Abstract
Expression of the alkane degradation pathway encoded by the OCT plasmid of Pseudomonas putida GPo1 is regulated by two control systems. One relies on the transcriptional regulator AlkS, which activates expression of the pathway in the presence of alkanes. The other, which is a dominant global regulation control, represses the expression of the pathway genes when a preferred carbon source is present in the growth medium in addition to alkanes. This catabolite repression control occurs through a poorly characterized mechanism that ultimately regulates transcription from the two AlkS-activated promoters of the pathway. To identify the factors involved, a screening method was developed to isolate mutants without this control. Several isolates were obtained, all of which contained mutations that mapped to genes encoding cytochrome o ubiquinol oxidase, the main terminal oxidase of the electron transport chain under highly aerobic conditions. Elimination of this terminal oxidase led to a decrease in the catabolic repression observed both in rich Luria-Bertani medium and in a defined medium containing lactate or succinate as the carbon source. This suggests that catabolic repression could monitor the physiological or metabolic status by using information from the electron transport chain or from the redox state of the cell. Since inactivation of the crc gene also reduces catabolic repression in rich medium (although not that observed in a defined medium), a strain was generated lacking both the Crc function and the cytochrome o terminal oxidase. The two mutations had an additive effect in relieving catabolic repression in rich medium. This suggests that crc and cyo belong to different regulation pathways, both contributing to catabolic repression.
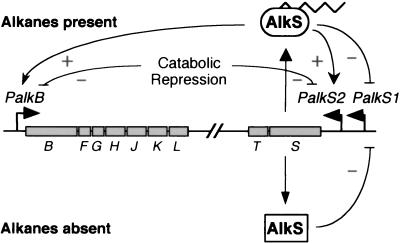
The expression of bacterial catabolic pathways for the assimilation of aliphatic and aromatic hydrocarbons is frequently subject to complex control which links induction of the corresponding genes not only to the presence of the substrates to be degraded but also to the proper physiological status of the cell (11). This is the case of the alkane degradation pathway encoded by the OCT plasmid of Pseudomonas putida GPo1 (previously named Pseudomonas oleovorans GPo1; see reference 53). The genes of this pathway are grouped into two clusters, alkBFGHJKL and alkST (Fig. 1) (53, 54). The alkBFGHJKL operon is transcribed from a promoter, PalkB, whose expression requires the transcriptional activator AlkS and the presence of alkanes (31, 42). In the absence of alkanes, the alkST genes are expressed from promoter PalkS1, which is recognized by σS-RNA polymerase (7, 8). Therefore, during exponential growth with a carbon source other than alkanes, the gene for the AlkS regulator is essentially silent. Upon entry into the stationary phase, when σS-RNA polymerase becomes available, transcription of alkS from this promoter increases. AlkS acts as a repressor of PalkS1, allowing for low expression of the alkST genes. When alkanes are present, the AlkS protein bound to PalkS1 activates the expression of promoter PalkS2, located 38 nucleotides (nt) downstream from PalkS1, and provides for high expression of the alkST genes (7). Therefore, the pathway is controlled by a positive feedback mechanism governed by AlkS (Fig. 1). The amount of AlkS protein in induced cells is, however, low since this protein seems to be very unstable (56, 57). The pathway is regulated by an additional control system, since the levels of the alkane degradation enzymes are modulated by catabolic repression depending on the carbon source being used (24, 47). This superimposed control occurs through a poorly characterized mechanism that ultimately regulates transcription from promoters PalkB and PalkS2 (7, 56, 57). Activation of these promoters by AlkS and the alkane inducer is very efficient when cells are grown in a minimal salts medium containing citrate as carbon source but shows a three- to fourfold reduction when organic acids such as lactate, pyruvate, or succinate are used as the carbon source. Repression is much stronger (ca. 50-fold repression) when cells grow exponentially in a rich medium such as Luria-Bertani (LB) medium or in minimal salts medium supplemented with Casamino Acids. Repression in rich medium abruptly disappears as cells enter the stationary phase of growth, which suggests the existence of elements that assure low-level expression of promoters PalkB and PalkS2 during exponential growth. The P. putida Crc protein plays an important role in the catabolic repression observed in rich medium, although not the one observed when cells grow in minimal salts medium containing lactate or succinate as a carbon source (57). Inactivation of the crc gene reduces catabolic repression in rich medium by about sixfold, although a fivefold repression still occurs in conditions of no catabolic repression, indicating that other elements besides Crc participate in this control (57). To identify additional factors involved in the modulation of this pathway, a screening method was developed to obtain mutants showing a reduced catabolic repression in rich medium. Nine mutants were isolated. In all cases, mutations mapped to genes encoding the cytochrome o ubiquinol oxidase, the main terminal oxidase of the electron transport chain under highly aerobic conditions. The aerobic respiratory chains of both Escherichia coli and P. putida include a number of membrane-bound dehydrogenases that transfer electrons to ubiquinone, reducing it to ubiquinol. This can then be oxidized by either of two respiratory ubiquinol oxidases: the cytochrome o complex or the cytochrome d complex (22). When cells grow exponentially with an ample supply of oxygen, cytochrome o oxidase (cyo) accommodates most of the electron flow. As the oxygen supply becomes limiting, cytochrome d oxidase (cyd) is synthesized as an alternative terminal oxidase (15, 16, 49). Strains that lack either cyo or cyd are fully capable of aerobic growth under normal laboratory conditions, since the function of the missing oxidase seems to be taken over by the one remaining (1). An analysis was made of the influence of cytochrome o ubiquinol oxidase in the AlkS-mediated activation of the PalkB and PalkS2 promoters. The results suggest that catabolic repression of the P. putida GPo1 alkane degradation pathway is linked to the activity of the electron transport chain and/or to the redox state of the cell.
FIG. 1.
