Abstract
In this study, we have characterized the in vitro binding of Neisseria gonorrhoeae Fur to several well-defined iron transport genes, as well as to additional genes involved in major catabolic, secretory, and recombination pathways of gonococci. The gonococcal Fur protein was recombinantly expressed in Escherichia coli HBMV119. Fur was isolated from inclusion bodies and partially purified by ion-exchange chromatography. Gonococcal Fur was found to bind to the promoter/operator region of a gene encoding the previously identified Fur-regulated periplasmic binding protein (FbpA) in a metal ion-dependent fashion, demonstrating that purified Fur is functional. In silico analysis of the partially completed gonococcal genome (FA1090) identified Fur boxes in the promoters of several genes, including tonB, fur, recN, secY, sodB, hemO, hmbR, fumC, a hypothetical gene (Fe-S homolog), and the opa family of genes. By using purified gonococcal Fur, we demonstrate binding to the operator regions of tonB, fur, recN, secY, sodB, hemO, hmbR, fumC, the Fe-S homolog gene, and the opa gene family as determined by an electrophoretic mobility shift assay. While gonococcal Fur was demonstrated to bind to the promoter regions of all 11 opa genes (opaA through -K), we did not detect binding of purified E. coli Fur with 8 of the 11 opa members, indicating that target DNA sequence specificities between these two closely related proteins exist. Furthermore, we observed differences in the relative strengths of binding of gonococcal Fur for these different genes, which most likely reflect a difference in affinity between gonococcal Fur and its DNA targets. This is the first report that definitively demonstrates the binding of gonococcal Fur to its own promoter/operator region, as well as to the opa family of genes that encode surface proteins. Our results demonstrate that the gonococcal Fur protein binds to the regulatory regions of a broad array of genes and indicates that the gonococcal Fur regulon is larger than originally proposed.
In several gram-negative pathogens, the ferric uptake regulator protein (Fur) functions as a general global regulator and controls the expression of not only genes required for iron transport but also genes required for virulence. The virulence-associated genes include those for the Shigella Shiga toxin, Escherichia coli hemolysin, the fimbriae of enterotoxigenic E. coli, and exotoxin A of Pseudomonas aeruginosa (8, 19, 21, 27, 41). Fur has also been demonstrated to repress the transcription of genes induced in response to oxidative stress, including the E. coli and P. aeruginosa sodA (superoxide dismutase) genes (13, 35). Furthermore, an E. coli fur mutant was shown to be sensitive to hydrogen peroxide and showed increased oxidative DNA damage (38). The P. aeruginosa Fur protein has been demonstrated to control the expression of other regulatory genes that encode transcriptional activators, which, in turn, regulate the expression of the toxA gene (1, 24, 27, 40, 41). In some organisms, Fur may also act as a positive regulator in controlling gene expression. In salmonellae, expression of the acid tolerance response is controlled positively by Fur (11), as is the expression of E. coli sodB (6). A recent study has shown that Fur enhances the expression of urease in enterohemorrhagic E. coli, thus contributing to the acid tolerance of the organism, similar to Salmonella enterica serovar Typhimurium (14). Bijlsma et al. have also demonstrated that Fur is involved in the urease-independent acid resistance of Helicobacter pylori (4).
Characterization of fur mutants isolated by a manganese selection procedure in several gram-negative organisms has also identified both Fur-repressed and Fur-activated genes (12, 21, 30). A Fur titration assay has also been utilized for the detection of novel Fur-regulated promoters in salmonellae and E. coli (2, 31, 39). The systematic evolution of ligands by exponential enrichment assay has been used to isolate novel Fur-dependent genes in P. aeruginosa. Expression of these new Fur-regulated Pseudomonas genes was derepressed under iron-depleted conditions, and this derepression ranged from highly stringent control to only partial control (25).
A Fur homolog has been identified in Neisseria gonorrhoeae (3). Although Neisseria genes involved in iron transport have been proposed to be regulated by Fur, binding of gonococcal Fur to the promoters of these genes has not been demonstrated. Furthermore, the gonococcal Fur regulon is not well defined; the only genes that appear to be regulated by Fur are those involved in iron transport. The characterization of a missense mutant of N. gonorrhoeae fur isolated following manganese selection demonstrated an alteration of the regulation of a broad range of genes (37), supporting the existence of a large set of genes that may be under Fur control. Nine iron-regulated genes have been identified in N. gonorrhoeae, all of which are involved in iron transport. Although these gonococcal genes have been proposed to be regulated by Fur, the Fur-dependent iron regulation of these proteins has been difficult to test directly because of the inability to isolate a gonococcal fur null mutant.
Previous studies in our laboratory have demonstrated that E. coli Fur binds to a 42-bp site within the gonococcal fbpA (ferric binding protein A) promoter (5). We have also shown that the fbpA promoter is regulated by Fur and iron in E. coli and that the level of transcriptional regulation of fbpA in the gonococcus is directly related to the degree of iron restriction. In order to determine additional components of the gonococcal Fur regulon, we used a 160-bp gonococcal fbpA promoter/operator fragment that has been shown to encompass the Fur box by footprinting analysis with E. coli Fur (5) and conducted an in silico search of the gonococcal genome for Fur box-like sequences. Our analysis identified additional gonococcal genes involved in major catabolic, secretory, and recombination pathways that contained putative iron boxes in their promoter regions. By using purified gonococcal Fur, we demonstrated binding to the promoter regions of several of these genes, including the genes that encode the gonococcal Opa (opacity) proteins, a family of antigenic and phase-variable outer membrane proteins known to promote Neisseria adherence to and invasion of epithelial and endothelial cells, as well as professional phagocytes. The in vitro analysis reported here demonstrates that the gonococcal Fur protein binds to the regulatory regions of the fur, tonB, and opa genes differentially and has a role similar to that of a global regulatory protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and medium.
The N. gonorrhoeae and E. coli strains and plasmids used in this study are indicated in Table 1. E. coli strains were grown on Luria-Bertani agar (Sigma, St. Louis, Mo.) or broth at 37°C. When necessary, ampicillin (100 μg/ml) was added to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
Nucleic acids.
