Abstract
The first committed step in the biosynthesis of heme, an important cofactor of two catalases and a number of cytochromes, is catalyzed by the hemA gene product. Salmonella enterica serovar Typhimurium hemA26::Tn10d (hemA26) was identified in a genetic screen of insertion mutants that were sensitive to hydrogen peroxide. Here we show that the hemA26 mutant respires at half the rate of wild-type cells and is highly susceptible to the effects of oxygen species. Exposure of the hemA26 strain to hydrogen peroxide results in extensive DNA damage and cell death. The chelation of intracellular free iron fully abrogates the sensitivity of this mutant, indicating that the DNA damage results from the iron-catalyzed formation of hydroxyl radicals. The inactivation of heme synthesis does not change the amount of intracellular iron, but by diminishing the rate of respiration, it apparently increases the amount of reducing equivalents available to drive the Fenton reaction. We also report that hydrogen peroxide has opposite effects on the expression of hemA and hemH, the first and last genes of heme biosynthesis pathway, respectively. hemA mRNA levels decrease, while the transcription of hemH is induced by hydrogen peroxide, in an oxyR-dependent manner. The oxyR-dependent induction is suppressed under conditions that accelerate the Fenton reaction by a mechanism that is not yet understood.
The Salmonella enterica serovar Typhimurium hemA gene encodes the enzyme glutamyl-tRNA reductase, which catalyzes the first committed step in heme biosynthesis. Because heme compounds are cofactors for a number of cytochromes and two catalases, heme is essential for respiration and defense against oxygen intermediates, including hydrogen peroxide (reviewed in reference 2). The heme biosynthetic pathway branches to produce two other tetrapyrroles: siroheme and cobalamin (vitamin B12). Siroheme is used in cysteine biosynthesis as a cofactor for sulfite reductase, and cobalamin is a cofactor utilized by several different enzymes (reviewed in references 2 and 21).
The hemA gene of serovar Typhimurium is the first gene in the operon hemA-prfA-dorf1. prfA encodes release factor 1, whereas the function of dorf1 (downstream open reading frame [ORF] 1) is unknown (5, 6). In Escherichia coli, an ORF designated HemK that shows 77% identity to Dorf1 was suggested initially to belong to the heme biosynthesis pathway (27). Recently, it was shown that the E. coli hemK gene encodes a methyltransferase that modifies peptide release factors and affects translation termination (14, 25). The activity of the HemA protein is regulated mainly at the level of protein stability in response to heme levels. HemA protein is more stable in heme-limited cells and is unstable in cells growing under normal conditions (38, 39). The proteolysis of HemA depends on Lon and ClpAP proteases, and the turnover is blocked in strains carrying mutations in both lon and clpP (38).
Heme synthesis is closely linked to iron metabolism. The final step of heme biosynthesis, which is catalyzed by hemH gene product, involves the insertion of ferrous iron (Fe2+) into protoporphyrin IX. The accumulation of porphyrins or iron is toxic to cells. Both porphyrins and iron stimulate the generation of highly reactive oxygen species, leading to damage of most biomolecules (26, 33). Thus, it may be useful for organisms to coordinate the cellular levels of iron and the biosynthesis of heme. The intimate relationship between iron and heme is highlighted by a syndrome displayed by patients with X-linked sideroblastic anemia. These patients display both heme deficiency and the accumulation of intracellular iron (reviewed in reference 8).
Being an essential element, iron has been the subject of many studies. Iron homeostasis is maintained by proteins that regulate iron acquisition, storage, and secretion (reviewed in references 28 and 35). In prokaryotes, Fur (ferric uptake repressor) is a central regulator for the maintenance of cytoplasmic iron levels. Fur associates with Fe2+ to repress transcription of genes involved in iron uptake and to activate expression of genes implicated in the defense against oxygen toxicity (reviewed in references 33 and 35). In low-iron conditions bacterial cells increase the expression of iron transporters and siderophores, whereas in high-iron conditions the expression of proteins involved in defense against oxidative stress is promoted (1, 35). The link between iron metabolism and oxidative stress is further demonstrated by the results that the primary regulators of the oxidative stress response in E. coli, OxyR and SoxRS, induce the synthesis of Fur and that the E. coli fur mutant is hypersensitive to oxidative stress (36, 44).
Although heme synthesis depends upon iron availability and heme products are essential for the defense against oxidative intermediates, very little is known about the coordination of heme synthesis, iron metabolism, and oxidative stress. We screened insertion mutants of S. enterica serovar Typhimurium for sensitivity to hydrogen peroxide and identified the hemA26::Tn10d-Tet mutant. In this mutant, Tn10 was inserted within the promoter region of hemA, leaving the gene intact. In contrast to hemA-null mutants that can hardly grow (40), the low expression of hemA26 by readthrough from the tetR gene made it possible to examine the phenotypes of this mutant. Here we show that exposure of hemA-deficient cells to hydrogen peroxide results in iron-dependent DNA damage and cell death.
MATERIALS AND METHODS
Strain construction and media.
The bacterial strains used in this study are listed in Table 1. Strains were routinely grown at 37°C in Luria-Bertani (LB) medium. Ampicillin (100 μg/ml) or tetracycline (10 μg/ml) were added where appropriate. Mutations were moved to SL1344 by P22 transductions. Where indicated, the strains were grown in NB medium (4) supplemented with 50 or 100 μg of 5-aminolevulinic acid (ALA) (Sigma)/ml. The generation time was calculated from semilogarithmic plots of A600 during exponential growth (23).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 | hisG46 | Lab collection |
| SA42 | SL1344 hemA26::Tn10d | This study |
| SA43 | SL1344 katE::Tn10 katG::pRR10ΔtrfA | This study |
| SA44 | SL1344 ΔoxyR::Tn10 | This study |
| FGP258 | SL1344 fur-1 zbf-5123::Tn10 | 9 |
| TT10423 | LT2 proAB47/F′pro+lac+zzf-1831:: Tn10d-Tet | 24 |
| TE719 | LT2 hemA60 | 38 |
| Plasmids | ||
| pSA31 | pGEM-4,5′hemA (Apr) | This study |
| pSA35 | pKK177-3, Ptac-dorf1-lacI | This study |
| pNK972 | Ptac-tnpA (Apr) | 24 |
| pKK177-3 | Ptac (Apr) | 3 |
| pTE367 | E. coli prfA+ (Apr) | 9 |
Apr, ampicillin resistant.
Isolation of the hemA26::Tn10d-Tet mutant.
The hemA26::Tn10d-Tet (hemA26) mutant was isolated in a search for insertion mutants sensitive to hydrogen peroxide as described below. A P22 phage lysate grown on TT10423 (zzf-1831::Tn10d-Tet) was transduced into a serovar Typhimurium SL1344 strain carrying the plasmid pNK972 expressing the transposase gene from the tac promoter (24). The tetracycline-resistant mutants selected on plates with LB medium and tetracycline (LB-Tet) were collected in two pools, and samples of each pool were used to grow P22 phage lysates. The resulting lysates carrying random chromosomal Tn10d-Tet insertions were used to infect overnight cultures of SL1344. The mutants were screened for sensitivity to hydrogen peroxide on LB plates containing 0.5, 1, or 2 mM hydrogen peroxide. We screened 3,000 mutants and selected 12 for further characterization. To locate the site of insertion in this specific mutant, a 3-kb EcoRI-KpnI fragment, previously identified on a Southern blot to carry part of the Tn10 and the flanking chromosomal DNA, was cloned and sequenced.