P. putida GPo1 alkane degradation pathway. The genes are grouped into two clusters, alkBFGHJKL and alkST, both of which are regulated by the AlkS protein. In the absence of alkanes, AlkS is expressed from promoter PalkS1; AlkS acts as a repressor of this promoter, allowing for low expression levels. This promoter is recognized by σS-RNA polymerase, being active only in the stationary phase of growth. In the presence of alkanes, AlkS activates expression from the PalkB and PalkS2 promoters, generating a positive amplification loop on alkS expression. Activation of these two promoters by alkanes is strongly repressed by catabolic repression when cells grow exponentially in rich LB medium. Growth in a minimal medium containing some organic acids (lactate or succinate) as a carbon source generates a milder catabolic repression effect.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used are listed in Table 1. P. putida strains were grown at 30°C in LB medium or in M9 minimal salts medium (45), the latter supplemented with trace elements (4), and 30 mM citrate, lactate, or succinate as the carbon source. Expression of the PalkB or PalkS2 promoters was induced was demontrated by the addition of 0.05% (vol/vol) dicyclopropylketone (DCPK), a nonmetabolizable inducer that mimics the effect of alkanes (24). E. coli cells were grown in LB medium at 37°C. Antibiotics were added when appropriate at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml; and tetracycline, 8 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description or relevant phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| CC118(λpir) | CC118 lysogenized with λpir phage | 25 |
| HB101 | Host for plasmid pRK600 | 5 |
| TG1 | Host for DNA manipulations | 45 |
| P. putida | ||
| KT2442 | hsdR Rif | 21 |
| PBS4 | KT2442 with PalkB::lacZ and alkS in the chromosome | 56 |
| PBS4B1 | PBS4 with an inactivated cyoB::tet allele | This work |
| PBS4C1 | PBS4 with an inactivated crc::tet allele | 57 |
| PBS4BC1 | PBS4 with an inactivated crc::xylE and cyoB::tet alleles | This work |
| RC12 | KT2442 with PalkB::kan in the chromosome | This work |
| RC12S8 | KT2442 with PalkB::kan and alkS in the chromosome | This work |
| RCM4 | RC12S8 with a mini-Tn5Sm insertion at the cyoC gene | This work |
| RCM18 | RC12S8 with a mini-Tn5Sm insertion at the cyoB gene | This work |
| RCM2A | RC12S8 with a mini-Tn5Sm insertion at the cyoB gene | This work |
| RCM3A | RC12S8 with a mini-Tn5Sm insertion at ORF2 of the cyo cluster | This work |
| RCM6A | RC12S8 with a mini-Tn5Sm insertion at the cyoA gene | This work |
| RCM22A | RC12S8 with a mini-Tn5Sm insertion at the cyoE gene | This work |
| RCM117A | RC12S8 with a mini-Tn5Sm insertion at the cyoB gene | This work |
| RCM137A | RC12S8 with a mini-Tn5Sm insertion at the cyoA gene | This work |
| RCM2B | RC12S8 with a mini-Tn5Sm insertion at the cyoE gene | This work |
| Plasmids | ||
| pALKRC1 | Apr Telr, PalkB::kan transcriptional fusion into pJMT6 | This work |
| pCRC5 | Apr; P. putida KT2442 crc gene cloned into pUC18 | 57 |
| pCRC5A | Apr; crc::xylE allele into pUC18; derives from pCRC5 | This work |
| pCRC20 | Smr; crc::xylE allele inserted at the BamHI site of pKNG101 | This work |
| pCYOB | P. putida KT2442 cyoB gene cloned into pUC18 | This work |
| pCYOBTc | cyoB::tet allele into pUC18 | This work |
| pJMT6 | Apr Telr; mini-Tn5 suicide donor plasmid | 46 |
| pKCYOBTc | cyoB::tet allele into pKNG101 | This work |
| pKNG101 | Smr; suicide vector for marker exchange mutagenesis | 29 |
| pPB7 | Apr; PalkB promoter into pUC19Ω | 56 |
| pPBK10 | Apr; PalkB::kan fusion into pUCKm | This work |
| pPBKJ1 | Apr; PalkB::kan fusion into pUJ8 | This work |
| pRK600 | Cmr Mob+ Tra+; donor of transfer functions | 30 |
| pTCS1 | Apr Tcr; alkS in mini-Tn5Tc | This work |
| pTS1 | Apr; alkS into pT7-12 | 56 |
| pUC18 | Apr; cloning vector | 55 |
| pUCKm | Apr; contains a promoterless Kmr cassette | 13 |
| pUJS1 | Apr; alkS gene cloned into pUJ8 | 56 |
| pUJ8 | Apr; vector for transcriptional fusions to lacZ | 18 |
| pUT-mini-Tn5Sm | Apr Smr; mini-Tn5 suicide donor | 18 |
| pUT-mini-Tn5Tc | Apr Tcr; mini-Tn5 suicide donor | 18 |
| pXYLE10 | Kmr; P. putida xylE gene as an excisable cassette | 48 |
Telr, tellurite resistant; Rifr, rifampin resistant.
P. putida RC12 is derived from P. putida KT2442 by insertion of a PalkB::kan transcriptional fusion into its chromosome by using the suicide donor plasmid pALKRC1. This plasmid was generated as follows. A transcriptional fusion of the PalkB promoter to a kanamycin resistance (Kmr) determinant was generated by excising a DNA fragment containing the PalkB promoter (positions −525 to +66 relative to the transcriptional start site) from plasmid pPB7 with KpnI (blunted with T4 DNA polymerase and deoxynucleoside triphosphates [dNTPs]) and HindIII and inserting it between the HincII and HindIII sites of plasmid pUCKm. The plasmid obtained was named pPBK10. The PalkB::kan fusion was excised from pPBK10 with KpnI (blunted with T4 DNA polymerase and dNTPs) and EcoRI and inserted between the HindIII (blunted with T4 DNA polymerase and dNTPs) and EcoRI sites of plasmid pUJ8. The plasmid obtained was named pPBKJ1. The PalkB::kan fusion was excised from pPBKJ1 as a NotI DNA fragment and inserted at the NotI site of the mini-Tn5 suicide donor plasmid pJMT6, generating plasmid pALKRC1. P. putida RC12S8 is derived from strain RC12 by insertion of the alkS gene into its chromosome by using the suicide delivery plasmid pTCS1. This plasmid is derived from mini-Tn5Tc by insertion at its NotI site of a NotI DNA fragment containing the alkS gene, which was obtained from plasmid pUJS1.