Double-stranded plasmid DNA was prepared by using the Qiaprep miniprep system (Qiagen, Valencia, Calif.). Restriction endonucleases, the Klenow fragment of DNA polymerase, and Taq DNA polymerase (Gibco BRL, Grand Island, N.Y.) were used in accordance with the manufacturer's specifications. Restriction endonuclease fragments were separated by electrophoresis in 1% agarose gels in Tris-acetate-EDTA buffer.
Expression and purification of gonococcal Fur.
Oligonucleotide primers (Table 2) F1 and R1 were designed to incorporate an NdeI restriction site at the 5′ end and a BamHI site at the 3′ end of the gene during the amplification of gonococcal fur from N. gonorrhoeae F62 (restriction sites are underlined in Table 2). The PCR-amplified fragment was cloned into the NdeI/BamHI site of expression vector pET11c (Novagen, San Diego, Calif.) to generate pKASH.1 (Table 1). The fur gene in pKASH.1 is under the control of a T7 promoter. The pKASH.1 construct was transformed into E. coli expression strain HBMV119 (Table 1). The overnight cultures of these transformants were used to inoculate 1 liter of fresh Luria-Bertani broth supplemented with ampicillin (100 μg/ml). At an optical density at 600 nm of 0.4, the cells were induced to express Fur by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma, St. Louis, Mo.) and growth continued at 39°C. Following induction, cells were harvested by centrifugation and resuspended in 20 ml of chilled STET buffer (50 mM Tris-Cl, 0.5 M EDTA, 8% sucrose, 5% Triton X-100). The cells were lysed by the addition of lysozyme (1 mg/ml), followed by sonication. The extract was then clarified by centrifugation at 13,000 rpm in a Sorvall RC5C Plus centrifuge for 25 min. The resulting pellet was enriched in the inclusion body fraction. The gonococcal Fur protein was purified from the inclusion body pellet by dissolving the pellet in 6 M urea in phosphate buffer (pH 7.5) at room temperature (RT) and then centrifuging it. The resulting supernatant, consisting of the solubilized inclusion body fraction, was dialyzed against refolding buffer (50 mM Tris-Cl, 50 mM NaCl, 5 mM reduced glutathione, 1 mM oxidized glutathione) supplemented with 500 μM manganese (Mn2+) to renature the protein. The sample was further dialyzed against 10 mM phosphate buffer (pH 7.5) supplemented with 100 μM Mn2+. The dialyzed sample was purified on an DEAE-Sepharose ion-exchange column equilibrated with 10 mM phosphate buffer, pH 7.5. The column was developed with a 0 to 0.5 M linear NaCl gradient in phosphate buffer. Fractions were collected, and then a protease inhibitor cocktail (Sigma) was immediately added. The presence of gonococcal Fur was confirmed by N-terminal sequencing. Rabbit polyclonal antibodies against recombinant gonococcal Fur were prepared in accordance with standard protocols (Lampire Biological Laboratories, Pipersville, Pa.) and used for immunoblot analysis.
TABLE 2.
Primers used in this study
| Primera | Description |
|---|---|
| F1: ACGTACGTCATATGGAAAAATTCAGCAACATTGCGAA | Amplifies 432-bp gonococcal Fur ORFb |
| R1: ACGTGGATCCTTAACGTTTGCCCTTGGCCTGACAGTC | |
| F2: CGGGATCCACAGGCGAGGATAGGGTTTGT | Amplifies 120-bp promoter fragment of gonococcal fur |
| R2: CCAAGCTTGACAGGGTAAAATACCGCTTA | |
| F3: CGGGATCCGAATTCCTATCCGATTTGCCG | Amplifies 100-bp promoter fragment of gonococcal rmp |
| R3: CCAAGCTTTTATTCCCTCATTAGATTTGTA | |
| F4: CGGGATCCCACGGTTTGCCTATGAATGG | Amplifies 100-bp promoter fragment of gonococcal tonB |
| R4: CCGGATCCGGGCATAATAATAGCAACAATTCC | |
| F5: CGGATCCGTTCCCTTTGAGCCGGGG | Amplifies 100-bp promoter fragment of gonococcal opa multigene family |
| R5: CGGATCCCGGCTCCTTATTCGGTTTGACC | |
| F6: CGGGATCCCGGTAAGGGATAAGGATGCGG | Amplifies 132-bp promoter fragment of gonococcal hemO |
| R6: CCGGATCCCCCGCTTAAAAATATAACGC | |
| F7: CGGGATCCCAACTCAAGATGTAATGGATTG | Amplifies 103-bp promoter fragment of gonococcal hmbR |
| R7: CGGGATCCCACAGATTTATTTGTACGGG | |
| F8: CGGGATCCGGCAGAGTCGGCATCCTGAATG | Amplifies 175-bp promoter fragment of hypothetical gonococcal Fe-S gene |
| R8: CCGGATCCTACTTTAAGTAATTGACCGTTGC | |
| F9: CGGGATCCGAATCGGCGGCTCACGCATTC | Amplifies 184-bp promoter fragment of gonococcal sod |
| R9: CCGGATCCCCGCCTCCATCGAATGCGGAG | |
| F10: CGGGATCCCTTTTCAGACGGCATTTTT | Amplifies 109-bp promoter fragment of gonococcal fumC |
| R10: CCAAGCTTCGTTTCTCCTTTTGAAATGT | |
| F11: CGGGATCCCGCCTACGGATGAAGAAGAGC | Amplifies 219-bp promoter fragment of gonococcal recN |
| R11: CCAAGCTTGGAATTCTACCGACAAAAGCAA | |
| F12: CCGGATCCCGTGTTGGTGGTAAGATTGAAA | Amplifies 193-bp promoter fragment of gonococcal secY |
| R12: CCAAGCTTGGCGCTTTCGTATAATTTAGC |
F, forward; R, reverse. Restriction sites are underlined.
ORF, open reading frame.