Plasmid construction.
To construct pGEM-5′hemA (pSA31), the hemA promoter fragment (838 nucleotides) was amplified from SL1344 chromosomal DNA by PCR by using the primers 452 (5′-CGG GAT CCA TCC TGT CCG GTC TG) and 453 (5′-GCG AAT TCT CCT GAT GCC AGT ACA) and subcloned into unique BamHI and EcoRI sites of pGEM-4 (Promega). To construct Ptac-dorf1-lacI (pSA35), the gene was amplified from SL1344 chromosomal DNA by PCR with the primers 586 (5′-TCC CCC GGG AGC CGC CTT ATC CGA GGA GGA A) and 478 (5′-ACG CGT CGA CGG AGA GCG CAA CAC AGA TA) and subcloned into unique SmaI and SalI sites of Ptac-lacI (pSA10 [31]). In this plasmid, dorf1 is transcribed from the tac promoter under the control of the LacI repressor. The sequence GCAG (five nucleotides upstream of the AUG) was replaced by GGAG to construct a better Shine-Dalgarno for the gene.
RNA isolation and primer extension assays.
To isolate total RNA, the cultures were pelleted and resuspended in 10 mM Tris (pH 7.5) and 1 mM EDTA. Lysozyme was added to 0.9 mg/ml, and the samples were subjected to three freeze-thaw cycles. Total RNA was isolated by using Ultraspec RNA according to the manufacturer's protocol (BIOTECX Laboratories), except that 1 ml of reagent was used for 12 to 16 OD600 units of the cells. The RNA samples (30 μg for the chromosomal gene, 3 μg for the plasmid-encoded gene, 2 μg for 5S RNA, and 30 μg for dps and himA) were subjected to primer extension (at 42°C for 45 min) by using AMV-RT (Boehringer) and the indicated end-labeled oligonucleotides: hemA (#655, 5′-GAT CAA GCG TGT CCG GCG), hemH (#531, 5′-TTA CCG CTT CAG GAG T), dps (#315, 5′-AGC AGA TTA GAC GCT TTT G), 5S RNA (#459, 5′-GAG ACC CCA CAC TAC CAT C), and himA (#2309, 5′-AAC AGA TCT TCT GAC ATT TCA GC). The extension products, together with sequencing reactions primed with an end-labeled primer, were separated on a 6% sequencing gel. To estimate the half-life of hemA mRNA, an SL1344 culture grown to mid-exponential phase in LB medium was split into four subcultures, and two of these were treated with 1 mM hydrogen peroxide. After 1 min, rifampin (0.2 mg/ml) was added to one treated and one nontreated culture. At selected time intervals after the rifampin addition, samples were withdrawn from treated and control cultures, and total RNA was extracted and subjected to a primer extension assay.
S1 nuclease assay.
The S1 nuclease protection assay was done as previously described (20) except that the annealing mixture (RNA and labeled probe) was treated with 70 U of S1 nuclease (MBI Fermentas) for 2 h. The probe was generated in a PCR machine with one end-labeled primer (#444, 5′-GGC ACC TGT ATC GCT GCG AG) and the hemA promoter fragment (844 nucleotides) as a template. The hemA promoter fragment was amplified from SL1344 chromosomal DNA by PCR with primers 452 and 453 (see plasmid construction). Total RNA (35 μg) and excess of labeled single-stranded DNA probe were mixed in 50 μl of hybridization buffer as described previously (20).
Hydrogen peroxide sensitivity assays.
Bacterial cultures grown to an A600 of 0.2 to 0.4 in LB were treated with 2.5 mM hydrogen peroxide. To determine viability, aliquots were taken at the indicated time points, diluted, and plated onto LB or LB-Tet plates. When indicated, the cells were exposed to 1 mM 2,2′-dipyridyl (Sigma) for 15 min or 3 mM potassium cyanide for 5 min prior to treatment with hydrogen peroxide. For ALA supplementation experiments, cells grown to an A600 of 0.2 to 0.4 in NB medium with or without 50 μg of ALA (4)/ml were treated with 1 mM hydrogen peroxide. Viability was assayed as described above.
Hydrogen peroxide degradation assay.
Hydrogen peroxide was determined by horseradish peroxidase-catalyzed oxidation of phenol red (22). Cultures grown to an A600 of 0.2 in LB medium were treated with 0.5 and 1 mM hydrogen peroxide for 10 min at 37°C. Then, 1 ml of each sample was pelleted, and 40 to 200 μl of the supernatants were mixed with phosphate-buffered saline (to give a final volume of 1 ml) containing horseradish peroxidase (8.4 purpurogallin units/ml) and phenol red (0.28 mM) and incubated at room temperature for 10 min before the addition of 10 μl of 5 M NaOH. The absorbance of the reaction mixtures was measured at 610 nm and correlated with values obtained by using appropriate dilutions of the reagent.
Respiration assay.
The respiration rate of exponentially (A600 = 0.2 to 0.3) growing cells in LB medium was measured by using an oxygen electrode (Rank Bros., Cambridge, United Kingdom).
SNG sensitivity assay.
Bacterial cultures of each strain were grown to an A600 of 0.18 in LB medium, and then half were treated with 1 mM 2,2′-dipyridyl (Sigma) for 20 min. Half of the iron chelator-treated cultures and half of the control untreated cultures were then treated with 1 μg of streptonigrin (SNG; Sigma)/ml. To determine viability, aliquots were taken at 0, 10, 20, and 40 min after the addition of SNG, diluted, and plated onto LB or LB-Tet plates.
Analysis of DNA topology.
Bacterial strains (SL1344 and SL1344 hemA26::Tn10) carrying pKK177-3 grown to mid-log phase in LB medium were split, and half were treated with 1 mM 2,2′-dipyridyl (Sigma) for 20 min. Thereafter, the treated and the untreated cultures were exposed to 0, 0.5, and 1 mM hydrogen peroxide for 15 min. To detect plasmid DNA strand breaks, the samples were analyzed on 1% agarose gels. The gels were run at 8 V/cm for 4 h in 40 mM Tris-acetate buffer. For documentation, the gels were stained with ethidium bromide (1 μg/ml). To detect changes in the negative supercoiling of the DNA, plasmid samples were analyzed on 1.4% agarose gels containing 10 μg of chloroquine/ml as described in Weinstein-Fischer et al. (41). The gels were run at 2.5 V/cm in 50 mM Tris phosphate buffer (pH 7.2) containing 10 μg of chloroquine/ml for 19 h with recirculated buffer. For documentation, the gels were soaked for 2 h in water and then stained with ethidium bromide (1 μg/ml) for 1 h.
Measurement of intracellular free iron.