To generate the ΔcyoB P. putida strain PBS4B1, a 1,200-bp DNA fragment containing the cyoB gene was PCR amplified from P. putida KT2442 with the primers 5′-CGGGATCCACGAAGAAGCAGGCAGCA-3′ and 5′-CGGGATCCAGAACCAGAAGGCAG-3′, which contain BamHI sites at their 5′ ends. The product was digested with BamHI and cloned at the BamHI site of plasmid pUC18, yielding plasmid pCYOB. A 2-kbp EcoRI fragment containing a tetracycline resistance (Tcr) determinant was obtained from plasmid pUT-mini-Tn5Tc and inserted at the EcoRI site of pCYOB, located inside the cyoB gene. The plasmid generated was named pCYOBTc. Finally, a 3.2-kb BamHI fragment from pCYOBTc containing the cyoB::tet allele was cloned into plasmid pKNG101, yielding pKCYOBTc. Plasmid pKNG101 is designed for marker exchange mutagenesis; it replicates in E. coli but not in P. putida and carries a streptomycin resistance (Smr) determinant and the sacB gene, which mediates sucrose sensitivity (29). Plasmid pKCYOBTc was transferred to P. putida PBS4 in triparental matings with plasmid pRK600 as donor of transfer functions, and Smr Tcr sucrose-sensitive (8% [wt/vol]) exconjugants were selected. These cells were expected to contain plasmid pKCYOBTc integrated into the chromosome by a single recombination event at the cyoB gene, which generates a mutant and a wild-type cyoB gene. Cells having undergone a second recombination event leading to loss of the wild-type cyoB allele were selected by screening for Tcr, streptomycin-sensitive (Sms), and sucrose-resistant colonies. The absence of the wild-type cyoB gene and the presence of the cyoB::tet allele were confirmed by PCR.
Inactivation of the crc gene in strain PBS4B1 was performed by marker exchange mutagenesis. The promoterless xylE reporter gene was excised from plasmid pXYLE10 with endonuclease SmaI and inserted at the NruI site of the crc gene present in plasmid pCRC5. The plasmid obtained was named pCRC5A. The crc::xylE allele was excised from pCRC5A by partial restriction with BamHI and cloned at the BamHI site of plasmid pKNG101, yielding plasmid pCRC20. This plasmid was transferred to strain PBS4B1 and exconjugants in which the wild-type crc gene had been substituted by the crc::xylE allele were isolated as described above. A representative isolate was selected and named PBS4BC1.
Generation of mutant P. putida strains showing reduced catabolic repression in LB medium.
P. putida RC12S8 (KT2442 with a PalkB::kan fusion and alkS in the chromosome) was mutagenized with minitransposon mini-Tn5Sm. This was delivered to the recipient cells in triparental matings as previously described (18) by using E. coli CC118λpir containing the plasmid pUT-mini-Tn5Sm as a donor and E. coli HB101 containing the plasmid pRK600 as a helper for transfer functions. The exconjugants were plated on LB plates containing streptomycin, kanamycin, and 0.05% (vol/vol) DCPK and then incubated at 30°C for 24 to 48 h. Kmr colonies were streaked onto LB plates with kanamycin in the absence or presence of DCPK. Colonies resistant to kanamycin in the absence of DCPK were discarded (ca. 20%). Those selected were further tested for DCPK-dependent resistnace to kanamycin in LB broth. To identify the insertion point of the minitransposon, two approaches were used. For the mutant strains RCM4, RCM18, RCM2A, RCM3A, and RCM137A, chromosomal DNA was purified and digested with either KpnI (strains RCM4 and RCM18), SacI (strains RCM2A and RCM3A), or KpnI+PstI (strain RCM137A), none of which cut inside the minitransposon. The digested fragments were ligated to pUC18 excised with the same endonucleases and transformed into E. coli TG1. Plasmid DNA was extracted from Smr ampicillin-resistant (Apr) colonies, and the DNA insert was sequenced in both strands. In all other mutant strains analyzed, the point of insertion was identified by arbitrary PCR (6, 41) and was later verified by PCR with specific primers.
Assay for β-galactosidase.
An overnight culture of the appropriate strain was diluted to a final turbidity (A 600) of ca. 0.04 in fresh LB medium, or in minimal salts M9 medium supplemented with the indicated carbon source. When turbidity reached to 0.08, the nonmetabolizable inducer DCPK (0.05% [vol/vol]) was added where indicated. Cultures were grown at 30°C. At different time points, aliquots were taken and β-galactosidase activity measured as described by Miller (33). At least three independent assays were performed in each case.
S1 nuclease protection assays.
Total RNA was isolated from bacterial cultures as previously described (34). S1 nuclease reactions were also performed as previously described (3) with 50 μg of total RNA and an excess of a 5′-end-labeled single-stranded DNA (ssDNA) hybridizing to the 5′ region of the mRNA. This allows detection of the transcription start sites, as well as detection of the amounts of transcript generated. The ssDNA probe was obtained by linear PCR as described previously (56) by using either plasmid pTS1 linearized with HindIII (contains alkS and its promoter region) or plasmid pPB7 linearized with PstI (contains the PalkB promoter) as a substrate. The primers used hybridized 70 nt downstream of promoter PalkS2 or 73 nt downstream of promoter PalkB (the start sites were as described elsewhere [7]).
RESULTS
Generation of mutant strains showing reduced catabolic repression in rich LB medium.
