homologies between pairs of genes are boxed. In icmR, the 10-nucleotide direct repeat is boxed. In icmV, icmW, and icmF, the 9-nucleotide regulatory element is boxed. Arrows (indicating the center of symmetry), potential inverted repeats. The distance to the first ATG of each gene is indicated. CONC., consensus.
Four icm::lacZ fusions that had low levels of expression (icmT, icmP, icmQ, and icmM fusions) were found to contain a 6-bp consensus sequence (TATACT) essential for their expression (Fig. 3). As can be seen in Fig. 8, these four genes can be divided into two pairs. In icmT and icmP there are four additional nucleotides (AGTA), located immediately upstream from the TATACT regulatory element, that can form an inverted repeat that overlaps the regulatory element found. In addition, these two genes were found to be expressed at similar levels in L. pneumophila, and both of them had higher levels of expression at stationary phase than at exponential phase (Fig. 1). The second pair of genes is icmQ and icmM. These two genes have only 2 bp (GT) of identical sequence immediately upstream from the TATACT site. In addition, they contain two identical sequences of 6 and 5 nucleotides (AGTGAT and TATAA, respectively) located upstream from the TATACT site. One of these sequences (TATAA) can form a potential inverted repeat that overlaps the regulatory element (Fig. 8). In addition, the expression of these two genes at both stationary and exponential phases was at least seven times higher in E. coli than in L. pneumophila (compare Fig. 1 and 2). The examination of a representative gene from each pair clearly indicated that the TATACT site constitutes the −10 promoter elements of these genes (Fig. 3 and 4). The other sites described might serve as binding sites for regulatory proteins. For icmQ and icmM the sites indicated might be used as repressor binding sites, a repressor which is probably missing in E. coli, and as a result of that the expression of the icmQ::lacZ and icmM::lacZ fusions in E. coli increases.
icmJ is the only icm gene that was found to have low expression levels, but the TATACT consensus was not found in its regulatory region (even when a 300-bp regulatory region was cloned upstream of the lacZ gene, the same low level of expression was observed [data not shown]). A potential site that has 2 nucleotides different from the consensus (TATCTT) was the closest match found, but, when the second and third nucleotides of this site were mutated, no change in the level of expression of the icmJ::lacZ fusion in L. pneumophila or E. coli was observed (data not shown).
icmR is the only gene that was found to have higher levels of expression in L. pneumophila than in E. coli. This gene was found to contain a −10 promoter element similar to those of icmT, icmP, icmM, and icmQ, as determined by primer extension (Fig. 5). In addition, we identified two identical regulatory elements (AAGATATATT) in the upstream region of this gene (Fig. 8) and showed that they play a role in icmR expression. These sites might constitute a binding site for an activator missing in E. coli. The 10-bp consensus sequence that was found in the icmR regulatory region was not found in any of the other icm regulatory regions. In addition, its −10 promoter element differs from all the other −10 promoter elements identified (the last nucleotide of the icmR promoter is G instead of T; Fig. 8). This result is in agreement with the observation that the icmR::lacZ fusion is the only fusion more highly expressed in L. pneumophila than in E. coli. This might indicate that the icmR gene is subjected to a unique regulation not present in the other icm genes.
By random mutagenesis and sequence alignment a 9-bp regulatory element (CTATAGTAT) (Fig. 8) was identified in the upstream regions of the three genes (icmF, icmV, and icmW) that were found to have high expression levels in both E. coli and L. pneumophila. This site was found to have an effect on the expression of these genes, but primer extension analysis indicated that it does not serve as a −10 promoter element. The −10 promoter elements that were determined by primer extension were found to be similar to the −10 promoter elements of the other icm genes (Fig. 8). The −10 promoter element and the regulatory elements identified in icmV and icmW were found to overlap one another in both genes and between the two genes (Fig. 7A). This organization might indicate that these two genes are subjected to complicated regulation.