Intracellular iron that is not incorporated in proteins was measured by whole-cell electron paramagnetic resonance (EPR) spectroscopy (19). Cultures were grown aerobically for at least five generations in 1 liter of LB medium to an A600 of 0.2 to 0.3. Cells were centrifuged and resuspended in 9 ml of LB. Then, 1 ml of 0.2 M desferrioxamine was added, and cells were incubated for 15 min at 37°C with shaking. The cells were then centrifuged, washed with 5 ml of cold 20 mM Tris buffer (pH 7.4), and resuspended in 400 μl of cold 20 mM Tris (pH 7.4)-10% glycerol. Then, 200 μl of the suspended cells was loaded into an EPR tube, frozen in dry ice, and stored at −80°C until analysis. Ferric sulfate standards were prepared in the same Tris-glycerol solution; the exact iron concentration was calculated by using a ɛmM value of 2.865 cm−1 at 420 nm. The EPR signals were averaged from 30 scans by using a Varian Century E-112 X-band spectrophotometer equipped with a Varian TE102 cavity and temperature controller. The spectrometer settings were as follows: field center, 1,570 G; receiver gain, 3,200; field sweep, 400 G; modulation amplitude, 12.5 G; temperature, −125°C; and power, 30 mW. The measured EPR signals were converted to approximate intracellular concentrations by normalization to the cell density by using the relation that 1 ml of a culture of LB medium-grown E. coli at an A600 of 1 comprises 0.47 μl of intracellular volume (16).
RESULTS
Mapping of hemA transcription start site and the insertion mutant.
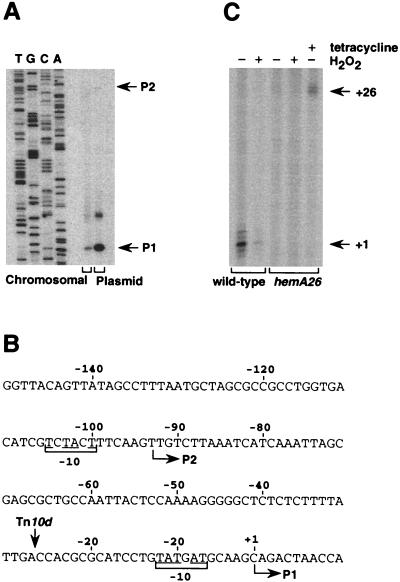
The hemA26::Tn10d-Tet (hemA26) mutant was isolated in a search for insertion mutants sensitive to hydrogen peroxide (see Materials and Methods for details). Analysis of the insertion site showed that the transposon is inserted 64 nucleotides upstream of the initiation codon of hemA (5). To locate the insertion within the hemA promoter, we analyzed hemA transcription by primer extension. To ensure detection of hemA mRNA we also cloned the 5′-end promoter region of the gene on a multicopy number plasmid. Samples of total RNA isolated from exponential-phase cultures of wild-type serovar Typhimurium with or without the plasmid were subjected to primer extension with a hemA primer. The transcript of the chromosomal-encoded gene revealed the presence of one major start site (Fig. 1A and B). The location of this start site corresponds to a site previously denoted P1 (4), as well as to a site that was identified by S1 analysis for the E. coli hemA gene (37). RNA analysis of the plasmid-encoded promoter region revealed two additional bands, of which the minor one corresponds to a site previously denoted P2 (4, 37). The third RNA product is presumably a degradation product originating from the P2 transcript. The mapping of the hemA transcripts indicated that the transposon was inserted within the promoter of hemA, 26 nucleotides upstream of P1 transcription start site (Fig. 1B).
FIG. 1.
Mapping of the hemA promoter and the insertion mutant. (A) Primer extension analysis of chromosomal (30 μg, total RNA) and plasmid-encoded (3 μg, total RNA) hemA promoter region. Total RNA was extracted from exponential-phase (A600 = 0.3) SL1344 and SL1344 cells carrying the plasmid encoding the hemA promoter region (pGEM-5′hemA). The sequencing reaction was carried out with the same primer. (B) Sequence of the hemA promoter region. The horizontal arrows indicate the start sites observed by primer extension. Brackets indicate the −10 regions of P1 and P2. The bases matching the −10 hexamers of the σ70 consensus are underlined. Both P1 and P2 promoters have no obvious −35 sequence. The vertical arrow indicates the site of insertion of the transposon Tn10d. (C) S1 mapping of the hemA transcripts. Total RNA was extracted from exponential cultures (A600 = 0.3) of SL1344 wild type and SL1344 hemA26 mutant prior to and after exposure to 1 mM hydrogen peroxide (15 min). To analyze hemA transcription from the tetR promoter, hemA26 strain was exposed to 10 μg of tetracycline/ml for 15 min. S1 mapping was carried out with an end-labeled single-stranded DNA fragment (612 bases) complementary to hemA. The fragments protected by hemA mRNA in wild-type and hemA26 cells were 93 (indicated as “+1”) and 119 (indicated as “+26”) nucleotides long, respectively. +1, Transcription start site of P1; +26, insertion site (i.e., the position at which transcription originating at the tetR promoter enters into hemA).
Characterization of hemA26 mutant.
It has been known since the 1970s that hemA mutants grow poorly, forming tiny colonies on LB plates (27). We noticed that the hemA26 mutant formed small colonies on LB plates but that the colony size increased substantially when the cells were grown on plates supplemented with tetracycline. The generation times of wild-type cells and of the mutant hemA26 grown in NB medium with or without tetracycline were 29.4, 70.8, and 31.7 min, respectively, indicating that the addition of tetracycline corrected the mutant phenotype (Table 2). Insertion of mini-Tn10 within a promoter region can block transcription of the downstream genes from the natural promoter. However, upon addition of tetracycline, the two divergent genes of the transposon, tetA and tetR, are induced, and the resulting transcriptions can extend beyond the ends of the element into a downstream operon (34). Mapping of the hemA transcripts by S1 further confirmed that the transposon insertion abolished transcription of hemA from its own promoter (Fig. 1C, first and third lanes) but enabled transcription from the tetR promoter upon addition of tetracycline (Fig. 1C, fifth lane). Because the hemA26 mutant was viable in the absence of tetracycline and not as sick as the previously isolated hemA mutants (generation time of >6 h; Table 2) (40), we concluded that the operon was not completely shut off.
TABLE 2.
ALA and tetracycline complementation of SL1344 wild-type and hemA26 mutant strains
| Strain | Tetracycline (μg/ml) | ALA (μg/ml) | Generation timea (min) |
|---|---|---|---|
| SL1344 | 0 | 29.4 | |
| SL1344 | 50 | 33.3 | |
| SL1344 | 100 | 33.9 | |
| SL1344/pTE367/(prfA) | 0 | 30.8 | |
| SL1344/Ptac-dorf1b | 0 | 32.1 | |
| LT2 hemA60 | 0 | 378.4 | |
| LT2 hemA60 | 100 | 55.0 | |
| SL1344 hemA26 | 0 | 70.8 | |
| SL1344 hemA26 | 10 | 0 | 31.7 |
| SL1344 hemA26 | 50 | 55.3 | |
| SL1344 hemA26 | 100 | 47.3 | |
| SL1344 hemA26/pTE367(prfA) | 0 | 72.1 | |
| SL1344 hemA26/pTE367(prfA) | 50 | 48.6 | |
| SL1344 hemA26/pTE367(prfA) | 100 | 44.8 | |
| SL1344 hemA26/Ptac-dorf1b | 0 | 67.9 | |
| SL1344 hemA26/Ptac-dorf1b | 50 | 50.8 | |
| SL1344 hemA26/Ptac-dorf1b | 100 | 47.7 |
Cultures were grown in NB. Tetracycline (10 μg/ml) or ALA (50 or 100 μg/ml) was added where indicated.