To obtain mutant strains showing a reduced catabolic repression on the PalkB promoter in LB medium, strain RC12S8 was constructed (see Materials and Methods). This strain, which derives from P. putida KT2442, contains in its chromosome a copy of the alkS gene and a PalkB::kan transcriptional fusion in which a Kmr determinant is expressed from the AlkS-dependent PalkB promoter. Strain RC12S8 was sensitive to kanamycin when grown in the absence of a positive effector of the AlkS regulator. In the presence of such an effector (an alkane, or the nonmetabolizable analogue DCPK), strain RC12S8 was resistant to kanamycin when grown in a medium generating no catabolic repression (e.g., minimal salts medium containing citrate as carbon source). However, this strain was sensitive to kanamycin when grown in LB medium in the presence of the inducer DCPK, as might be expected owing to the strong catabolic repression that the LB medium imposes on PalkB induction. To search for mutants showing a reduced repression effect, strain RC12S8 was mutagenized with minitransposon mini-Tn5Sm. Nine mutant strains were isolated that were resistant to kanamycin in LB medium in the presence of inducer but not in its absence (see Materials and Methods). They were named RCM4, RCM18, RCM2A, RCM2B, RCM3A, RCM6A, RCM22A, RCM117A, and RCM137A. The sequence of the PalkB and PalkS promoters in all of these strains was found to be identical to that of the parental strain.
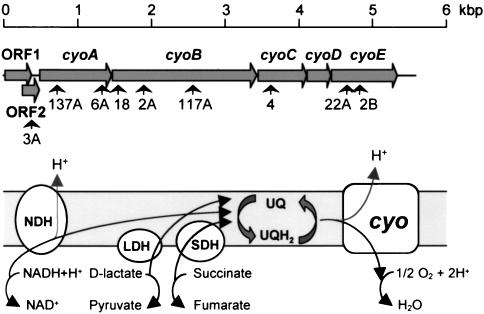
The point of insertion of the mini-Tn5Sm in these strains was localized either by cloning and sequencing of the chromosomal DNA segment containing the Smr determinant or by arbitrary PCR (see Materials and Methods). In mutant strain RCM4, the minitransposon was found inserted at an open reading frame showing 88% amino acid similarity to the cyoC gene of P. putida IH-2000 (28), which codes for subunit III (also known as subunit C) of cytochrome o ubiquinol oxidase (Fig. 2). In mutant strains RCM18, RCM2A, and RCM117A, the minitransposon was found to interrupt an open reading frame showing 96% amino acid similarity to the cyoB gene of P. putida IH-2000 encoding subunit I (or subunit B) of the same cytochrome (28). These two genes form part of the cyoABCDE cluster, which encodes subunits II, I, III, and IV of the P. putida cytochrome o oxidase complex (also known as the cyo complex) and the heme o synthase (28, 36) (see Fig. 2). Cytochrome o ubiquinol oxidase is one of the two terminal ubiquinol oxidases in the P. putida respiratory chain (36). In the other mutant strains analyzed, the minitransposon was found to map to other genes of the cyo cluster (Fig. 2), namely, the cyoA gene (strains RCM6A and RCM137A), and the cyoE gene (strains RCM22A and RCM2B), or at ORF2 (strain RCM3A).
FIG. 2.
Insertion points of mini-Tn5Sm in the P. putida catabolic repression mutants isolated. The upper scheme shows the genes encoding the P. putida cyo complex (28) and the positions of the mini-Tn5Sm insertions found in mutant strains RCM4, RCM18, RCM2A, RCM2B, RCM3A, RCM6A, RCM22A, RCM117A, and RCM137A (indicated by arrows). The cyoA, cyoB, cyoC, and cyoD genes encode subunits II, I, III, and IV of the oxidase complex, respectively; cyoE encodes the heme o synthase. The bottom scheme represents the electron transport chain. NDH, NADH dehydrogenase; LDH, lactate dehydrogenase; SDH, succinate dehydrogenase; UQ, oxidized ubiquinone; UQH2, reduced ubiquinone; cyo, cytochrome o ubiquinol oxidase.
Expression of the PalkB and PalkS promoters upon inactivation of the cyoB gene.
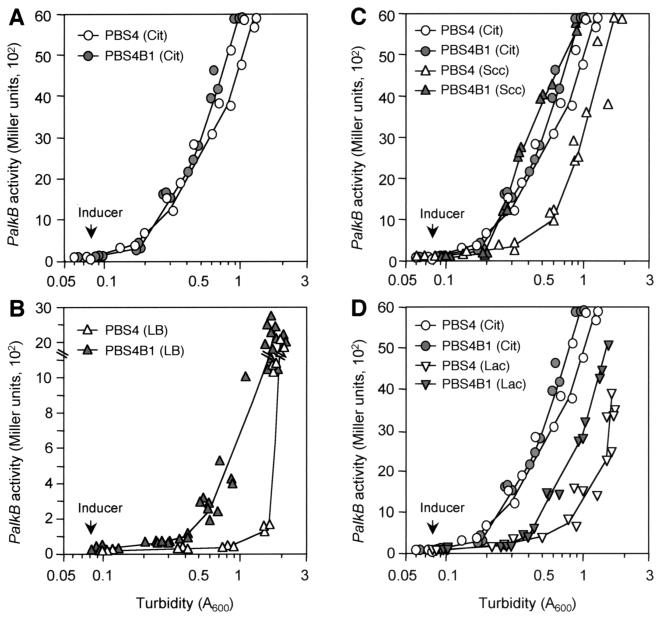
To ascertain that the reduction of catabolic repression observed in the above mutant strains was due to the inactivation of cytochrome o ubiquinol oxidase rather than to another unknown mutation and to have a reliable system to measure the effect of this terminal oxidase on catabolic repression, a knockout mutation was performed at the cyoB gene of P. putida PBS4. This strain contains a PalkB::lacZ transcriptional fusion and the alkS gene inserted into the chromosome and was the host used in previous studies to analyze the catabolic repression of the PalkB promoter. Extensive work has shown that β-galactosidase levels in strain PBS4 faithfully reproduce the transcriptional activity of the PalkB promoter (56). The PBS4 cyoB gene was inactivated by marker exchange mutagenesis by using a plasmid containing a cyoB gene interrupted by a Tcr determinant (the cyoB::tet allele; see Materials and Methods); the strain obtained was named PBS4B1. This strain showed normal growth in rich medium under the conditions used above and had essentially the same growth rate as the wild type. At least in E. coli, cells containing mutations in the cyo complex grow aerobically using the alternative cyd complex (2), which probably explains the normal growth of strain PBS4B1. When grown in defined medium with citrate, a compound that does not generate catabolic repression (56), activation of the PalkB promoter after the addition of the inducer was equally efficient in strains PBS4 and PBS4B1 (Fig. 3A). However, when cells were grown in LB medium, PalkB induction was different in the two strains (see Fig. 3B). As previously described (56), activation of PalkB in the wild-type strain PBS4 was very low during the exponential phase of growth and strongly increased at the onset of the stationary phase. This occurred at turbidity values (A600) of ca. 1.2 to 1.4. In the case of the cyo-deficient strain PBS4B1, although PalkB activity was poor at the start of the exponential phase, repression was clearly relieved as growth proceeded. The β-galactosidase levels increased almost fourfold at mid-exponential phase (A600 of 0.5) and more than ninefold at late exponential phase (A600 of 1.0) compared to those with the wild-type strain PBS4 (Fig. 3B). If we consider the repression values observed for the wild-type strain, inactivation of cytochrome o ubiquinol oxidase therefore reduced the catabolic repression exerted by the LB medium by ∼3.4-fold at the mid-exponential phase and by 7.5-fold at the late exponential phase of growth (Table 2). Since repression in the wild type is in the range of 50- to 70-fold (Table 2), it is clear that inactivation of cytochrome o ubiquinol oxidase only provides partial relief of the catabolic repression observed in LB medium.