Gel electrophoresis and immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (20), with 12% polyacrylamide gels. Proteins were either stained with Coomassie brilliant blue R-250 (Bio-Rad, Hercules, Calif.) or electrophoretically transferred to polyvinylidene difluoride membranes (Bio-Rad) for immunoblot analysis. Immunoblots were probed with either P. aeruginosa anti-Fur serum (1:1,000) or N. gonorrhoeae F62 anti-Fur serum (1:1,000), and then goat anti-rabbit immunoglobulin G antibody (1:10,000; Sigma) was added. Immunoreactive proteins were detected by the addition of nitroblue tetrazolium and 5-bromo-4-chloro-3-indoylphosphate substrate in accordance with the directions of the manufacturer (Sigma).
EMSA.
The different promoter fragments for electrophoretic mobility shift assays (EMSA) were PCR amplified from N. gonorrhoeae FA1090, with the exception of fur and tonB, which were amplified from N. gonorrhoeae F62 with the oligonucleotides shown in Table 2 (restriction sites are underlined). Restriction-digested and gel-purified DNA fragments were end labeled by a Klenow fill-in reaction with [α-32P]dATP and a nucleoside triphosphate mixture (dCTP, dTTP, and dGTP). For the opa probes used (see Fig. 8), 100-bp forward and reverse oligonucleotides specific for each opa member were generated (see Fig. 7), annealed, and similarly labeled. Unincorporated nucleotides were removed with MicroSpin G-25 columns (Amersham Pharmacia Biotech, Piscataway, N.J.). The fragments were diluted to a final concentration of 0.1 nM and incubated with the indicated amounts of E. coli or N. gonorrhoeae Fur protein in binding buffer. Where indicated, competitor DNA (a 50- to 1,200-fold excess) was added to the reaction mixture. Samples were incubated at RT for 20 min, and the reaction mixture was loaded immediately onto a native 6% polyacrylamide gel (acrylamide/bisacrylamide ratio, 30:1 [wt/wt]). The gel was subjected to electrophoresis for 2 h at RT and 100 V, placed on filter paper, dried for 2 h at 80°C, and exposed overnight with an intensifying screen at −70°C. Radioactive bands were visualized by autoradiography. To quantitate binding, the gels were exposed to a PhosphorImager screen (Molecular Dynamics) and band intensity was measured with a Phosphorimager. Sigma Plot software was then used, data were graphed, and the Kd values of gonococcal Fur for the tonB, fur, and opa promoters were determined.
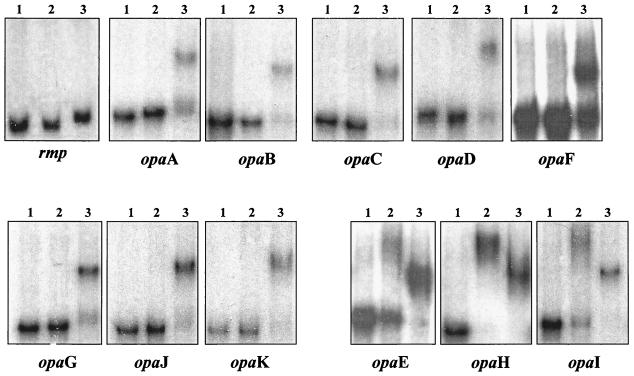
FIG. 8.
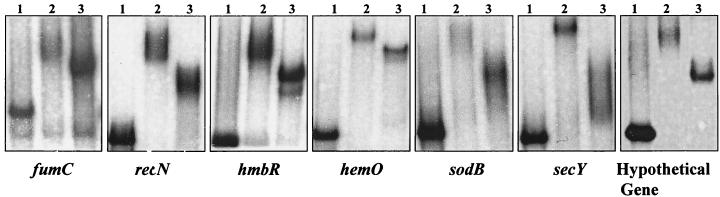
Differential binding of E. coli and gonococcal Fur proteins to the gonococcal opa multigene family (opaA to -K). Lanes: 1, free probe; 2, with E. coli Fur; 3, with gonococcal Fur. A 500-ng sample of each Fur protein and 0.1 nmol of each probe were used. The probe used is indicated at the bottom of each panel.
FIG. 7.
ClustalW multiple-sequence alignment of the 100-bp opa promoter region from N. gonorrhoeae FA1090. Sequences encompassing the opa promoters were provided by Janne Cannon (University of North Carolina at Chapel Hill). The gonococcal consensus sequence, as previously defined, is shown (9). Identical nucleotides in the putative Fur box in the opa promoters and the neisserial consensus are in bold. The reverse oligonucleotide binding site used in the generation of the opa multigene family probe is indicated by the arrows.
RESULTS
Overexpression, purification, and functionality of gonococcal Fur.
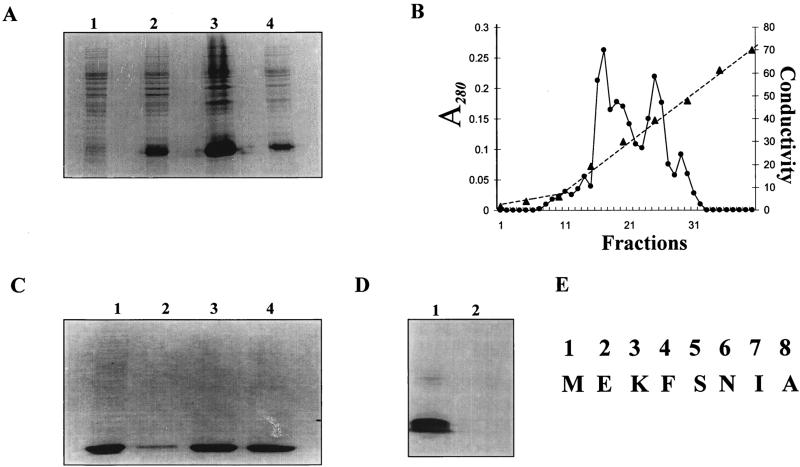
To study the interaction of gonococcal Fur with the operator regions of gonococcal genes, we overexpressed and purified gonococcal Fur by using an E. coli expression strain (HBMV119). E. coli strain HBMV119 is a fur mutant (QC1732) that was infected with lambda DE3 to create an expression strain (Table 1). Expression of the gonococcal Fur protein was examined by SDS-PAGE. As shown in Fig. 1A, a 16-kDa protein representing gonococcal Fur was overexpressed by using the T7 expression system and subsequently purified from the inclusion body pellet by dissolving the pellet in 6 M urea in phosphate buffer. The solubilized inclusion body fraction was dialyzed against refolding buffer supplemented with manganese (100 μM) to renature the protein, and the dialyzed sample was purified by ion-exchange chromatography. Figure 1B shows the A280 profile of fractions obtained by 0 to 0.5 M NaCl gradient elution. As shown in Fig. 1C, most of the gonococcal Fur protein was found in fractions 15 and 16. Western blot analysis with polyclonal anti-P. aeruginosa Fur sera (data not shown) and anti-N. gonorrhoeae Fur sera (Fig. 1D) confirmed that the purified protein with an apparent molecular mass of 16 kDa was indeed gonococcal Fur. N-terminal sequencing confirmed the identity of the first eight amino acids of gonococcal Fur (Fig. 1E).