To induce expression of dorf1 from Ptac-dorf1 plasmid, the cells were diluted in NB medium supplemented with IPTG (0.1 μg/ml).
Null mutations in hemA cause an auxotrophy for ALA, and supplementing the medium with ALA can rescue the growth defects of these mutants (5, 40). Addition of ALA had no effect on the growth rate of wild-type cells; however, it diminished the generation time of hemA26 cells substantially (Table 2). The finding that ALA can suppress the growth defects of hemA26 further confirmed that the transposon insertion within the upstream region of hemA resulted in inactivation of heme synthesis.
The hemA gene is the first gene in the operon hemA-prfA-dorf1. The gene prfA, encoding release factor 1, is essential, and mutations in this gene lead to impaired cell division and sometimes to cell lysis due to inhibition of septation (29). The function of dorf1 is unknown (5, 6). In E. coli, an ORF designated HemK that shows 77% identity to Dorf1 was recently shown to encode a methyltransferase that modifies peptide release factors and affects translation termination (14, 25). Because transcription of the operon in hemA26 strain is impaired, we examined whether plasmids expressing the genes prfA and dorf1 could complement the growth defects of this mutant. To test the effect of prfA, the hemA26 cells were transformed with a plasmid expressing the E. coli prfA gene, which was previously used to complement a null mutant of prfA of S. enterica serovar Typhimurium (7). To examine dorf1, the gene was cloned downstream of the tac promoter under the control of the LacI repressor. We found that plasmids expressing prfA or dorf1 had no effect on the doubling time of hemA26 (Table 2). In addition, ALA supplements improved the growth of these strains exactly as it did that of their hemA26 parent (Table 2). We conclude that the growth deficiency of the hemA mutant is primarily the result of inadequate heme biosynthesis.
S. enterica serovar Typhimurium hemA26 is highly sensitive to hydrogen peroxide.
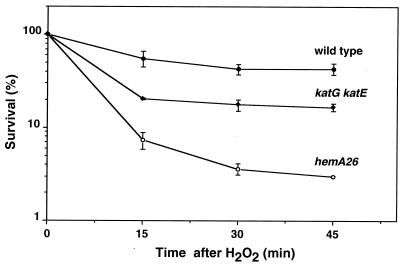
To quantify the sensitivity of the hemA26 mutant to hydrogen peroxide, we examined the viability of exponential phase cells after exposure to 2.5 mM hydrogen peroxide. The assays demonstrated that only 7% of the hemA26 mutant cells survived the first 15 min of treatment (Fig. 2). In contrast, 60% of wild-type cells survived. We also examined hydrogen peroxide sensitivity of hemA26 cells carrying prfA or dorf1 plasmids. We found that, although the prfA plasmid increased the total number of CFU (three- to fourfold), both hemA26/pprfA and hemA26/pdorf1 cells were as sensitive to hydrogen peroxide as were hemA26 cells in the absence of the plasmids (not shown).
FIG. 2.
Hydrogen peroxide killing assay. Bacterial cultures (SL1344 wild-type, SL1344 katG katE, and SL1344 hemA26 strains) grown to A600 = 0.2 to 0.4 in LB medium were treated with 2.5 mM hydrogen peroxide. Viability was assayed at the indicated time points by plating the cells on LB or LB-Tet plates.
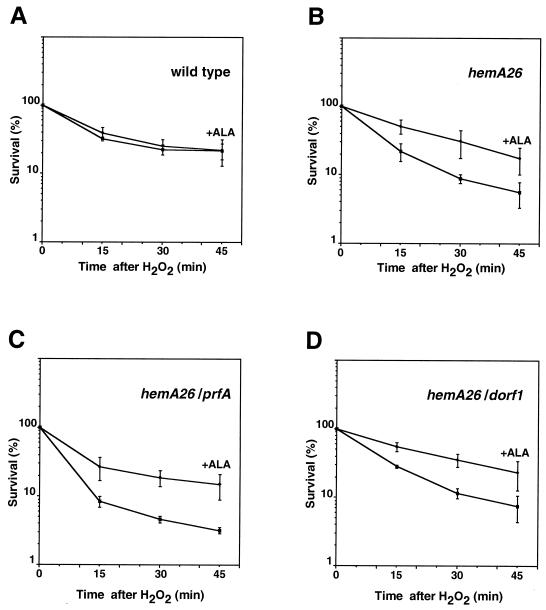
To test whether the increased sensitivity to hydrogen peroxide was caused by the limited flow through the heme pathway, we examined the viability of hemA26 cells grown in medium supplemented with ALA and exposed to hydrogen peroxide. We found that the addition of ALA rendered the hemA26 mutant as resistant as the wild type (Fig. 3A and B). ALA supplementation of hemA26 cells carrying prfA or dorf1 plasmids had a similar corrective effect (Fig. 3C and D). We sought to clone the hemA gene on a plasmid, for inducible expression of HemA protein, but were unable to do so. Others have previously reported difficulties in the cloning of this gene on plasmids (27).
FIG. 3.
ALA supplementation in hydrogen peroxide killing. Bacterial cultures (i.e., SL1344 wild type [A], SL1344 hemA26 mutant [B], SL1344 hemA26 mutant carrying pTE367 [prfA] [C], and SL1344 hemA26 mutant carrying pSA35 [Ptac-dorf1 lacI] [D]) grown to A600 = 0.2 to 0.4 in NB in the absence or in the presence of ALA (at 50 μg/ml) were treated with 1 mM hydrogen peroxide. Viability was assayed at the indicated time points by plating the cells on LB or LB-Tet plates. To induce expression of dorf1 from the Ptac-dorf1 plasmid, the cells were dilute in NB medium supplemented with IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 μg/ml).
Heme is a cofactor of two catalases that degrade hydrogen peroxide, hydroperoxidase I and hydroperoxidase II, encoded by the genes katG and katE, respectively (30). Thus, heme deficiency may diminish the ability of cells to scavenge hydrogen peroxide. We assayed elimination of hydrogen peroxide by wild-type, hemA26, and katG katE strains. The katG katE mutant was largely deficient in hydrogen peroxide degradation (Table 3); the small residual activity was likely due to a manganese-dependent nonheme catalase (30). The hemA26 mutant was also substantially deficient in the degradation of hydrogen peroxide. However, the deficiency was not as great as that of the katG katE mutant. This point is important, since viability assays demonstrated that the katG katE mutant was less sensitive to killing (Fig. 2). These results suggested that the sensitivity of the hemA26 mutant to hydrogen peroxide was not due solely to decreased activity of catalases.
TABLE 3.