FIG. 3.
Effect of the cyoB mutation on induction of the PalkB promoter in cells growing with different carbon sources. P. putida strains PBS4 and PBS4B1 (PBS4 with a knockout mutation at the cyoB gene) were grown in duplicate flasks either in LB medium (B) or in minimal salts medium containing either citrate (Cit) (A), succinate (Scc) (C), or lactate (Lac) (D) as the carbon source. At an A600 of 0.08, the nonmetabolizable inducer DCPK was added to one of the flasks; the other was left as a noninduced control. Aliquots were taken at different times, and the β-galactosidase activity was measured. The plots show the values observed for induced cultures (noninduced cultures had very low β-galactosidase activities [30 to 100 Miller units] and are not represented). The values shown correspond to three to six independent assays, all of which are represented on the same plot.
TABLE 2.
Catabolic repression of the PalkB promoter in strains PBS4 (wild type for cyoB) and PBS4B1 (cyoB::tet)
| Medium | Activity (repression value)a in strain:
|
Repression reliefb
|
||||
|---|---|---|---|---|---|---|
| PBS4
|
PBS4B1
|
|||||
| ME phase | LE phase | ME phase | LE phase | ME phase | LE phase | |
| Citrate | 2,500 (1) | 4,800 (1) | 2,800 (1) | 6,000 (1) | 1 | 1 |
| LB medium | 50 (51) | 70 (68) | 190 (14.7) | 670 (9) | 3.4 | 7.5 |
| Lactate | 450 (5.5) | 1,350 (3.5) | 1,000 (2.8) | 3,100 (1.9) | 2 | 1.8 |
| Succinate | 750 (3.3) | 3,000 (1.6) | 3,900 (0.7) | 6,000 (1) | 3.3 | 1.6 |
β-Galactosidase activities (in Miller units) in strains PBS4 and PBS4B1 at the mid-exponential (ME; A600 of 0.5) or late-exponential (LE; A600 of 1) phase of growth, growing in either LB medium or in a defined medium with citrate, lactate, or succinate as the carbon source, were obtained from Fig. 3. Repression values (indicated in parentheses) were obtained by dividing the activity observed when cells used citrate as a carbon source by that observed when LB medium, lactate, or succinate was used.
That is, the quotient between the repression observed in strain PBS4 and that observed in the cyo-deficient strain PBS4B1.
The influence of the cyoB::tet mutation on the catabolic repression observed in cells growing in a minimal salts medium containing organic acids as the carbon source was also analyzed. In the case of the wild-type strain, the use of succinate or lactate as the carbon source reduced PalkB induction at mid-exponential phase by about threefold in the case of succinate and by fivefold in the case of lactate versus the values observed when citrate was the carbon source (Fig. 3C and D; see also Table 2). Repression declined as cells approached the stationary phase of growth. The picture was different in the strain lacking a functional cytochrome o ubiquinol oxidase. In this case, succinate allowed an induction of the PalkB promoter as efficiently as citrate (Fig. 3C), showing that the cyoB::tet mutation had totally eliminated catabolic repression. When lactate was the carbon source, a reduction in repression was also evident, but the increase in PalkB expression was only of about twofold (Fig. 3D). Inactivation of cytochrome o, therefore, seems to affect not only the catabolic observed in LB medium but also the repression exerted by organic acids in a defined medium.
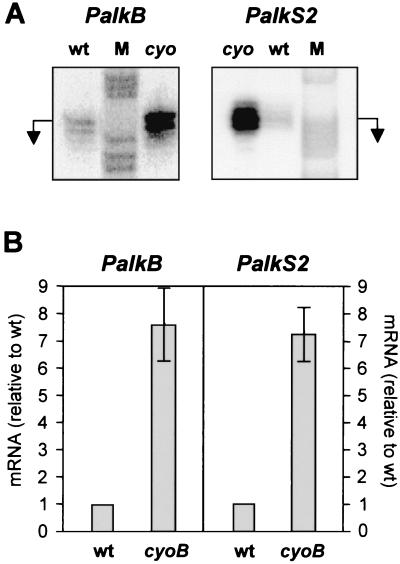
It was previously shown that the promoters PalkB and PalkS2 are similarly regulated by the AlkS protein and are subject to the same catabolic repression effect (7, 57). To analyze whether inactivation of cytochrome o also eliminated catabolic repression of the PalkS2 promoter, the activity of this promoter in cells growing in LB medium in the presence of the inducer DCPK was analyzed by S1 nuclease protection assays. As a control, the activity of the PalkB promoter was analyzed in parallel. As shown in Fig. 4, the levels of the transcripts originated at the PalkS2 and PalkB promoters at the late exponential phase of growth were approximately sevenfold higher in strain PBS4B1 than in the wild-type strain PBS4. This is direct evidence that inactivation of cytochrome o ubiquinol oxidase diminishes catabolic repression at both the PalkS2 and the PalkB promoters in cells growing in LB medium. It should be noted that when cells grow exponentially in the presence of DCPK, promoter PalkS1 is inactive and is not influenced by catabolic repression (7).