FIG. 1.
Expression of gonococcal Fur in E. coli HBMV119. (A) SDS-PAGE analysis of recombinant Fur. Lanes: 1, before IPTG addition; 2, 2 h after IPTG addition; 3, inclusion body fraction; 4, supernatant fraction. (B and C) Purification of gonococcal Fur by ion-exchange chromatography. (B) A280 (•) of fractions obtained from the NaCl gradient (▴). (C) SDS-PAGE analysis of fractions 14 (lane 2), 15 (lane 3), and 16 (lane 4). Lane 1, inclusion body fraction used for purification. (D) Western blot analysis. The E. coli and gonococcal Fur proteins (5 μg) were subjected to SDS-PAGE, transferred to polyvinylidene difluoride, and probed with N. gonorrhoeae Fur antiserum. Lanes: 1, N. gonorrhoeae Fur; 2, E. coli Fur. (E) N-terminal sequence of gonococcal Fur.
Anti-N. gonorrhoeae Fur sera cross-reacted weakly with the E. coli Fur protein (Fig. 1D), indicating that N. gonorrhoeae Fur harbors at least one immunodominant domain that differs from that in E. coli Fur. Gonococcal Fur lacks the potential metal binding motif present in the C-terminal portion of E. coli Fur (HCX4-C-X4HXH) (15). Western blot results shown here are in agreement with similar observations obtained with anti-P. aeruginosa Fur sera (24) and E. coli Fur.
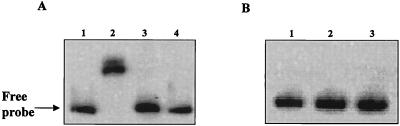
To demonstrate the functionality of a partially purified repressor, we examined the interaction of gonococcal Fur with the gonococcal fbpA promoter/operator (5), a well-established fur-regulated gene, by EMSA. Following incubation of gonococcal Fur with the fbpA probe, we detected a shift in the DNA probe (Fig. 2). These results demonstrated that the recombinant gonococcal Fur protein was able to bind to the fbpA probe. To define the role of divalent metal ions in the activity of gonococcal Fur, we preincubated gonococcal Fur with either 1 mM EDTA or 1 mM 2,2′-dipyridyl and then examined the ability of the repressor to bind to the fbpA promoter by EMSA. As expected, preincubation with either of these chelators resulted in a lack of shifting of the fbpA probe (Fig. 2). In the same set of experiments, the mobility of an internal fragment of the gonococcal fur gene was not altered (Fig. 2B). These results demonstrate that the partially purified gonococcal Fur protein was functional and that binding of the repressor to the fbpA promoter was metal ion dependent and specific.
FIG. 2.
EMSA analysis of gonococcal Fur binding. (A) fbpA operator. Lanes 1 through 4 contained the 160-bp fbpA operator. Lanes: 1, free DNA; 2 through 4, 250 nM gonococcal Fur protein without preincubation (lane 2) or preincubated with 1 mM EDTA (lane 3) or 1 mM 2,2′-dipyridyl (lane 4). (B) Internal fur fragment. Lanes 1 through 3 contained the 200-bp internal fragment of fur. Lanes: 1, free DNA; 2, DNA plus 250 nM gonococcal Fur protein without preincubation; 3, DNA plus 250 nM gonococcal Fur protein preincubated with 1 mM EDTA.
Identification of gonococcal contigs containing putative Fur boxes in their promoters.