Remaining hydrogen peroxide
| H2O2 (mM) | Mean hydrogen peroxide remaining (% of initial amt) ± SD in straina:
|
||
|---|---|---|---|
| SL1344 wt | SL1344 katG katE mutant | SL1344 hemA26 mutantb | |
| 0.5 | 1.3 ± 0.9 | 55.3 ± 2.7 | 34.0 ± 2.8 |
| 1.0 | 9.6 ± 3.9 | 76.8 ± 8.0 | 52.3 ± 8.4 |
Hydrogen peroxide was measured 10 min after exposure to either 0.5 or 1 mM H2O2. wt, wild type
hemA26 cells were introduced with the prfA plasmid (pTE367) to adjust the total number of CFU of the hemA26 strain.
The effect of catalase activity upon hydrogen peroxide toxicity is minimal when cultures are dilute, since the collective catalase activity is very small (18, 22). However, the hemA26 mutant remained more susceptible to hydrogen peroxide than the wild type even when the cells were diluted prior to challenge (A600 = 0.025 instead of 0.2) (data not shown).
The hemA26 mutant is prone to iron-mediated DNA damage.
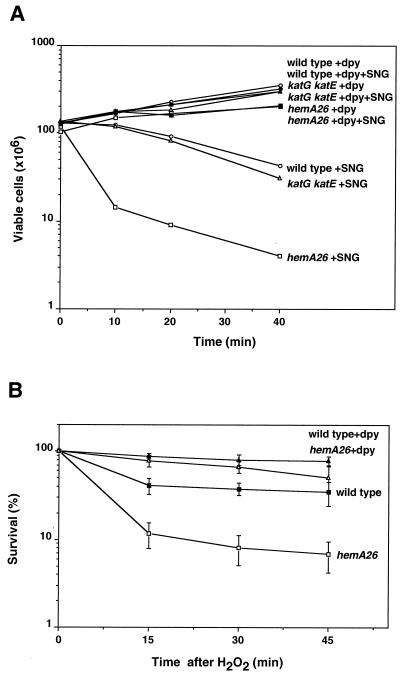
The result that the hemA26 mutant was more sensitive to hydrogen peroxide than were mutants of catalases led us to suspect that inactivation of heme synthesis might affect iron homeostasis. In higher organisms interruptions in heme synthesis can trigger massive import of iron into the cell, presumably because the lack of heme is interpreted by regulatory controls as an indication of iron deficiency (8). SNG has been previously used to assess free iron levels in bacterial cells (43). SNG is an aminoquinone that is cyclically reduced and oxidized inside bacteria, producing superoxide and hydroxyl radicals (10, 13). It has been found that the antibiotic activity of SNG is enhanced by iron and that an increase in the levels of cellular free ferrous iron leads to the production of higher levels of reactive oxygen species and to more damage (42). We used SNG to assess iron-mediated DNA damage in the hemA26 mutant. Exposure of the hemA26 mutant to a low concentration of SNG (1 μg/ml) was sufficient to kill 90% of the cells within the first 10 min (Fig. 4A). Both wild-type cells and the katG katE double mutant were much less susceptible, with only 20 to 30% of the cells killed. 2,2′-Dipyridyl is an iron chelator that penetrates cells and chelates intracellular ferrous iron as well as iron outside the cells. The addition of 2,2′-dipyridyl rendered the hemA26 strain tolerant to SNG, indicating that the effects of SNG observed with the hemA26 mutant were related to free ferrous iron (Fig. 4A). Similarly, both 2,2′-dipyridyl and desferrioxamine, a cell-permeable iron chelator that is structurally unrelated to dipyridyl, fully protected the mutant against 2.5 mM hydrogen peroxide (Fig. 4B and data not shown).
FIG. 4.
Effect of iron chelator. (A) SNG survival assay. Bacterial cultures of each strain carrying the prfA plasmid were grown to an A600 of 0.18 in LB medium, and then half were treated with 1 mM 2,2′-dipyridyl (dpy) for 20 min. Thereafter, the iron chelator-treated cultures and the control untreated cultures were treated with 1 μg of SNG/ml. Viability was assayed at 0, 10, 20, and 40 min after the addition of SNG by plating the bacterial cells onto LB or LB-Tet plates. The average of five independent experiments is shown. (B) Hydrogen peroxide killing. Wild-type and hemA26 strains carrying the prfA plasmid were examined for hydrogen peroxide sensitivity (2.5 mM) with or without prior treatment with 1 mM 2,2′-dipyridyl for 15 min. In both experiments, the cells were introduced with the prfA plasmid (pTE367) to adjust the total number of hemA26 CFU.
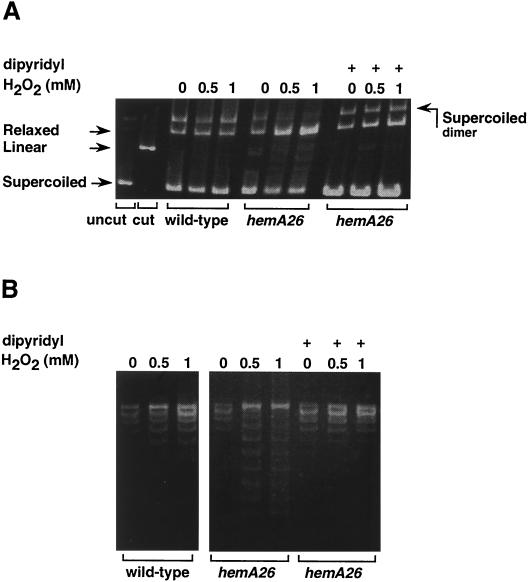
Hydroxyl radicals are generated when ferrous iron transfers an electron to hydrogen peroxide (the Fenton reaction). These radicals then directly oxidize DNA, producing strand breaks and changes in DNA topology (15, 41). We monitored the levels of oxidative DNA lesions in hemA26 cells by looking at a small reporter plasmid after treatment with hydrogen peroxide. Wild-type and mutant cultures at mid-log phase were treated with 0.5 and 1 mM hydrogen peroxide for 15 min, and then plasmid samples were extracted and analyzed on 1% agarose gels. The gel shows that much of the plasmid in the hemA26 mutant is in a nicked circular form (relaxed), while the supercoiled form decreases, indicating that upon exposure to hydrogen peroxide the reporter plasmid in hemA26 mutant is subjected to high levels of hydroxyl radicals (Fig. 5A). In contrast, little damage was evident in the wild-type strain. Treatment of the cultures with 2,2′-dipyridyl prior to the hydrogen peroxide treatment eliminated the effects of hydrogen peroxide and the DNA remained intact (Fig. 5A). Analysis of the plasmid samples on chloroquine gels showed that exposure of the hemA26 strain to hydrogen peroxide resulted in a dramatic decrease in the negative supercoiling of the DNA (Fig. 5B). Hydrogen peroxide treatment in the presence of the iron chelator had no effect on the level of the negative supercoiling of the DNA. Taken together, the results show that exposure of hemA-deficient cells to hydrogen peroxide leads to extensive, iron-mediated DNA damage and to cell death.
FIG. 5.