FIG. 4.
Effect of the cyoB mutation on expression of the PalkB and PalkS2 promoters. Strains PBS4 and PBS4B1 were grown in LB medium supplemented with the inducer DCPK (0.05% [vol/vol]). At an A600 of 0.8, cells were collected and the total RNA was obtained. The levels of mRNA originated at the PalkB and PalkS2 promoters were measured by S1 nuclease protection assays in the presence of a large excess of the probe. Promoter PalkS1 is inactive under these conditions (7) and gave no signal (results not shown). The cDNA resistant to S1 nuclease was resolved in a denaturing polyacrylamide gel, in parallel with a DNA size ladder obtained by chemical sequencing of the ssDNA used as probe.
Inactivation of the crc and cyoB genes has an additive effect in reducing catabolic repression.
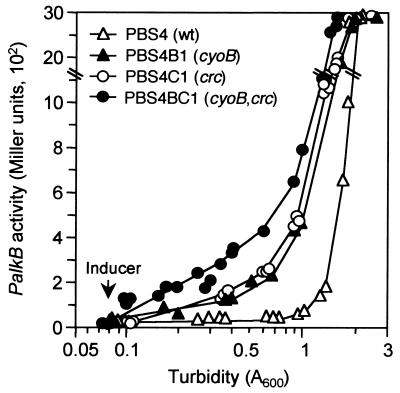
Inactivation of the crc gene decreases the catabolic repression of promoters PalkB and PalkS2 in rich LB medium (57). If crc and cyo form part of independent regulation pathways, their simultaneous inhibition should have an additive effect. On the contrary, if they are components of the same regulation pathway, their simultaneous inhibition should cause a decrease in catabolic repression not greater than that exerted by any of the two elements alone. To distinguish between these two possibilities, a P. putida strain was constructed in which both the crc and cyoB genes had been inactivated by marker exchange mutagenesis (strain PBS4BC1). As shown in Fig. 5, at mid-exponential phase the β-galactosidase activity in the wild-type strain was of 50 Miller units; inactivation of the crc gene (strain PBS4C1) increased this activity to 220 U, whereas inactivation of the cyoB gene (strain PBS4B1) increased the activity to 190 U. Simultaneous inactivation of the crc and cyoB genes (strain PBS4BC1) led to β-galactosidase activities of 400 Miller units, a value that is about twice that observed in strains PBS4C1 or PBS4B1. This indicates that the catabolic repression effect exerted by the Crc and cyo functions is additive. It is likely, therefore, that Crc and cyo modulate induction of the PalkB promoter through different pathways.
FIG. 5.
Effect of the simultaneous inactivation of the crc and cyo genes on catabolic repression of the PalkB promoter. P. putida strains PBS4, PBS4B1 (PBS4 with a knockout mutation at the cyoB gene), PBS4C1 (PBS4 with a knockout mutation at the crc gene), and PBS4BC1 (PBS4 with knockout mutations at the cyoB and crc genes) were grown in duplicate flasks in LB medium. At an A600 of 0.08, the nonmetabolizable inducer DCPK was added to one of the flasks, whereas the other flask was left as a noninduced control. Aliquots were taken at different times, and the β-galactosidase activity was measured. The levels of β-galactosidase are represented as a function of cell growth. The plot shows the values observed for induced cultures (noninduced cultures had very low β-galactosidase activities [30 to 90 Miller units] and are not represented). The values shown correspond to several independent assays, all represented on the same plot.
DISCUSSION
The results presented here show that inactivation of cytochrome o ubiquinol oxidase reduces the catabolic repression that occurs at the PalkB and PalkS2 promoters when cells grow exponentially in rich LB medium or in a defined medium with succinate or lactate as the carbon source. Repression reduction was only partial, since a clear repression effect was still observed. Inactivation of cyo did not affect growth rate significantly under the conditions used. Therefore, the lesser catabolic repression cannot be attributed to a decrease in the growth rate. Further, it is known that growth rate per se does not explain the catabolic repression of the alkane degradation pathway (56). Finally, mutations in many genes could have caused a decrease in growth rate, but the mutations obtained in subsequent screenings repeatedly mapped to the cyo cluster.
The observed link between a component of the electron transport chain and catabolic repression opens new ways to understand this global regulation process. There are some interesting examples of regulatory systems that monitor the flow of electrons through the electron transport chain and which use that information to regulate the activity of specific genes and integrate different metabolic activities. For example, in Rhodobacter sphaeroides the information obtained at two points of the electron transport chain is used to regulate the expression of photosynthesis genes (37, 38). A signal generated at the cbb3 branch of the electron transport chain is transduced to a two-component activation system which directly regulates gene expression. In addition, the redox state of the quinone pool (the ubiquinol/ubiquinone ratio) is monitored by a redox-active antirepressor protein, which determines the functional state of a transcriptional repressor. The redox state of quinones is also used to regulate the transition from aerobic to anaerobic metabolism in E. coli by means of the Arc two-component system (23). In this case, the oxidized form of the quinones serves as a specific signal for the ArcB sensor kinase, silencing it and impeding phosphorylation of the ArcA global transcriptional regulator. It has also been proposed that the Aer protein responds to the cellular redox state to regulate aerotaxis, which guides cells to oxygen-rich environments (44). Therefore, it is clear that bacterial cells can monitor the activity of the electron transport chain and use this information to regulate gene expression. The results presented in the present study suggest that the flow of electrons through the electron transport chain, the redox state of the cell, or the amounts of cyo ubiquinol oxidase (the levels of which are known to depend on the oxygen tension [15, 16, 49]) could be one of the signals that P. putida uses to modulate catabolic repression of the alkane degradation pathway. It cannot be ascertained at present whether the cyo terminal oxidase has a direct or indirect role in this signal transmission process.