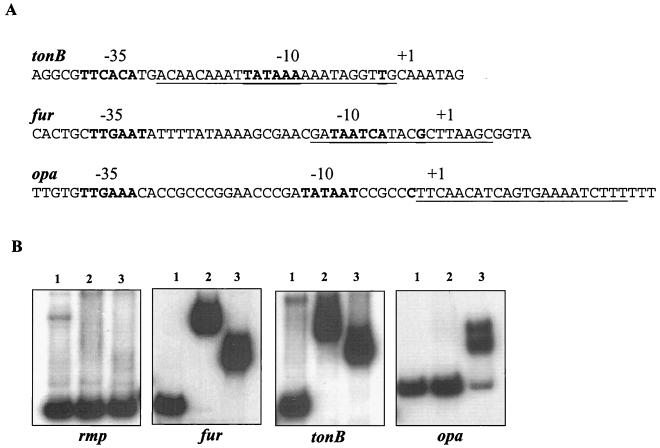
Potential Fur boxes that have previously been identified by in silico analysis in neisseriae include those in the fur, tbpB, lbpB, fbpA, frpC, hmbR, and fetA gene promoter regions (9). Alignment of these promoters with the ClustalW multiple-sequence alignment program yields a putative Neisseria Fur box consensus sequence that has ∼80% homology with the E. coli Fur box consensus binding sequence (9). An initial analysis of the N. gonorrhoeae FA1090 and N. meningitidis MC58 genome databases with the Neisseria Fur box consensus sequence in untranslated regions within 200 bp upstream of an open reading frame revealed matches to several gene promoters. Some of these were upstream of the previously identified iron transport genes. However, other regions were identified in the promoter regions of genes that have not previously been reported to be iron or Fur regulated in gonococci (data not shown). Recognizing that this analysis could provide new information on the gonococcal Fur regulon, we reexamined the meningococcal MC58 and gonococcal FA1090 databases by using a 160-bp sequence comprising the fbpA Fur binding sequence as defined by DNase I footprinting with E. coli Fur (5). This analysis identified several gonococcal and meningococcal genes that contain putative Fur boxes in their promoter regions and included genes encoding well-defined gonococcal virulence factors (Table 3). Our analysis revealed several genes that we anticipated could contain the Fur box sequence in the promoter/operator region. This included the tonB gene, which encodes a cytoplasmic membrane protein that, in association with the ExbB and ExbD proteins, provides energy for the transport of iron from host iron binding receptors across the outer membrane into the periplasmic space in a number of gram-negative organisms. The Fur box in the tonB promoter is 61% identical to the gonococcal Fur consensus binding sequence and overlaps the −10 region of the promoter (Table 3; Fig. 3A). The presence of a Fur box-like sequence 40 bp upstream of the tonB start codon has previously been suggested (33). A Fur binding sequence located in the promoter region of the gonococcal fur gene, and overlapping the −10 region of the fur promoter (Table 3; Fig. 3A), was also identified, in agreement with a previous report (36). Fur box-like sequences were also identified upstream of the iron-regulated outer membrane hemoglobin binding receptor gene hmbR (32) and the heme oxygenase-encoding gene hemO (42). Comparison of the promoter regions of hmbR and hemO with the gonococcal consensus Fur box revealed identities of 80% (17 of 21 bp) and 71% (15 of 21 bp), respectively (Table 3). In silico analysis further revealed 61% identity in the Fur binding site upstream of the gonococcal fumC gene, which encodes a tricarboxylic acid cycle fumarase, and 66% identity in the putative Fur binding site upstream of the gonococcal recN gene. The gonococcal RecN protein has recently been demonstrated to be involved in DNA repair and DNA transformation (29).
TABLE 3.
Neisserial genes with putative Fur boxesa
| Gene | Function | Comparison with neisserial consensus (GATAATATAATAA-TTATCTTT)b | No. of nucleotides identical | Binding of Gc/Ec Furc |
|---|---|---|---|---|
| tonB | Energy transducer | ACAACAAATTATAAAAAATAGG | 13 | Yes/Yes |
| opa | Intergonococcal adherence | TTCAACATCAGTGAAAATCTTT | 13 | Yes/Nod |
| fur | Global regulator | GATAATCATACGCTTAAGCGC | 12 | Yes/Yes |
| sod | Superoxide dismutase | TATAATAGAAGATTGCAATTT | 13 | Yes/Yes |
| hemO | Heme oxygenase | AAAAATATCATTTGCGTTATATTT | 15 | Yes/Yes |
| hmbR | Hemoglobin receptor | GTTAATTCATAATAAATTTTCTGA | 17 | Yes/Yes |
| secY | Preprotein translocase | GAGATCTTAAGAAACGTCTTT | 13 | Yes/Yes |
| recN | Recombination protein | AAAAATCTTAGTTATAATTCGTAT | 14 | Yes/Yes |
| fumC | Fumarate hydratase | GCGAATCACTCTTATTCACATTT | 13 | Yes/Yes |
| Hypothetical gene | TGTAATGATAATTATTATCGAA | 15 | Yes/Yes |
Partial listing of Neisseria genes containing putative Fur binding sequence (Fur box) in the promoter/operator. Consensus homology was determined by comparison with the previously published 21-bp gonococcal Fur box consensus (9).
Identical nucleotides are in boldface type.
Binding of gonococcal (Gc) and E. coli (Ec) Fur proteins to the promoter regions of these genes was determined by EMSA with 500 ng of Fur protein.
opaH, opaI, and opaE do bind to E. coli Fur.
FIG. 3.
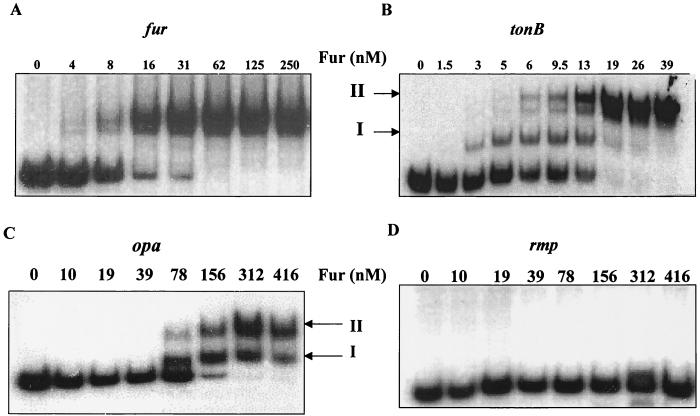
(A) Schematic representation of the relative position of the putative iron box (underlined) with respect to the promoter/operator regions of the tonB, fur, and opa genes. (B) Differential binding of E. coli and gonococcal Fur proteins to gonococcal rmp, tonB, fur, and opa operator DNA probes. Lanes: 1, free probe; 2, with E. coli Fur; 3, with gonococcal Fur. A 500-ng sample of each Fur protein and 0.1 nmol of each probe were used. The probe used is indicated at the bottom of each panel.
From a pathogenesis standpoint, it was of interest that we identified putative Fur binding sequences in the promoter regions of the opa gene family. In N. gonorrhoeae, the opa family of genes encodes 11 related outer membrane proteins (OpaA through -K). ClustalW multiple-sequence alignment results revealed that 8 of the 11 N. gonorrhoeae FA1090 opa genes have a highly conserved promoter region, with the exception of opaE, opaH, and opaI (Fig. 3B). We observed 61% identity between the putative Fur box upstream of the opa promoter/operator regions and the gonococcal consensus sequence (Table 3; Fig. 3A). In contrast to the positions of the fur and tonB Fur boxes, it was interesting that the Fur box-like sequence in the opa multigene family was positioned downstream of the −10 site of the opa promoter (Fig. 3A).
Differential binding of the E. coli and gonococcal Fur proteins to the operator regions of fur, tonB, and opa.