Analysis of plasmid DNA topology. (A) Detection of single-strand breaks. Cultures of wild-type SL1344 and hemA26 mutant carrying pKK177-3 were grown to an A600 of 0.25 in LB mutant and were then exposed to 0, 0.5, and 1 mM hydrogen peroxide for 15 min. Where indicated, the iron chelator 2,2′-dipyridyl (1 mM) was added to the cells 15 min prior to the treatment with hydrogen peroxide. Plasmid DNA samples were separated on a 1% agarose gel. DNA was visualized by ethidium bromide staining. uncut, pKK177-3 DNA; cut, pKK177-3 DNA digested with EcoRI. (B) Detection of changes in the negative supercoiling of the DNA. The plasmid samples from above were analyzed on 1.4% agarose gels containing 10 μg of chloroquine/ml. At this chloroquine concentration, the most relaxed molecules migrate most rapidly through the gel (41). DNA was visualized by ethidium bromide staining.
The hemA26 mutant exhibits no increase in iron levels.
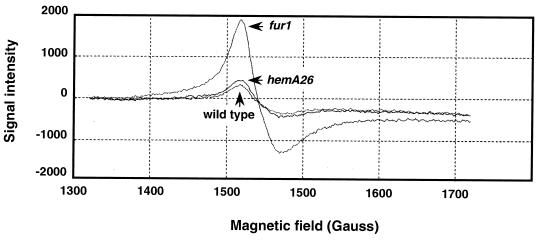
It has previously been demonstrated that the intracellular accumulation of free iron renders bacteria unusually vulnerable to being killed by hydrogen peroxide. Examples of this include E. coli fur mutants, which import more iron than can be used or stored (36), and superoxide dismutase-deficient mutants, in which superoxide releases iron from the [4Fe-4S] clusters of oxidant-sensitive dehydratases (19). To test whether hemA26 mutant hypersensitivity to hydrogen peroxide was due to iron accumulation, we quantified free-iron levels inside intact cells by EPR spectroscopy. This method detects chelator-accessible iron, including iron loosely bound to biomolecules such as cells membranes and nucleic acids. Because desferrioxamine does not extract iron atoms from metalloproteins, they do not contribute to the iron signal (19). The hemA26 mutant had only 1.2-fold more iron than did wild-type cells (Fig. 6). In contrast, the fur mutant contained 5.1-fold more iron. Thus, the hypersensitivity of the hemA26 strain to hydrogen peroxide was not due to an increase in iron levels.
FIG. 6.
EPR analysis of intracellular free iron. Samples for EPR analysis were taken from exponential-phase cultures of SL1344 wild-type, hemA26, and fur-1 strains. The iron concentration was calculated based on the following ferric sulfate standards: wild type, 36 μM intracellular chelatable iron; hemA26, 45 μM iron; fur-1, 190 μM iron.
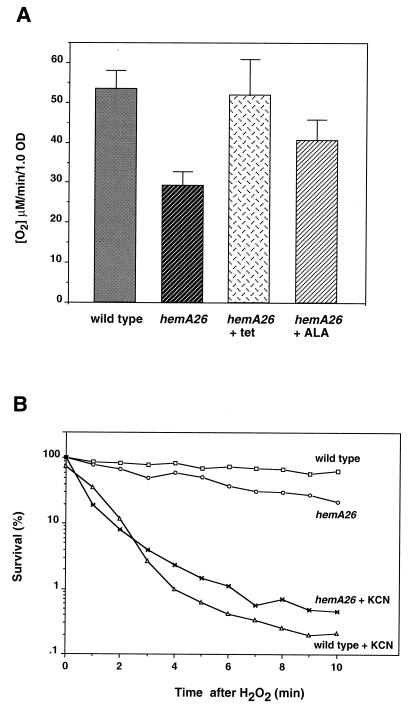
A respiratory defect in the hemA mutant causes sensitivity to hydrogen peroxide.
Measurements of respiratory rates during exponential growth revealed that the hemA26 mutant respired at only half the rate of wild-type cells (Fig. 7), indicating that the cytochrome oxidases were insufficiently charged with heme. Respiration was largely restored when hemA expression was stimulated by tetracycline or when ALA was added to the medium (Fig. 7). This observation suggested an explanation for the vulnerability to hydrogen peroxide, since respiration-deficient strains of E. coli were shown to be hypersensitive to DNA damage by hydrogen peroxide (17, 32). Cyanide directly inhibits cytochrome oxidases, and its addition greatly enhances the killing of wild-type E. coli by hydrogen peroxide. We found that the same was true of the wild-type Salmonella strain (Fig. 7). Notably, the hemA mutant cells were no more sensitive than the wild-type cells when both were challenged with hydrogen peroxide in the presence of cyanide. This result suggests that it is the diminished cytochrome oxidase activity that sensitizes hemA26 mutant. The basis of this effect is believed to be that respiratory deficiency causes the accumulation of NADH, which ultimately provides to free iron the electrons that drive the Fenton reaction.
FIG. 7.
Respiratory blocks sensitize Salmonella to killing by hydrogen peroxide. (A) Oxygen consumption was measured in exponential phase cultures grown in LB medium. Tetracycline (10 μg/ml), or ALA (50 μg/ml) were added where indicated. (B) Killing by hydrogen peroxide in respiration deficient cells. Where indicated, 3 mM potassium cyanide was added to cells 5 min before challenge with 2.5 mM hydrogen peroxide. Viability was assayed at the indicated time points. The addition of cyanide alone did not diminish cell viability.
Regulation of hemA and hemH expression by hydrogen peroxide.
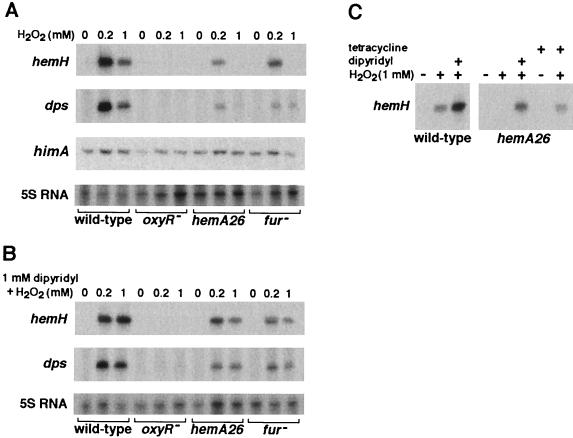
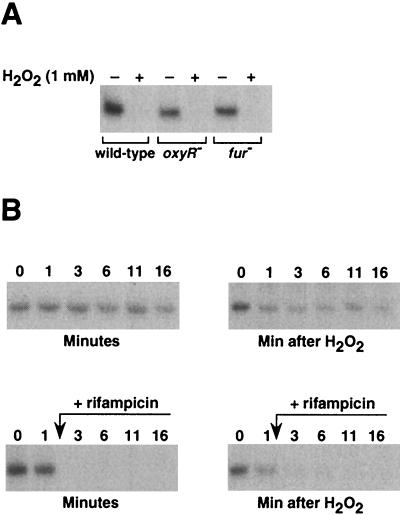
To learn more about the coordination between heme biosynthesis, iron metabolism, and oxidative stress, we examined the effects of hydrogen peroxide and iron deficiency on hemA and hemH, the first and the last genes in heme pathway, respectively. We found that iron deficiency, imposed by iron chelators, had no effect on the transcription of these genes (Fig. 9B and data not shown). However, exposure to hydrogen peroxide led to a decrease in hemA mRNA levels (Fig. 8A), whereas hemH levels were increased (Fig. 9A). The decrease in the transcript levels of the hemA gene was also detected by S1 analysis (Fig. 1C, first and second lanes), and we found that this decrease was independent of both Fur and OxyR (Fig. 8A).