Inactivation of cytochrome o ubiquinol oxidase reduced, but did not eliminate, catabolic repression (except that generated by succinate). This suggests that catabolic repression depends on more factors than just the electron transport chain. It has recently been reported that the Crc protein has an important role in the catabolic repression of the alkane degradation pathway (57). Inactivation of the crc gene substantially reduced (although did not entirely eliminate) catabolic repression in rich medium but had no effect on the catabolic repression exerted by lactate or succinate in a defined medium. Crc appears to be an element of a signal transmission pathway connecting cell physiology to carbon metabolism, participating in catabolic repression of some sugar and amino acid pathways (14, 26, 27), as well as in cell adhesion (40). Simultaneous inactivation of the cyoB and crc genes generated a greater decrease in catabolic repression in rich medium than the individual inactivation of either crc or cyoB. This additive effect suggests that Crc and cyo form part of different signal transduction pathways, both of them contributing to catabolic repression. In both cases, the signal leads to a decrease in the AlkS-mediated induction of the PalkB and PalkS2 promoters (57; the present study). The final consequence would be a decrease in the levels of the AlkS regulator, which appears to be present in the cell in limiting amounts (57). Keeping AlkS levels below those required for full induction of the pathway allows the cell to downmodulate expression of the alkane degradation genes when cells grow exponentially in medium containing a preferred carbon source in addition to alkanes. In agreement with this idea, overexpression of the alkS gene from a strong heterologous promoter totally eliminates catabolic repression (57).
Many other pathways for the degradation of linear or aromatic hydrocarbons are subject to physiological control (global regulation) in Pseudomonas spp. (17, 19, 32, 35, 39, 51), and the mechanisms involved often differ. Comparison of the factors implicated in the physiological control of the pWW0 toluene degradation pathway and of the P. putida CF600 phenol degradation pathway provides a remarkable example of how similar regulatory outcomes can be achieved by using different strategies (50). Both pathways are controlled by σ54-dependent regulator-promoter pairs that are mechanistically and functionally similar. In the two cases, activation of the corresponding promoters is modulated by a dominant physiological control that generates a similar final effect. The host factors responsible for this physiological control differ in the two pathways (9, 12, 13, 50, 51, 52). The effect of the cyo terminal oxidase on the regulation of these pathways has not been specifically addressed. However, it is worth noting that the catabolic repression generated by excess succinate on expression of the pWW0 toluene degradation pathway decreases considerably when O2 is limited (19, 20). In such conditions, the cyo terminal oxidase should decrease in favor of the cyd oxidase. This suggests the cyo terminal oxidase could also have some role in the repression of this pathway. Interestingly, the catabolic repression exerted by succinate on the expression of the phenol degradation pathway encoded in the P. putida H plasmid pPGH1 is reduced by inactivation of the cyo terminal oxidase (43). However, the effect of LB medium on the expression of this pathway has not been not reported. The role of the cyo terminal oxidase on the expression of other pathways has, to our knowledge, not been analyzed. The available data do not allow a unified picture of catabolic repression in Pseudomonas spp. to be drawn. However, the mechanisms used in each case seem to rely heavily on factors dictated by the promoters and regulators of the catabolic pathway (e.g., sigma factors involved in promoter recognition, regulator stability, etc.). It is likely that the physiological status of the cell is connected to gene expression in more than one way. Although certain mechanisms will probably be more suited to a particular pathway (or promoter-regulator pair) than to others, the evolutionary history of each pathway may be important in determining which factors participate in each case (10, 50). This could provide substantial diversity to physiological control mechanisms. However, more knowledge on the mechanisms mediating global control of the catabolic pathways of Pseudomonas sp. is required before final conclusions can be drawn.
Acknowledgments
We are grateful to M. Marín for helpful discussions and to L. Yuste for technical assistance.
This work was supported by grants BIO2000-0939 from the Comisión Interministerial de Ciencia y Tecnología and 07 M/0120/2000 from Comunidad Autónoma de Madrid. M.A.D. was the recipient of fellowships from the Instituto de Cooperación Iberoamericana and the Comisión Nacional de Investigación Científica y Tecnológica/Banco Interamericano de Desarrollo (Chile). A.R.-M. was the recipient of a fellowship from Gobierno Vasco.