Gonococcal fur is 50% identical to E. coli fur and has been shown to partially complement an E. coli fur mutant (3). In order to determine whether DNA binding sequence specificities between the two fur homologs exist, we compared the abilities of the purified E. coli and gonococcal Fur proteins to bind to the operator regions of the fur, tonB, opa, recN, secY, sodB, hemO, hmbR, and fumC genes and that of a hypothetical gene (Fe-S homolog) by EMSA. Both the E. coli and gonococcal Fur proteins resulted in a shift of the fur, tonB, recN, secY, sodB, hemO, hmbR, fumC, and hypothetical gene (Fe-S homolog) probes (Fig. 3B and 4). Greater retardation of the complex was observed when E. coli Fur was used than when gonococcal Fur was incubated with the fur gene probe. In marked contrast, when the opa probe was used (the opa probe used in these assays represents the entire opa multigene family), a shift was observed when the gonococcal Fur protein was used but not when the E. coli Fur protein was used (Table 3; Fig. 3B). No shift was observed when either of the Fur proteins was incubated with the promoter of the constitutively expressed gonococcal rmp gene (10) (Fig. 3B).
FIG. 4.
Differential binding of E. coli and gonococcal Fur to gonococcal fumC, recN, hmbR, hemO, sodB, and secY and a hypothetical gene. Lanes: 1, free probe; 2, with E. coli Fur; 3, with gonococcal Fur. A 500-ng sample of each Fur protein and 0.1 nmol of each probe were used. The probe used is indicated at the bottom of each panel.
Relative strengths of binding of gonococcal Fur to the tonB, fur, and opa promoter/operators.
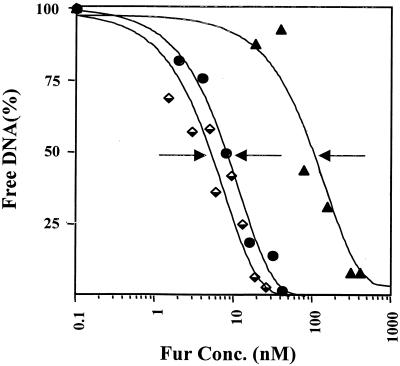
A hallmark of a global regulator is differential regulation of genes that the regulator controls. For gonococcal Fur to function as a global regulator, we postulated that the protein should recognize both high- and low-affinity operator sites. To begin to understand the relative strengths of binding of gonococcal Fur to different Neisserial iron-regulated genes, we performed EMSA with candidate genes from different categories (regulatory protein, transport protein, and adhesion-invasion protein), including the fur, tonB, and opa genes. We first observed a shift in the fur probe with 4 nM gonococcal Fur, and this was complete at 62 nM (Fig. 5A). Interestingly, we found that gonococcal Fur bound to the tonB promoter in several steps. At low Fur concentrations (1.5 to 3.0 nM), a type I complex was formed (Fig. 5B). When higher concentrations of Fur were used (5 to 13 nM), two slowly migrating complexes (types I and II) accumulated. At a Fur concentration of 39 nM, complex II was the major Fur-DNA complex observed (Fig. 5B). An apparent complex was observed with the opa multigene family probe at 78 nM gonococcal Fur, and shifting of the probe was complete at 312 nM Fur (Fig. 5C). Two distinct complexes were also identified with the opa probe. The two complexes formed with the tonB and opa probes may correspond to one or two dimers of Fur bound to DNA at one or multiple binding sites. We did not observe a shift with the gonococcal rmp probe, even with 416 nM Fur (Fig. 5D). The apparent repressor-operator dissociation constants [Kd(app)] for the tonB, fur, and opa probes were calculated to be approximately 6, 10, and 100 nM, respectively (Fig. 6). Collectively, these results suggest that gonococcal Fur has a lower affinity for the opa promoters than for the tonB or fur promoters. These differences most likely reflect the different affinities of gonococcal Fur-target operator interactions. To demonstrate the specificity of binding of gonococcal Fur to the putative Fur boxes identified upstream of the tonB, fur, and opa promoter regions, EMSA were performed with an excess of unlabeled probes (unlabeled probe contained the same sequence as the labeled probe). Gradual loss of each of the complexes was observed with a concomitant fold increase in unlabeled probes (data not shown). It is important to note that the opa probe used in this assay is representative of the entire opa multigene family and that, thus, the Kd of opa, as shown here, would represent a range of responses for the opa promoters.
FIG. 5.
EMSA analysis of gonococcal Fur binding to the gonococcal fur, tonB, opa, and rmp operator DNA probes. A 0.1-nmol sample of each probe was used in each reaction mixture with the indicated increasing concentrations of gonococcal Fur. I, high-affinity complex; II, low-affinity complex. The probe used is indicated above each EMSA.
FIG. 6.
Determination of the Kd(app) of the Fur repressor for the tonB, fur, and opa operators. Binding reactions were performed with 0.1 nmol of DNA and increasing amounts of the Fur repressor. Symbols: ♦, tonB; •, fur; ▴, opa. Each arrow points to the position of half-maximal binding and corresponds to the Kd(app).
To further delineate specifically which opa gene of the multigene family could be bound by gonococcal Fur, we performed EMSA with 100-bp oligonucleotides based on the promoter sequence of each specific opa gene (Fig. 7). ClustalW analysis of the 100-bp promoter regions of the 11 opa genes revealed a high degree of homology among 8 of the 11 opa promoter regions. The promoter regions of opaE, opaH, and opaI were more homologous to each other than to other opa promoters (Fig. 7). Gonococcal Fur was demonstrated to bind to the promoter regions of all of the opa genes tested (Fig. 8). However, when E. coli Fur was used in these studies, we only observed a shift with the opaE, opaH, and opaI promoter/operator-specific probes. It was interesting that in studies performed with the opa multigene family probe, we did not observe a shift with E. coli Fur. One explanation would be that the opa multigene family probe did not represent the opaE, opaH, and opaI promoter probes, since the reverse oligonucleotide used in the generation of this probe differed at 10, 11, and 12 nucleotides in the opaH, opaI, and opaE promoter regions, respectively, compared to the remainder of the opa family members (Fig. 7).