As can be seen in Fig. 8, all the sites that were identified as −10 promoter elements have extensive homology to one another and are probably recognized by one of the L. pneumophila sigma factors. As most of the genome of L. pneumophila has already been sequenced (http://genome3.cpmc.columbia.edu/∼legion/index.html), we looked for all the genes that have homology to known sigma factors and found that L. pneumophila contains homologs to at least six sigma factors (RpoD, RpoH, RpoF, RpoE, RpoS, and RpoN). The promoter sequence recognized by two of these sigma factors in L. pneumophila is already known (2, 22). RpoH and RpoF recognize sequences similar to the ones recognized by their E. coli homologs (CTTGAAA[11 to 16]CCCATnT for RpoH [n = A, G, T, or C] and TAAA[15]GCCGATAA for RpoF). These sequences are different from the ones found in the icm gene promoter regions, which might indicate that these two sigma factors are not involved in the recognition of the icm promoters. The sequence recognized by the L. pneumophila RpoE and RpoN sigma factors is not known. But it was shown that both the E. coli RpoE and its Pseudomonas aeruginosa homolog recognize the same promoter sequence (16) (GAACTT[16 to 17]TCTGA), and the sequence recognized by the RpoN sigma factor is known for many bacteria (TGGCAC[5]TTGC) (6). Because all these bacteria are evolutionarily closely related to one another (they are all γ purple bacteria), it is most likely that these two sigma factors recognize similar promoter sequences in L. pneumophila. However, as these sequences were not found in the regulatory regions of the icm genes, we concluded that neither of these sigma factors is involved in the recognition of the icm promoters. Both the vegetative sigma factor, RpoD, and the stationary-phase sigma factor, RpoS, of E. coli recognize very similar −10 promoter elements (TATAAT for RpoD and CTATACT for RpoS) (53). This sequence is also similar to the sequence of the −10 promoter element that we found in the icm gene regulatory regions (TATAYT). However, as we recently demonstrated (55), a knockout of the rpoS gene moderately influenced the expression of only one (icmP::lacZ) out of the nine icm::lacZ fusions described. Taking all this information together we believe that the −10 promoter element of the icm and dot genes is recognized by the vegetative sigma factor (RpoD) of L. pneumophila. Further support for this assumption comes from the analysis of the icmQ −10 promoter element (Fig. 4). This analysis revealed that the first 2 nucleotides and the last nucleotide of this −10 promoter element are more important for expression then the other nucleotides. A similar situation was also found with the E. coli −10 promoter element of the vegetative sigma factor (31).
It is well known that promoters recognized by the vegetative sigma factor contain a conserved −35 promoter element in addition to the conserved −10 promoter element (53). Examination of the sequence located at the region where the −35 promoter element should have been located revealed that no obvious consensus sequence can be found in this region (beside the sequences found in the icmM and icmQ regulatory regions) (Fig. 8). However, this is not the first case where a −10 promoter element is present and a −35 element is missing. For two other bacteria, Helicobacter pylori (17) and Agrobacterium tumefaciens (14), a similar situation, which also involve virulence-related genes, was described. In A. tumefaciens it is known that a two-component regulatory system (virA and virG) is involved in the expression of the virulence-related genes (48).
However, two additional sites might be involved in the expression of genes beside the −10 and −35 promoter elements. An extended −10 promoter element was described for several systems (7, 12, 30). The sequence of the extension is composed of 2 nucleotides (usually TG), located 1 bp upstream from the −10 promoter element, and this sequence was shown to interact with region 2.5 of the vegetative sigma factor (5). A sequence identical to that of the known extended −10 promoter element was found only in the icmR promoter (Fig. 8), but it might be that in L. pneumophila other sequences constitute an extended −10 promoter that functions instead of a −35 promoter element in the icm genes. A second sequence that was found to be part of several promoters recognized by the vegetative sigma factor is called the UP element (19, 37). This sequence was found to be located upstream from the −35 promoter element in the region between nucleotides −59 and −42 with respect to the transcription start site, and it is an AT-rich sequence (AAA[A/T][A/T]T[A/T]TTTTNNAAAA) (15) that interacts with the alpha subunit of RNA polymerase (36). No sequence similar to the one presented was found in any of the icm promoter regions.