REFERENCES
- 1.Anraku, Y., and R. B. Gennis. 1987. The aerobic respiratory chain of Escherichia coli. Trends Biochem. Sci. 12:262-266. [Google Scholar]
- 2.Au, D. C., R. M. Lorence, and R. B. Gennis. 1985. Isolation and characterization of an Escherichia coli mutant lacking the cytochrome o terminal oxidase. J. Bacteriol. 161:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bauchop, T., and S. R. Eldsen. 1960. The growth of microorganisms in relation to their energy supply. J. Gen. Microbiol. 23:457-569. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 7.Canosa, I., J. M. Sánchez-Romero, L. Yuste, and F. Rojo. 2000. A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Mol. Microbiol. 35:791-799. [DOI] [PubMed] [Google Scholar]
- 8.Canosa, I., L. Yuste, and F. Rojo. 1999. Role of the alternative sigma factor sigma-S in expression of the AlkS regulator of the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol. 181:1748-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmona, M., M. J. Rodriguez, O. Martinez-Costa, and V. De Lorenzo. 2000. In vivo and in vitro effects of (p)ppGpp on the sigma-54 promoter Pu of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 182:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cases, I., and V. de Lorenzo. 1998. Expression systems and physiological control of promoter activity in bacteria. Curr. Opin. Microbiol. 1:303-310. [DOI] [PubMed] [Google Scholar]
- 12.Cases, I., V. de Lorenzo, and J. Pérez-Martín. 1996. Involvement of sigma-54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol. Microbiol. 19:7-17. [DOI] [PubMed] [Google Scholar]
- 13.Cases, I., J. Pérez-Martín, and V. de Lorenzo. 1999. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the sigma-54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem. 274:15562-15568. [DOI] [PubMed] [Google Scholar]
- 14.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in Pseudomonads. Res. Microbiol. 147:551-561. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotter, P. A., S. B. Melville, J. A. Albrecht, and R. P. Gunsalus. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25:605-615. [DOI] [PubMed] [Google Scholar]
- 17.de Lorenzo, V., I. Cases, M. Herrero, and K. N. Timmis. 1993. Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol. 175:6902-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 19.Duetz, W. A., S. Marqués, B. Wind, J. L. Ramos, and J. G. van Andel. 1996. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol. 62:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duetz, W. A., B. Wind, M. Kamp, and J. G. van Andel. 1997. Effect of growth rate, nutrient limitation, and succinate on expression of TOL pathway enzymes in response to m-xylene in chemostat cultures of Pseudomonas putida (pWW0). Microbiology 143:2331-2338. [DOI] [PubMed] [Google Scholar]
- 21.Franklin, F. C., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhart, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 23.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 24.Grund, A., J. Shapiro, M. Fennewald, P. Bacha, J. Leahy, K. Markbreiter, M. Nieder, and M. Toepfer. 1975. Regulation of alkane oxidation in Pseudomonas putida. J. Bacteriol. 123:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hester, K. L., K. T. Madhusudhan, and J. R. Sokatch. 2000. Catabolite repression control by crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 182:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirayama, H., H. Takami, A. Inoue, and K. Horikoshi. 1998. Isolation and characterization of toluene-sensitive mutants from Pseudomonas putida IH-2000. FEMS Microbiol. Lett. 169:219-225. [DOI] [PubMed] [Google Scholar]
- 29.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 30.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 31.Kok, M., R. Oldenhuis, M. P. van der Linden, P. Raatjes, J. Kingma, P. H. van Lelyveld, and B. Witholt. 1989. The Pseudomonas oleovorans alkane hydroxylase gene: sequence and expression. J. Biol. Chem. 264:5435-5441. [PubMed] [Google Scholar]
- 32.McFall, S. M., B. Abraham, C. G. Narsolis, and A. M. Chakrabarty. 1997. A tricarboxylic acid cycle intermediate regulating transcription of a chloroaromatic biodegradative pathway: fumarate-mediated repression of the clcABD operon. J. Bacteriol. 179:6729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Monsalve, M., M. Mencía, F. Rojo, and M. Salas. 1995. Transcription regulation in Bacillus subtilis phage φ29: expression of the viral promoters throughout the infection cycle. Virology 207:23-31. [DOI] [PubMed] [Google Scholar]
- 35.Müller, C., L. Petruschka, H. Cuypers, G. Burchhardt, and H. Herrmann. 1996. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 178:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura, H., K. Saiki, T. Mogi, and Y. Anraku. 1997. Assignment and functional roles of the cyoABCDE gene products required for the Escherichia coli bo-type quinol oxidase. J. Biochem. 122:415-421. [DOI] [PubMed] [Google Scholar]
- 37.Oh, J. I., and S. Kaplan. 2001. Generalized approach to the regulation and integration of gene expression. Mol. Microbiol. 39:1116-1123. [DOI] [PubMed] [Google Scholar]
- 38.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 19:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Leary, N. D., K. E. O'Connor, W. Duetz, and A. D. Dobson. 2001. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology 147:973-979. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 42.Panke, S., A. Meyer, C. M. Huber, B. Witholt, and M. G. Wubbolts. 1999. An alkane-responsive expression system for the production of fine chemicals. Appl. Environ. Microbiol. 65:2324-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petruschka, L., G. Burchhardt, C. Müller, C. Weihe, and H. Herrmann. 2001. The cyo operon of Pseudomonas putida is involved in catabolic repression of phenol degradation. Mol. Gen. Genomics 266:199-206. [DOI] [PubMed] [Google Scholar]
- 44.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Sánchez-Romero, J. M., R. Dí az-Orejas, and V. De Lorenzo. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 64:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staijen, I. E., R. Marcionelli, and B. Witholt. 1999. The PalkBFGHJKL promoter is under carbon catabolite repression control in Pseudomonas oleovorans but not in Escherichia coli alk+ recombinants. J. Bacteriol. 181:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein, D. C. 1992. Plasmids with easily excisable xylE cassettes. Gene 11:157-158. [DOI] [PubMed] [Google Scholar]
- 49.Sweet, W. J., and J. A. Peterson. 1978. Changes in cytochrome content and electron transport patterns in Pseudomonas putida as a function of growth phase. J. Bacteriol. 133:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sze, C. C., L. M. D. Bernardo, and V. Shingler. 2002. Integration of global regulation of two aromatic-responsive σ54-dependent systems: a common phenotype by different mechanisms. J. Bacteriol. 184:760-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sze, C. C., T. Moore, and V. Shingler. 1996. Growth phase-dependent transcription of the σ54-dependent Po promoter controlling the Pseudomonas-derived (methyl)phenol dmp operon of pVI150. J. Bacteriol. 178:3727-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sze, C. C., and V. Shingler. 1999. The alarmone (p)ppGpp mediates physiological-responsive control at the σ54-dependent Po promoter. Mol. Microbiol. 31:1217-1228. [DOI] [PubMed] [Google Scholar]
- 53.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 54.van Beilen, J. B., M. G. Wubbolts, and B. Witholt. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174. [DOI] [PubMed] [Google Scholar]
- 55.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 56.Yuste, L., I. Canosa, and F. Rojo. 1998. Carbon-source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol. 180:5218-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuste, L., and F. Rojo. 2001. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 183:6197-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]