FIG. 9.
Primer extension assays. (A) Cultures at exponential phase were exposed to 0, 0.2, and 1 mM of hydrogen peroxide for 5 min. (B) The cultures shown in panel A were exposed to the iron chelator (1 mM) for 15 min prior to the treatment with hydrogen peroxide. (C) Cultures of wild-type and hemA26 strains grown to an A600 of 0.25 in LB medium were split. One part of each culture was treated with the iron chelator 2,2′-dipyridyl (1 mM) for 15 min. Thereafter, the dipyridyl treated and the untreated cultures were exposed to 1 mM hydrogen peroxide for 5 min. In addition, two parts of hemA26 culture were exposed to 10 μg of tetracycline/ml for 10 min, and then one was further exposed to hydrogen peroxide (1 mM) for 5 min.
FIG. 8.
Primer extension assays. (A) Cultures of SL1344 wild-type, ΔoxyR, and fur1 strains were grown to an A600 of 0.3 to 0.4 in LB medium, and then half of each culture was treated with 1 mM hydrogen peroxide for 15 min. P1 of hemA mutant is shown. No expression could be detected from P2. (B) A wild-type culture was split into four subcultures, and two were treated with 1 mM hydrogen peroxide. Rifampin (0.2 mg/ml) was added after 1 min to one treated and one untreated culture. Samples were taken from control untreated and treated cultures 1, 3, 6, 11, and 16 min after the addition of hydrogen peroxide. Approximately 80% of the hemA mRNA in the cells exposed to hydrogen peroxide and 95% of the nonexposed mRNA were degraded within the first 2 min of rifampin treatment, as measured with a BioImaging Analyzer.
To test whether the decrease in hemA mRNA level is due to an effect of hydrogen peroxide on RNA stability, we examined hemA transcript degradation with rifampin. Much of the RNA was degraded within the first 2 min after the addition of rifampin, indicating that hemA mRNA is highly unstable (Fig. 8B). Examination of the stability of hemA in cells exposed to hydrogen peroxide, however, revealed no increase in the degradation rate of this mRNA (Fig. 8B), indicating that the decrease in hemA mutant steady-state levels in response to the treatment is due an effect of transcriptional repression.
The activation of hemH transcription by hydrogen peroxide was oxyR dependent (Fig. 9A). A recent study has documented such an induction for the E. coli hemH gene (45). Interestingly, activation of hemH by oxidative stress in the hemA26 mutant was greatly reduced, particularly at millimolar concentrations of hydrogen peroxide. Likewise, hemH induction in fur mutant cells was reduced. The addition of 2,2′-dipyridyl to both hemA26 and fur strains restored regulation, resulting in activation of hemH in response to the treatment (Fig. 9B). Similarly, the addition of tetracycline to restore hemA transcription and thereby heme synthesis also restored hemH activation by hydrogen peroxide (Fig. 9C). These results suggested that conditions that accelerate oxidative DNA damage simultaneously suppress the hydrogen peroxide dependent transcription activation of hemH.
To find out whether hemA26 and fur mutations only reduce the expression of genes in the heme pathway, we analyzed the transcription of dps, which encodes a nonspecific DNA-binding protein that is known to be induced by OxyR in response to hydrogen peroxide (reviewed in reference 33). We noticed that hydrogen peroxide- dependent activation of dps in hemA26 or fur mutant strains was reduced and that the removal of iron restored efficient induction of dps (Fig. 9A and B). In contrast, analysis of mRNA levels of himA encoding the α subunit of IHF protein in hemA26 and fur mutants showed that hydrogen peroxide did not suppress its expression, indicating that this reduction in expression by hydrogen peroxide is specific to oxyR-regulated genes.
DISCUSSION
We have isolated a hemA26 mutant of S. enterica serovar Typhimurium in a genetic screen of insertion mutants that were sensitive to hydrogen peroxide. We show that hemA26 mutant is partially defective at heme biosynthesis, diminishing the rates at which it can both scavenge hydrogen peroxide and respire. Both effects are likely to have contributed to its sensitivity in the initial screen. When bacteria are incubated on plates laden with hydrogen peroxide, they only form colonies if they can maintain viability until they can reduce hydrogen peroxide to subinhibitory levels. Both the diminished rate of scavenging and the accelerated rate of DNA damage would compromise their effectiveness at doing so.
While the reduced scavenging activity is an obvious consequence of diminished catalase activity, the connection between poor respiration and accelerated DNA damage is less obvious. The present study shows that exposure of a heme-deficient mutant with reduced respiration to hydrogen peroxide results in extensive iron-mediated DNA damage and cell death. The inactivation of heme synthesis does not change the amount of intracellular iron, and we propose that it is the respiratory deficiency that causes the accumulation of NADH, which ultimately provides to free iron the electrons that drive the Fenton reaction. Previous work demonstrated that inhibition of any step in the respiratory chain dramatically accelerates the rate at which hydrogen peroxide kills E. coli (17, 32). It was proposed that, by blocking the oxidation of NADH, respiratory inhibitors would increase the amount of cytosolic reductants available to reduce free ferric iron. In the presence of hydrogen peroxide, the ferrous iron thus formed will donate an electron and generate a hydroxyl radical that can damage DNA. Although NADH can directly reduce free iron at a low rate, recent data indicate that it does so in vivo much more efficiently through the agency of an NADH:flavin oxidoreductase (A. N. Woodmansee and J. Imlay, unpublished data). Reduced flavins are the direct electron donors.
Interestingly, control of heme biosynthesis is attuned more to oxidative stress than to iron levels. We found that neither iron loading nor iron depletion had a significant effect on hemA or hemH expression in S. enterica serovar Typhimurium. Mutations in fur appeared not to affect transcription. This result contrasts with the example of Bradyrhizobium japonicum, in which iron-dependent regulation of heme biosynthesis involves both Fur and a Fur-like protein Irr: Fur regulates hemA and Irr regulates hemB in response to iron (11, 12). Since free-iron levels are unaffected by the hemA26 allele, we infer that deficiencies in heme biosynthesis are not compensated for by accelerated iron loading of the cell. Thus, iron metabolism and heme biosynthesis seem not to be coordinated in Salmonella.
However, heme biosynthesis is affected on multiple levels by oxidative stress. Low levels of hydrogen peroxide activate OxyR and induce hemH, which encodes the ultimate enzyme in heme biosynthesis. Yet the transcription of hemA, which encodes the initial pathway enzyme, is reduced. This arrangement is unusual and warrants some speculation. Induction of heme synthesis during oxidative stress seems reasonable because OxyR also induces the synthesis of a major heme-requiring enzyme, hydroperoxidase I. Nevertheless, the reduction in hemA mRNA levels while the hemH mutant is activated suggests that during oxidative stress it may be important to simultaneously shut off the heme synthesis pathway at its first step, while completing the conversion of potentially toxic intermediate products into heme. Furthermore, the induction of hemH-encoded ferrochetalase may be particularly useful in allowing the heme biosynthetic pathway to scavenge intracellular iron before it is oxidized by hydrogen peroxide.