It is highly possible that additional regulatory elements, like the sequences described, that form potential inverted repeats with the −10 promoter element (Fig. 8) participate in the regulation of the icm genes. In addition, since we used translational fusions (for each gene, the codons for the first seven amino acids were included in the fusion) to determine the expression levels of the different icm genes, we cannot rule out contributions by posttranscriptional events to the differences in the expression observed.
We believe that the icm and dot genes are subjected to complicated regulation, which is required for the correct and successful function of their gene products. The fact that genes that are part of the same virulence system are expressed at different levels and contain similar as well as different DNA regulatory elements might indicate that a regulatory network that includes factors that respond to different environmental and growth conditions participates in their regulation. We began here to explore this regulatory network by finding 12 regulatory elements involved in the regulation of eight icm and dot genes and operons. The identification of these sites will allow us to find additional sites and regulatory factors that respond to changes in environmental signals and growth conditions and that participate in the regulation of the icm/dot virulence machinery.
Acknowledgments
We are grateful to Howard A. Shuman for carefully reading the manuscript.
The research was supported by the Charles H. Revson Foundation of the Israel Science Foundation (grant 45/00). G. Segal was supported by the Alon fellowship awarded by the Israeli Ministry of Education.
REFERENCES
- 1.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amemura-Maekawa, J., and H. Watanabe. 1997. Cloning and sequencing of the dnaK and grpE genes of Legionella pneumophila. Gene 197:165-168. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 5.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the "extended-10′' motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashyam, M. D., and A. K. Tyagi. 1998. Identification and analysis of “extended −10” promoters from mycobacteria. J. Bacteriol. 180:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger, K. H., J. J. Merriam, and R. R. Isberg. 1994. Altered intracellular targeting properties associated with mutations in the Legionella dotA gene. Mol. Microbiol. 14:809-822. [DOI] [PubMed] [Google Scholar]
- 9.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand, B. C., A. B. Sadosky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. [DOI] [PubMed] [Google Scholar]
- 11.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camacho, A., and M. Salas. 1999. Effect of mutations in the “extended-10” motif of three Bacillus subtilis sigmaA-RNA polymerase-dependent promoters. J. Mol. Biol. 286:683-693. [DOI] [PubMed] [Google Scholar]
- 13.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 14.Das, A., S. Stachel, P. Ebert, P. Allenza, A. Montoya, and E. Nester. 1986. Promoters of Agrobacterium tumefaciens Ti-plasmid virulence genes. Nucleic Acids Res. 14:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 95:9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firoved, A. M., J. C. Boucher, and V. Deretic. 2002. Global genomic analysis of AlgU σE-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184:1057-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsyth, M. H., and T. L. Cover. 1999. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J. Bacteriol. 181:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromant, M., S. Blanquet, and P. Plateau. 1995. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal. Biochem. 224:347-353. [DOI] [PubMed] [Google Scholar]
- 19.Gourse, R. L., W. Ross, and T. Gaal. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 20.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 22.Heuner, K., J. Hacker, and B. C. Brand. 1997. The alternative sigma factor σ28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J. Bacteriol. 179:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell. 36:27-33. [DOI] [PubMed] [Google Scholar]
- 27.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 60:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komano, T., T. Yoshida, K. Narahara, and N. Furuya. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35:1348-1359. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 31.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 89:9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 35.Purcell, M. W., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross, W., A. Ernst, and R. L. Gourse. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: alpha subunit binding to the UP element minor groove. Genes Dev. 15:491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 38.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segal, G., and E. Z. Ron. 1993. Heat shock transcription of the groESL operon of Agrobacterium tumefaciens may involve a hairpin-loop structure. J. Bacteriol. 175:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal, G., and H. A. Shuman. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6:253-255. [DOI] [PubMed] [Google Scholar]
- 45.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilize the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal, G., and H. A. Shuman. 1999. Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii. Mol. Microbiol. 33:669-670. [DOI] [PubMed] [Google Scholar]
- 47.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stachel, S. E., and P. C. Zambryski. 1986. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325-333. [DOI] [PubMed] [Google Scholar]
- 49.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 51.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2:30-34. [DOI] [PubMed] [Google Scholar]
- 52.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella pneumophila. Infect. Immun. 66:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and the roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]