DISCUSSION
It has been hypothesized that the expression of a number of gonococcal iron-regulated proteins is under the control of the transcriptional regulatory protein Fur. However, conclusive demonstration that the expression of these iron-regulated genes is controlled by Fur has been hampered by the inability to construct a gonococcal fur null mutant. In this study, we took an in vitro approach and demonstrated binding of purified gonococcal Fur to the promoter regions of the fur, opa, recN, secY, tonB, sodB, hemO, hmbR, and fumC genes and the gene encoding a hypothetical protein (homolog of Fe-S protein). In a separate study, we have shown that the expression of fur, tonB, recN, hemO, fumC, and hmbR in neisseriae is down regulated during growth with a high iron concentration while the expression of secY and sodB is upregulated under these conditions (unpublished data). Taken together, these results indicate that the gonococcal Fur regulon is larger than originally proposed and encompasses a broad array of genes.
The association of Fur and iron in regulation of the expression of genes involved in acid tolerance has been previously reported in S. enterica serovar Typhimurium (11) and E. coli (6, 7). Niederhoffer et al. have reported that the expression of sodB in E. coli is modulated by Fur, although not in a classical manner of Fe-dependent repression (23). In this study, we identified a Fur box-like element in the promoter region of sodB and demonstrated binding of Fur to the N. gonorrhoeae sodB promoter region. Furthermore, the transcript level of sodB was upregulated under iron-sufficient conditions in neisseriae (unpublished data), and this is in agreement with the positive regulation of sodB in E. coli (6). The physiological purpose of the Fur-mediated increase in gonococcal sodB transcription may be to prevent the damaging effects of iron overload, eventually leading to oxidative stress. A putative Fur consensus sequence was also identified in the promoter region of secY, a gene encoding the preprotein translocase subunit. In vitro analysis further confirmed the binding of gonococcal Fur to the secY promoter. As observed with sodB, the transcript level of secY was found to be upregulated in neisseriae under iron-sufficient conditions (manuscript in preparation), suggesting a positive regulatory role of Fur, as proposed for sodB.
A recent report demonstrated that growth of N. gonorrhoeae under iron limitation resulted in increased recombinational events, resulting in an increased frequency of pilin antigenic variation (28). Studies to understand the role of recombination genes that might contribute to pilin antigenic variation have revealed conflicting results. Siefert et al. (29) have shown that recJ, but not recN, in N. gonorrhoeae FA1090 and N. gonorrhoeae MS11 is involved in pilin antigenic variation. However, studies by Hill et al. showed that an N. gonorrhoeae MS11 recJ knockout did not alter the antigenic variation at the pilin (pilE) locus (16). Nevertheless, the observation that iron limitation resulted in an increased frequency of pilin antigenic variation, together with our identification of a Fur box upstream of recN that was shown to bind gonococcal Fur, is noteworthy, since this may allow insights into the role of iron and Fur in recombination of N. gonorrhoeae.
As expected, we identified a putative Fur binding sequence in the promoter regions of several gonococcal genes required for iron transport. Although putative Fur binding sequences have been previously identified in the promoter regions of the hemO, hmbR, and tonB genes, experimental evidence of gonococcal Fur binding to these sequences has not been provided. That iron and Fur may modulate the level of the tonB transcript in gram-negative bacteria has been further substantiated by the findings that the level of tonB is upregulated 2.5-fold under low-iron conditions in Pasteurella multocida (26). Furthermore, expression of HmbR-mediated hemoglobin binding activity has been previously demonstrated to be repressed by iron (32). In vitro studies, such as those described here, with purified gonococcal Fur further support the role of Fur and iron in the regulation of hmbR, hemO, and tonB in N. gonorrhoeae.
Our results obtained with a tonB probe, a fur probe, and an opa multigene family probe indicate that a hierarchy exists in the binding affinities of gonococcal Fur for different operators. The Kd(app) of gonococcal Fur for the tonB and fur promoter/operator is consistent with the high-affinity binding range of E. coli Fur previously reported for the fhuA and fbpA operators (3). The Kd(app) of gonococcal Fur protein to the opa multigene family probe was indicative of a lower binding affinity, similar to the reported Kd(app) of E. coli Fur protein binding to the sodB operator (22). The DNA sequence of a given operator is likely to determine the affinity of Fur for a specific target. This would allow for differential expression of Fur-sensitive genes that is dependent, in part, on the ratio of apo-Fur to the active Fur-Fe2+ complex. These types of Fur-DNA interactions would allow for differential repression in response to iron among promoters to which Fur binds. Our in vitro analysis shows that gonococcal Fur binds the gene promoters analyzed in this study with various degrees of affinity and thus suggests that Fur may be involved in fine tuning of the expression of the genes under its control.
The finding that gonococcal Fur binds to the opa promoters may have direct implications for pathogenesis. The importance of Opa expression for neisserial infection is suggested by the finding that gonococci recovered after urogenital, cervical, or rectal infection are typically Opa+, as are bacteria recovered after the inoculation of human volunteers with transparent (Opa−) bacteria (18, 34). Interestingly, the only exception to such in vivo selection is the observation that Opa− bacteria predominate during menses (17). Our in vitro and in silico analyses suggest the potential of the entire FA1090 opa multigene family to be regulated by Fur. ClustalW analysis of the promoter regions of the 11 members of the N. gonorrhoeae FA1090 opa multigene family revealed that 8 of the 11 opa members have similar levels of identity in the Fur box (13 of 21 bp). A 12- of 21-bp level of identity was revealed in the putative Fur box of the opaH promoter region, while opaE and opaI had an identity level of 9 of 21 bp. EMSA analysis revealed that gonococcal Fur bound to the promoters of all of the 11 opa genes. In contrast to the binding of gonococcal Fur, only the mobility of opaE, opaH, and opaI promoter probes was altered by E. coli Fur binding. ClustalW alignment of the opaE, opaH, and opaI promoter sequence with the previously published E. coli Fur box consensus revealed an identity level of 10 of 21 bp in these promoter regions. Interestingly, the putative Fur box of the opa multigene family was positioned downstream of the −10 promoter box and found to be AT rich, similar to the Fur box found within the promoter of the E. coli sodB gene. Furthermore, the Kd(app) of the opa multigene family probe was more closely related to the Kd(app) of the sodB promoter. In E. coli, the expression of sodB has been shown to be activated by Fur and iron (6). We cannot speculate about whether the opa multigene family is positively or negatively regulated by Fur and iron. While the repressive mechanism of Fur has been thoroughly investigated, the mechanism of positive regulation by this protein has not been well elucidated. Studies are under way to define the mechanism of regulation of the opa multigene family in vitro and in vivo.