We have shown that higher (millimolar) concentrations of hydrogen peroxide affect the ability of OxyR to induce expression of the its target genes. This effect was exacerbated by high levels of free iron (in the fur mutant) or the inhibition of respiration (in the hemA26 mutant), conditions which accelerate Fenton chemistry. Whether the accelerated Fenton reaction renders the OxyR protein inactive or we observed a general effect on protein synthesis and/or degradation is not clear. It is interesting that the mRNA levels of the two non-oxyR-regulated genes we tested (himA of IHF and Z2519) seem to remain unchanged, except for in the fur mutant (Fig. 9A and data not shown). Further study will be needed to clarify transcriptional responses to the rate of Fenton chemistry.
Acknowledgments
We thank F. Fang and T. Elliott for strains and plasmid.
This study was supported by The Bruno Goldberg Endowment Fund; by the Zetner Family Fund for Research in Cancer, Heart Disease, and Pharmaceutical Chemistry; and by The Israel Science Foundation, founded by The Academy of Sciences and Humanities-Centers of Excellence Program (S.A.). This work was also supported by grant GM59030 from the National Institutes of Health to J.I.
REFERENCES
- 1.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 2.Beale, S. I. 1996. Biosynthesis of hemes, p. 731-748. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella:cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 3.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, P., L. Wang, C. D. Archer, and T. Elliott. 1996. Transcription of the glutamyl-tRNA reductase (hemA) gene in Salmonella typhimurium and Escherichia coli: role of the hemA P1 promoter and the arcA gene product. J. Bacteriol. 178:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott, T. 1989. Cloning, genetic characterization, and nucleotide sequence of the hemA-prfA operon of Salmonella typhimurium. J. Bacteriol. 171:3948-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, T., and X. Wang. 1991. Salmonella typhimurium prfA mutants defective in release factor 1. J. Bacteriol. 173:4144-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzsimons, E. J., and A. May. 1996. The molecular basis of the sideroblastic anemias. Curr. Opin. Hematol. 3:167-172. [DOI] [PubMed] [Google Scholar]
- 9.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory, E. M., and I. Fridovich. 1973. Oxygen toxicity and the superoxide dismutase. J. Bacteriol. 114:1193-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamza, I., S. Chauhan, R. Hassett, and M. R. O'Brian. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669-21674. [DOI] [PubMed] [Google Scholar]
- 12.Hamza, I., Z. Qi, N. D. King, and M. R. O'Brian. 2000. Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum. Microbiology 146:669-676. [DOI] [PubMed] [Google Scholar]
- 13.Hassett, D. J., B. E. Britigan, T. Svendsen, G. M. Rosen, and M. S. Cohen. 1987. Bacteria form intracellular free radicals in response to paraquat and streptonigrin: demonstration of the potency of hydroxyl radical. J. Biol. Chem. 262:13404-13408. [PubMed] [Google Scholar]
- 14.Heurgué-Hamard, V., S. Champ, A. Engström, M. Ehrenberg, and R. H. Buckingham. 2002. The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imlay, J. A., S. M. Chin, and S. Linn. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640-642. [DOI] [PubMed] [Google Scholar]
- 16.Imlay, J. A., and I. Fridovich. 1991. Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem. 266:6957-6965. [PubMed] [Google Scholar]
- 17.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1308. [DOI] [PubMed] [Google Scholar]
- 18.Imlay, J. A., and S. Linn. 1987. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 169:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 93:13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella:cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 22.Ma, M., and J. W. Eaton. 1992. Multicellular oxidant defense in unicellular organisms. Proc. Natl. Acad. Sci. USA 89:7924-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Microbial growth, p. 137-139. In M. T. Madigan, J. M. Martinko, and J. Parker (ed.), Brock biology of microorganisms, 9th ed. Prentice-Hall, Upper Saddle River, N.J.
- 24.Maloy, S. R., and V. J. Stewart, and R. K. Taylor (ed.). 1996. Genetic analysis of pathogenic bacteria, p. 265-272. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Nakahigashi, K., N. Kubo, S-i. Narita, T. Shimaoka, S. Goto, T. Oshima, H. Mori, M. Maeda, C. Wada, and H. Inokuchi. 2002. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. USA 99:1473-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakahigashi, K., K. Nishimura, K. Miyamoto, and H. Inokuchi. 1991. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 88:10520-10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayashiki, T., K. Nishimura, and H. Inokuchi. 1995. Cloning and sequencing of a previously unidentified gene that is involved in the biosynthesis of heme in Escherichia coli. Gene 153:67-70. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, N. 1999. Metal ion transporters and homeostasis. EMBO J. 18:4361-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olafsson, O., J. U. Ericson, R. VanBogelen, and G. R. Bjork. 1996. Mutation in the structural gene for release factor 1 (RF-1) of Salmonella typhimurium inhibits cell division. J. Bacteriol. 178:3829-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbe-Saule, V., C. Coynault, M. Ibanez-Ruiz, D. Hermant, and F. Norel. 2001. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σS). Mol. Microbiol. 39:1533-1545. [DOI] [PubMed] [Google Scholar]
- 31.Schlosser-Silverman, E., M. Elgrably-Weiss, I. Rosenshine, R. Kohen, and S. Altuvia. 2000. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 182:5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soballe, B., and R. K. Poole. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146:787-796. [DOI] [PubMed] [Google Scholar]
- 33.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 34.Takiff, H. E., T. Baker, T. Copeland, S. M. Chen, and D. L. Court. 1992. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJ operon. J. Bacteriol. 174:1544-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verkamp, E., and B. K. Chelm. 1989. Isolation, nucleotide sequence, and preliminary characterization of the Escherichia coli K-12 hemA gene. J. Bacteriol. 171:4728-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, L., M. Elliott, and T. Elliott. 1999. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J. Bacteriol. 181:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, L., S. Wilson, and T. Elliott. 1999. A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J. Bacteriol. 181:6033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, L. Y., L. Brown, M. Elliott, and T. Elliott. 1997. Regulation of heme biosynthesis in Salmonella typhimurium: activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism, which increases abundance of the protein. J. Bacteriol. 179:2907-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein-Fischer, D., M. Elgrably-Weiss, and S. Altuvia. 2000. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 35:1413-1420. [DOI] [PubMed] [Google Scholar]
- 42.White, J. R., and H. N. Yeowell. 1982. Iron enhances the bactericidal action of streptonigrin. Biochem. Biophys. Res. Commun. 106:407-411. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, T. J., N. Bertrand, J. L. Tang, J. X. Feng, M. Q. Pan, C. E. Barber, J. M. Dow, and M. J. Daniels. 1998. The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol. Microbiol. 28:961-970. [DOI] [PubMed] [Google Scholar]
- 44.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]