Acknowledgments
This study was supported by Public Health Service grants U19AI 38515 and AI 48611.
We thank Hazel A. Barton for providing E. coli strain HBMV119 and Mark Coy for providing purified E. coli Fur. We also thank Janne Cannon for providing the N. gonorrhoeae FA1090 opa promoter sequences.
REFERENCES
- 1.Barton, H. A., Z. Johnson, C. D. Cox, A. I. Vasil, and M. L. Vasil. 1996. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol. Microbiol. 21:1001-1017. [DOI] [PubMed] [Google Scholar]
- 2.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 3.Berish, S. A., S. Subbarao, C. Y. Chen, D. L. Trees, and S. A. Morse. 1993. Identification and cloning of a fur homolog from Neisseria gonorrhoeae. Infect. Immun. 61:4599-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma, J. J., B. Waidner, A. H. Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, P. J., A. Angerer, and C. A. Genco. 1996. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J. Bacteriol. 178:5020-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubrac, S., and D. Touati. 2002. Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology 148:147-156. [DOI] [PubMed] [Google Scholar]
- 8.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genco, C. A., and P. J. Desai. 1996. Iron acquisition in the pathogenic Neisseria. Trends Microbiol. 4:179-184. [DOI] [PubMed] [Google Scholar]
- 10.Gotschlich, E. C., M. Seiff, and M. S. Blake. 1987. The DNA sequence of the structural gene of gonococcal protein III and the flanking region containing a repetitive sequence. Homology of protein III with enterobacterial OmpA proteins. J. Exp. Med. 165:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hantke, K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 210:135-139. [DOI] [PubMed] [Google Scholar]
- 13.Hassett, D. J., M. L. Howell, U. A. Ochsner, M. L. Vasil, Z. Johnson, and G. E. Dean. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 179:1452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heimer, S. R., R. A. Welch, N. T. Perna, G. Posfai, P. S. Evans, J. B. Kaper, F. R. Blattner, and H. L. Mobley. 2002. Urease of enterohemorrhagic Escherichia coli: evidence for regulation by fur and a trans-acting factor. Infect. Immun. 70:1027-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennecke, H. 1990. Regulation of bacterial gene expression by metal-protein complexes. Mol. Microbiol. 4:1621-1628. [DOI] [PubMed] [Google Scholar]
- 16.Hill, S. A. 2000. Neisseria gonorrhoeae recJ mutants show defects in recombinational repair of alkylated bases and UV-induced pyrimidine dimers. Mol. Gen. Genet. 264:268-275. [DOI] [PubMed] [Google Scholar]
- 17.James, J. F., and J. Swanson. 1978. Studies on gonococcus infection. XIII. Occurrence of color/opacity colonial variants in clinical cultures. Infect. Immun. 19:332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jerse, A. E., M. S. Cohen, P. M. Drown, L. G. Whicker, S. F. Isbey, H. S. Seifert, and J. G. Cannon. 1994. Multiple gonococcal opacity proteins are expressed during experimental urethral infection in the male. J. Exp. Med. 179:911-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karjalainen, T. K., D. G. Evans, D. J. Evans, Jr., D. Y. Graham, and C. H. Lee. 1991. Iron represses the expression of CFA/I fimbriae of enterotoxigenic E. coli. Microb. Pathog. 11:317-323. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K., F. Beguin, and G. Gujer-Kellenberger. 1970. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J. Mol. Biol. 47(1):69-85. [DOI] [PubMed] [Google Scholar]
- 21.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niederhoffer, E. C., and J. A. Fee. 1990. Novel effect of aromatic compounds on the iron-dependent expression of the Escherichia coli K12 manganese superoxide dismutase (sodA) gene. Biol. Met. 3:237-241. [DOI] [PubMed] [Google Scholar]
- 23.Niederhoffer, E. C., C. M. Naranjo, K. L. Bradley, and J. A. Fee. 1990. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J. Bacteriol. 172:1930-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochsner, U. A., A. I. Vasil, and M. L. Vasil. 1995. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177:7194-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince, R. W., C. D. Cox, and M. L. Vasil. 1993. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J. Bacteriol. 175:2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serkin, C. D., and H. S. Seifert. 2000. Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol. Microbiol. 37:1075-1086. [DOI] [PubMed] [Google Scholar]
- 29.Skaar, E. P., M. P. Lazio, and H. S. Seifert. 2002. Roles of the recJ and recN Genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J. Bacteriol. 184:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staggs, T. M., J. D. Fetherston, and R. D. Perry. 1994. Pleiotropic effects of a Yersinia pestis fur mutation. J. Bacteriol. 176:7614-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 32.Stojiljkovic, I., J. Larson, V. Hwa, S. Anic, and M. So. 1996. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J. Bacteriol. 178:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojiljkovic, I., and N. Srinivasan. 1997. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J. Bacteriol. 179:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson, J., O. Barrera, J. Sola, and J. Boslego. 1988. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J. Exp. Med. 168:2121-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tardat, B., and D. Touati. 1993. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol. Microbiol. 9:53-63. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, C. E., and P. F. Sparling. 1994. Identification and cloning of a fur homologue from Neisseria meningitidis. Mol. Microbiol. 11:725-737. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, C. E., and P. F. Sparling. 1996. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J. Bacteriol. 178:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsolis, R. M., A. J. Baumler, I. Stojiljkovic, and F. Heffron. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 41.Vasil, M. L., U. A. Ochsner, Z. Johnson, J. A. Colmer, and A. N. Hamood. 1998. The Fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator PtxR in Pseudomonas aeruginosa. J. Bacteriol. 180:6784-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, W., D. J. Hunt, A. R. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]